Abstract
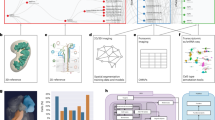
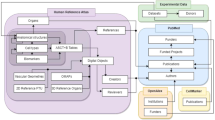
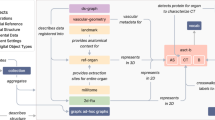
The Human Reference Atlas (HRA) aims to map all of the cells of the human body to advance biomedical research and clinical practice. This Perspective presents collaborative work by members of 16 international consortia on two essential and interlinked parts of the HRA: (1) three-dimensional representations of anatomy that are linked to (2) tables that name and interlink major anatomical structures, cell types, plus biomarkers (ASCT+B). We discuss four examples that demonstrate the practical utility of the HRA.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
27,99 € / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
209,00 € per year
only 17,42 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Chen, G., Ning, B. & Shi, T. Single-cell RNA-seq technologies and related computational data analysis. Front. Genet. https://doi.org/10.3389/fgene.2019.00317 (2019).
Rozenblatt-Rosen, O., Stubbington, M. J. T., Regev, A. & Teichmann, S. A. The human cell atlas: from vision to reality. Nature 574, 187–192 (2017).
Regev, A. et al. The Human Cell Atlas. eLife 6, e27041 (2017).
Ding, S. L. et al. Comprehensive cellular-resolution atlas of the adult human brain. J. Comp. Neurol. 524, 3127–3481 (2016).
Devor, A. et al. The challenge of connecting the dots in the B.R.A.I.N. Neuron 80, 270–274 (2013).
Moghe, I., Loupy, A. & Solez, K. The human cell atlas project by the numbers: relationship to the Banff classification. Am. J. Transpl. 18, 1830 (2018).
Lonsdale, J. et al. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 45, 580–585 (2013).
McMahon, A. P. et al. GUDMAP: the genitourinary developmental molecular anatomy project. J. Am. Soc. Nephrol. 19, 667–671 (2008).
Elmentaite, R. et al. Cells of the human intestinal tract mapped across space and time. Nature 597, 250–255 (2021).
Srivastava, S. et al. The making of a PreCancer Atlas: promises, challenges, and opportunities. Trends Cancer 4, 523–536 (2018).
Snyder, M. P. et al. The human body at cellular resolution: the NIH Human Biomolecular Atlas Program. Nature 574, 187–192 (2019).
Himmelstein, D. S. et al. Systematic integration of biomedical knowledge prioritizes drugs for repurposing. eLife 6, e26726 (2017).
El-Achkar, T. M. et al. A multimodal and integrated approach to interrogate human kidney biopsies with rigor and reproducibility: guidelines from the Kidney Precision Medicine Project. Physiol. Genomics 53, 1–11 (2021).
Ardini-Poleske, M. E. et al. LungMAP: the molecular atlas of lung development program. Am. J. Physiol. Lung Cell. Mol. Physiol. 313, L733–L740 (2013).
Oxburgh, L. et al. (Re)building a kidney. J. Am. Soc. Nephrol. 28, 1370–1378 (2017).
Stimulating Peripheral Activity to Relieve Conditions (SPARC) (NIH, 2020); https://commonfund.nih.gov/sparc
Heng, H. H. Q. Cancer genome sequencing the challenges ahead. Bioessays 29, 783–794 (2007).
TGCA Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455, 1061–1068 (2008).
Freire, P. et al. Exploratory analysis of the copy number alterations in glioblastoma multiforme. PLoS ONE 3, e4076 (2008).
Aumann, S., Donner, S., Fischer, J. & Muller, F. in High Resolution Imaging in Microscopy and Ophthalmology: New Frontiers in Biomedical Optics (ed. Bille, J. F.) 59–85. Springer (2019).
Yin, R., Burnum-Johnson, K. E., Sun, X., Dey, S. K. & Laskin, J. High spatial resolution imaging of biological tissues using nanospray desorption electrospray ionization mass spectrometry. Nat. Protoc. 14, 3445–3470 (2019).
Goltsev, Y. et al. Deep profiling of mouse splenic architecture with CODEX multiplexed imaging. Cell 174, 968–981 (2018).
Uhlén, M. et al. Tissue-based map of the human proteome. Science 347, 1260419 (2015).
Buenrostro, J. D. et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523, 486–490 (2015).
Stoeckius, M. et al. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 14, 865–868 (2017).
Chen, K. H., Boettiger, A. N., Moffitt, J. R., Wang, S. & Zhuang, X. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090 (2015).
Moffitt, J. R. et al. High-throughput single-cell gene-expression profiling with multiplexed error-robust fluorescence in situ hybridization. Proc. Natl Acad. Sci. USA 113, 11046–11051 (2016).
Rodriques, S. G. et al. Slide-seq: a scalable technology for measuring genome-wide expression at high spatial resolution. Science 363, 1463–1467 (2019).
Asp, M., Bergenstrahle, J. & Lundeberg, J. Spatially resolved transcriptomes-next generation tools for tissue exploration. Bioessays 42, e1900221 (2020).
Zollinger, D. R., Lingle, S. E., Sorg, K., Beechem, J. M. & Merritt, C. R. GeoMx RNA Assay: high multiplex, digital, spatial analysis of RNA in FFPE tissue. Methods Mol. Biol. 2148, 331–345 (2020).
Visium Spatial Gene Expression (10x Genomics, 2021); https://www.10xgenomics.com/products/spatial-gene-expression
Miller, J. A. et al. Common cell type nomenclature for the mammalian brain. eLife 9, e59928 (2020).
Mungall, C. J., Torniai, C., Gkoutos, G. V., Lewis, S. E. & Haendel, M. A. Uberon, an integrative multi-species anatomy ontology. Genome Biol. 13, R5 (2012).
Golbreich, C., Grosjean, J. & Darmoni, S. J. The foundational model of anatomy in OWL 2 and its use. Artif. Intell. Med. 57, 119–132 (2013).
Rosse, C. & Mejino, J. L. V. A reference ontology for biomedical informatics: the Foundational Model of Anatomy. J. Biomed. Inform. 36, 478–500 (2003).
Meehan, T. F. et al. Logical development of the Cell Ontology. BMC Bioinform. 12, 6 (2011).
Ding, S. L. et al. Allen Human Reference Atlas—3D, 2020 (2021); http://download.alleninstitute.org/informatics-archive/allen_human_reference_atlas_3d_2020/version_2021
Fonseca, C. G. et al. The Cardiac Atlas Project: an imaging database for computational modeling and statistical atlases of the heart. Bioinformatics 27, 2288–2295 (2011).
Géron, A., Werner, J., Wattiez, R., Lebaron, P. & Mattallana-Surget, S. Deciphering the functioning of microbial communities: shedding light on the critical steps in metaproteomics. Front. Microbiol. 10, 2395 (2019).
Manz, M. G., Miyamoto, T., Akashi, K. & Weissman, I. L. Prospective isolation of human clonogenic common myeloid progenitors. Proc. Natl Acad. Sci. USA 99, 11872–11877 (2002).
Fajtova, M., Kovarikova, A., Svec, P., Kankuri, E. & Sedlak, J. Immunophenotypic profile of nucleated erythroid progenitors during maturation in regenerating bone marrow. Leuk. Lymphoma 54, 2523–2530 (2013).
Kawamura, S. et al. Identification of a human clonogenic progenitor with strict monocyte differentiation potential: a counterpart of mouse cMoPs. Immunity 54, 2523–2530 (2017).
Mousset, C. M. et al. Comprehensive phenotyping of T cells using flow cytometry. Cytom. A 95, 647–654 (2019).
Mello, F. V. et al. Maturation-associated gene expression profiles along normal human bone marrow monopoiesis. Br. J. Haematol. 176, 464–474 (2017).
Tomer, A. Human marrow megakaryocyte differentiation: multiparameter correlative analysis identifies von Willebrand factor as a sensitive and distinctive marker for early (2N and 4N) megakaryocytes. Blood 104, 2722–2727 (2004).
Doulatov, S. et al. Revised map of the human progenitor hierarchy shows the origin of macrophages and dendritic cells in early lymphoid development. Nat. Immunol. 11, 585–593 (2010).
Elghetany, M. T., Ge, Y., Patel, J., Martinez, J. & Uhrova, H. Flow cytometric study of neutrophilic granulopoiesis in normal bone marrow using an expanded panel of antibodies: correlation with morphologic assessments. J. Clin. Lab. Anal. 18, 36–41 (2004).
Szabo, P. A. et al. Single-cell transcriptomics of human T cells reveals tissue and activation signatures in health and disease. Nat. Commun. 10, 4706 (2019).
Kaminski, D. A., Wei, C., Qian, Y., Rosenberg, A. F. & Sanz, I. Advances in human B cell phenotypic profiling. Front. Immunol. 3, 302 (2012).
Hay, S. B., Ferchen, K., Chetal, K., Grimes, H. L. & Salomonis, N. The Human Cell Atlas bone marrow single-cell interactive web portal. Exp. Hematol. 68, P51–P61 (2018).
Clavarino, G. et al. Novel strategy for phenotypic characterization of human B lymphocytes from precursors to effector cells by flow cytometry. PLoS ONE 11, e0162209 (2016).
Popescu, D. M. et al. Decoding human fetal liver haematopoiesis. Nature 574, 365–371 (2019).
Hodge, R. D. et al. Conserved cell types with divergent features in human versus mouse cortex. Nature 573, 61–68 (2019).
Hawrylycz, M. J. et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 489, 391–399 (2012).
Litviňuková, M. et al. Cells of the adult human heart. Nature 588, 466–472 (2020).
Tucker, N. R. et al. Transcriptional and cellular diversity of the human heart. Circulation 142, 466–482 (2020).
Giannasca, P. J., Giannasca, K. T., Leichtner, A. M. & Neutra, M. R. Human intestinal M cells display the sialyl Lewis A antigen. Infect. Immun. 67, 946–953 (1999).
Buettner, M. & Lochner, M. Development and function of secondary and tertiary lymphoid organs in the small intestine and the colon. Front. Immunol. 7, 342 (2016).
Hoyle, C. H. & Burnstock, G. Neuronal populations in the submucous plexus of the human colon. J. Anat. 166, 7–22 (1989).
Westerhoff, M. & Greeson, J. in Histology for Pathologists (ed. Mills, S.) Ch. 24, 640–663 (Wolters Kluwer, 2019).
Azzali, G. Structure, lymphatic vascularization and lymphocyte migration in mucosa-associated lymphoid tissue. Immunol. Rev. 195, 178–189 (2003).
Furness, J. B., Callaghan, B. P., Rivera, L. R. & Cho, H. J. The enteric nervous system and gastrointestinal innervation: integrated local and central control. Adv. Exp. Med. Biol. 817, 39–71 (2014).
Arai, T. & Kino, I. Morphometrical and cell kinetic studies of normal human colorectal mucosa. Comparison between the proximal and the distal large intestine. Acta Pathol. Jpn 39, 725–730 (1989).
Fenton, T. M. et al. Immune profiling of human gut-associated lymphoid tissue identifies a role for isolated lymphoid follicles in priming of region-specific immunity. Immunity 52, 557–570 (2020).
Habowski, A. N. et al. Transcriptomic and proteomic signatures of stemness and differentiation in the colon crypt. Commun. Biol. 3, 453 (2020).
Lundqvist, C., Baranov, V., Hammarström, S., Athlin, L. & Hammarström, M. L. Intra-epithelial lymphocytes. Evidence for regional specialization and extrathymic T cell maturation in the human gut epithelium. Int. Immunol. 7, 1473–1487 (1995).
Lockyer, M. G. & Petras, R. E. in Histology for Pathologists (ed. Mills, S.) Ch. 25, 664–676 (Wolters Kluwer, 2019).
Pittman, M. E. & Yantiss, R. K. in Histology for Pathologists (ed. Mills, S.) Ch. 26, 677–691 (Wolters Kluwer, 2019).
Kriz, W. & Bankir, L. A standard nomenclature for structures of the kidney. The Renal Commission of the International Union of Physiological Sciences (IUPS). Kidney Int. 33, 1–7 (1988).
Lake, B. B. et al. A single-nucleus RNA-sequencing pipeline to decipher the molecular anatomy and pathophysiology of human kidneys. Nat. Commun. 10, 2832 (2019).
Barry, D. M. et al. Molecular determinants of nephron vascular specialization in the kidney. Nat. Commun. 10, 5705 (2019).
Menon, R. et al. Single cell transcriptomics identifies focal segmental glomerulosclerosis remission endothelial biomarker. JCI Insight 5, e133267 (2020).
Ransick, A. et al. Single-cell profiling reveals sex, lineage, and regional diversity in the mouse kidney. Dev. Cell 51, 399–413 (2019).
Limbutara, K., Chou, C. L. & Knepper, M. A. Quantitative proteomics of all 14 renal tubule segments in rat. J. Am. Soc. Nephrol. 31, 1255–1266 (2020).
Kuppe, C. et al. Decoding myofibroblast origins in human kidney fibrosis. Nature 589, 281–286 (2021).
Stewart, B. J. et al. Spatiotemporal immune zonation of the human kidney. Science 365, 1461–1466 (2019).
Kirita, Y., Wu, H., Uchimura, K., Wilson, P. C. & Humphreys, B. D. Cell profiling of mouse acute kidney injury reveals conserved cellular responses to injury. Proc. Natl Acad. Sci. USA 117, 15874–15883 (2020).
Standring, S. Gray’s Anatomy: The Anatomical Basis of Clinical Practice (Elsevier, 2016).
Haefeli-Bleuer, B. & Weibel, E. R. Morphometry of the human pulmonary acinus. Anat. Rec. 220, 401–414 (1988).
Whitsett, J. A., Kalin, T. V., Xu, Y. & Kalinichenko, V. V. Building and regenerating the lung cell by cell. Physiol. Rev. 99, 513–554 (2019).
Plasschaert, L. W. et al. A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature 560, 377–381 (2018).
Xu, Y. et al. Single-cell RNA sequencing identifies diverse roles of epithelial cells in idiopathic pulmonary fibrosis. JCI Insight 1, e90558 (2016).
Adams, T. S. et al. Single-cell RNA-seq reveals ectopic and aberrant lung-resident cell populations in idiopathic pulmonary fibrosis. Sci. Adv. 6, eaba1983 (2020).
Wang, A. et al. Single-cell multiomic profiling of human lungs reveals cell-type-specific and age-dynamic control of SARS-CoV2 host genes. eLife 9, e62522 (2020).
Travaglini, K. J. et al. A molecular cell atlas of the human lung from single-cell RNA sequencing. Nature 587, 619–625 (2020).
Deprez, M. et al. A single-cell atlas of the human healthy airways. Am. J. Respir. Crit. Care Med. 202, 1636–1645 (2019).
Medeiros, L. J. et al. in Tumors of the Lymph Node and Spleen Ch. 1 (American Registry of Pathology, 2017).
Medeiros, L. J. et al. Tumors of the Lymph Node and Spleen (American Registry of Pathology, 2017).
O’Malley, D. P., George, T. I., Orazi, A. & Abbondanzo, S. L. in Benign and Reactive Conditions of Lymph Node and Spleen Ch. 1 (American Registry of Pathology, 2009).
Angel, C. E. et al. Distinctive localization of antigen-presenting cells in human lymph nodes. Blood 113, 1257–1267 (2009).
James, K. R. et al. Distinct microbial and immune niches of the human colon. Nat. Immunol. 21, 343–353 (2020).
Link, A. et al. Association of T-zone reticular networks and conduits with ectopic lymphoid tissues in mice and humans. Am. J. Pathol. 178, 1662–1675 (2011).
Park, S. M. et al. Mapping the distinctive populations of lymphatic endothelial cells in different zones of human lymph nodes. PLoS ONE 9, e106814 (2014).
Xiang, M. et al. A single-cell transcriptional roadmap of the mouse and human lymph node lymphatic vasculature. Front. Cardiovasc. Med. 7, 52 (2020).
Takeda, A. et al. Single-cell survey of human lymphatics unveils marked endothelial cell heterogeneity and mechanisms of homing for neutrophils. Immunity 51, 561–572 (2019).
Kunicki, M. A., Hernandez, L. C. A., Davis, K. L., Bacchetta, R. & Roncarolo, M. G. Identity and diversity of human peripheral Th and T regulatory cells defined by single-cell mass cytometry. J. Immunol. 200, 336–346 (2018).
Pusztaszeri, M. P., Seelentag, W. & Bosman, F. T. Immunohistochemical expression of endothelial markers CD31, CD34, von Willebrand factor, and Fli-1 in normal human tissues. J. Histochem. Cytochem. 54, 385–395 (2006).
Reynolds, G. et al. Developmental cell programs are co-opted in inflammatory skin disease. Science 371, eaba6500 (2021).
Dyring-Anderson, B. et al. Spatially and cell-type resolved quantitative proteomic atlas of healthy human skin. Nat. Commun. 11, 5587 (2020).
Fuchs, E. Keratins and the skin. Annu. Rev. Cell Dev. Biol. 11, 123–153 (1995).
Nestle, F. O., Meglio, P. D., Qin, J. Z. & Nickoloff, B. J. Skin immune sentinels in health and disease. Nat. Rev. Immunol. 9, 679–691 (2009).
Laverdet, B. et al. Skin innervation: important roles during normal and pathological cutaneous repair. Histol. Histopathol. 30, 875–892 (2015).
Ryan, T. J. The blood vessels of the skin. J. Invest. Dermatol. 67, 110–118 (1976).
Popescu, D. M. A Single Cell Atlas of Adult Healthy, Psoriatic and Atopic Dermatitis Skin (2021); https://developmentcellatlas.ncl.ac.uk/datasets/hca_skin_portal
Bos, J. D. et al. The skin immune system (SIS): distribution and immunophenotype of lymphocyte subpopulations in normal human skin. J. Invest. Dermatol. 88, 569–573 (1987).
Schweizer, J. et al. New consensus nomenclature for mammalian keratins. J. Cell Biol. 174, 169–174 (2006).
Eberl, G., Colonna, M., Di Santo, J. P. & McKenzie, A. N. J. Innate lymphoid cells: a new paradigm in immunology. Science 348, aaa6566 (2015).
Ali, N. & Rosenblum, M. D. Regulatory T cells in skin. Immunology 152, 372–381 (2017).
Huber, W. E. et al. A tissue-restricted cAMP transcriptional response: SOX10 modulates alpha-melanocyte-stimulating hormone-triggered expression of microphthalmia-associated transcription factor in melanocytes. J. Biol. Chem. 278, 45224–45230 (2003).
Haniffa, M., Gunawan, M. & Jardine, L. Human skin dendritic cells in health and disease. J. Dermatol. Sci. 77, 85–92 (2015).
Cesta, M. F. Normal structure, function, and histology of the spleen. Toxicol. Pathol. 34, 455–465 (2006).
Madissoon, E. et al. scRNA-seq assessment of the human lung, spleen, and esophagus tissue stability after cold preservation. Genome Biol. 21, 1 (2019).
Pack, M. et al. DEC-205/CD205+ dendritic cells are abundant in the white pulp of the human spleen, including the border region between the red and white pulp. Immunology 123, 438–446 (2008).
Steiniger, B. S. Human spleen microanatomy: why mice do not suffice. Immunology 145, 334–346 (2015).
Steiniger, B. S., Seiler, A., Lampp, K., Wilhelmi, V. & Stachniss, V. B lymphocyte compartments in the human splenic red pulp: capillary sheaths and periarteriolar regions. Histochem. Cell Biol. 141, 507–518 (2014).
Steiniger, B. S., Stachniss, V., Schwarzbach, H. & Barth, P. J. Phenotypic differences between red pulp capillary and sinusoidal endothelia help localizing the open splenic circulation in humans. Histochem. Cell Biol. 128, 391–398 (2007).
Qiu, J. et al. The characteristics of vessel lining cells in normal spleens and their role in the pathobiology of myelofibrosis. Blood Adv. 2, 1130–1145 (2018).
Lewis, S. M., Williams, A. & Eisenbarth, S. C. Structure and function of the immune system in the spleen. Sci. Immunol. 4, eaau6085 (2019).
Mittag, D. et al. Human dendritic cell subsets from spleen and blood are similar in phenotype and function but modified by donor health status. J. Immunol. 186, 6207–6217 (2011).
Cheng, H. W. et al. Origin and differentiation trajectories of fibroblastic reticular cells in the splenic white pulp. Nat. Commun. 10, 1739 (2019).
Van Krieken, J. H. J. M. & Te Velde, J. Immunohistology of the human spleen: an inventory of the localization of lymphocyte subpopulations. Histopathology 10, 285–294 (1986).
Van Krieken, J. H. J. M., Te Velde, J., Leenheers-Binnendijk, L. & Van de Velde, C. J. H. The human spleen; a histological study in splenectomy specimens embedded in methylmethacrylate. Histopathology 9, 571–585 (1985).
Bautista, J. L. et al. Single-cell transcriptional profiling of human thymic stroma uncovers novel cellular heterogeneity in the thymic medulla. Nat. Commun. 12, 1096 (2021).
Haynes, B. F. The human thymic microenvironment. Adv. Immunol. 36, 87–142 (1984).
Park, J. E. et al. A cell atlas of human thymic development defines T cell repertoire formation. Science 367, eaay3224 (2020).
Pearse, G. Normal structure, function and histology of the thymus. Toxicol. Pathol. 34, 504–514 (2006).
Suster, S. & Rosai, J. Histology of the normal thymus. Am. J. Surg. Pathol. 14, 284–303 (1990).
Mignini, F. et al. Neuro-immune modulation of the thymus microenvironment (Review). Int. J. Mol. Med. 33, 1392–1400 (2014).
Stoeckle, C. et al. Isolation of myeloid dendritic cells and epithelial cells from human thymus. J. Vis. Exp. 79, e50951 (2013).
Marcovecchio, G. E. et al. Thymic epithelium abnormalities in DiGeorge and Down syndrome patients contribute to dysregulation in T cell development. Front. Immunol. 10, 447 (2019).
Wakimoto, T. et al. Identification and characterization of human thymic cortical dendritic macrophages that may act as professional scavengers of apoptotic thymocytes. Immunobiology 213, 837–847 (2008).
Lavaert, M. et al. Integrated scRNA-seq identifies human postnatal thymus seeding progenitors and regulatory dynamics of differentiating immature thymocytes. Immunity 52, 1088–1104 (2020).
Nuñez, S. et al. The human thymus perivascular space is a functional niche for viral-specific plasma cells. Sci. Immunol. 1, eaah4447 (2016).
Bendriss-Vermare, N. et al. Human thymus contains IFN-α-producing CD11c−, myeloid CD11c+, and mature interdigitating dendritic cells. J. Clin. Invest. 107, 835–844 (2001).
Netter, F. H. Atlas of Human Anatomy 7th edn (Elsevier, 2019).
Kandathil, A. & Chamarthy, M. Pulmonary vascular anatomy & anatomical variants. Cardiovasc. Diagn. Ther. 8, 201–207 (2018).
Perlmutter, D. & Rhoton, A. L. Jr Microsurgical anatomy of the distal anterior cerebral artery. J. Neurosurg. 49, 204–228 (1978).
Hacein-Bey, L. et al. The ascending pharyngeal artery: branches, anastomoses, and clinical significance. AJNR Am. J. Neuroradiol. 23, 1246–1256 (2002).
Vuong, S. M., Jeong, W. J., Morales, H. & Abruzzo, T. A. Vascular diseases of the spinal cord: infarction, hemorrhage, and venous congestive myelopathy. Semin. Ultrasound CT MR 37, 466–481 (2016).
Picel, A. C., Hsieh, T. C., Shapiro, R. M., Vezeridis, A. M. & Isaacson, A. J. Prostatic artery embolization for benign prostatic hyperplasia: patient evaluation, anatomy, and technique for successful treatment. Radiographics 39, 1526–1548 (2019).
Vummidi, D. et al. Pseudolesion in segment IV A of the liver from vein of Sappey secondary to SVC obstruction. Radiol. Case Rep. 5, 394 (2015).
Tang, F. et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods 6, 377–382 (2009).
Moffitt, J. R. et al. Molecular, spatial, and functional single-cell profiling of the hypothalamic preoptic region. Science https://doi.org/10.1126/science.aau5324 (2018).
International Human Genome Sequencing (IHGS) Consortium Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001).
Venter, J. C. et al. The sequence of the human genome. Science 291, 1304–1351 (2001).
Visible Human Project (VHP) Data Sets (NLM, 2020); https://www.nlm.nih.gov/databases/download/vhp.html
Fair Principles (Go Fair, 2021); https://www.go-fair.org/fair-principles
CCF Anatomical Structures, Cell Types and Biomarkers (ASCT+B) Tables (HuBMAP Consortium, 2021); https://hubmapconsortium.github.io/ccf/pages/ccf-anatomical-structures.html
CCF 3D Reference Object Library (HuBMAP Consortium, 2021); https://hubmapconsortium.github.io/ccf/pages/ccf-3d-reference-library.html
Graphics Language Transmission Format (glTF) a File Format Specification for 3D Scenes and Models (Khronos Group, 2021); https://www.khronos.org
CCF Registration User Interface (HuBMAP Consortium, 2021); https://hubmapconsortium.github.io/ccf-ui/rui
CCF Exploration User Interface (HuBMAP Consortium, 2021); https://portal.hubmapconsortium.org/ccf-eui
Human Biomolecular Atlas Program (HuBMAP) Data Portal (NIH, 2020); https://portal.hubmapconsortium.org
CCF User Interfaces (RUI, EUI) (HuBMAP Consortium, 2021); https://github.com/hubmapconsortium/ccf-ui
Jardine, L. et al. Blood and immune development in human fetal bone marrow and Down syndrome. Nature 598, 327–331 (2021).
Stephenson, E. et al. Single-cell multi-omics analysis of the immune response in COVID-19. Nat. Med. 27, 904–916 (2021).
Lake, B. B. et al. An atlas of healthy and injured cell states and niches in the human kidney. Preprint at bioRxiv https://doi.org/10.1101/2021.07.28.454201 (2021).
Zhuo, L., Huang, L., Yang, Z., Li, G. & Wang, L. A comprehensive analysis of NPHS1 gene mutations in patients with sporadic focal segmental glomerulosclerosis. BMC Med. Genet. 20, 111 (2019).
Ong, E. et al. Modelling kidney disease using ontology: insights from the Kidney Precision Medicine Project. Nat. Rev. Nephrol. 16, 686–696 (2020).
Barwinska, D. et al. Molecular characterization of the human kidney interstitium in health and disease. Sci. Adv. 7, eabd3359 (2021).
ASCT+B Reporter (HuBMAP Consortium, 2021); https://hubmapconsortium.github.io/ccf-asct-reporter
Majumder, P. P., Mhlanga, M. M. & Shalek, A. K. The Human Cell Atlas and equity: lessons learned. Nat. Med. 26, 1509–1511 (2020).
Spatial Biology Europe: Online (HuBMAP Consortium, 2021); https://cns-iu.github.io/workshops/2021-2004-2014_spatial_biology_europe_online
Azimuth App for reference-based single-cell analysis (Satija Lab, 2021); https://azimuth.hubmapconsortium.org/
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587 (2021).
Osumi-Sutherland, D. et al. Cell type ontologies of the Human Cell Atlas. Nat. Cell Biol. https://doi.org/10.1038/s41556-021-00787-7 (2021).
Radtke, A. J. et al. IBEX: an open and extensible method for high content multiplex imaging of diverse tissues. Preprint at https://arxiv.org/abs/2107.11364 (2021).
Hickey, J. W. et al. Spatial mapping of protein composition and tissue organization: a primer for multiplexed antibody-based imaging. Preprint at https://arxiv.org/abs/2107.07953 (2021).
Manz, T. et al. Viv: multiscale visualization of high-resolution multiplexed bioimaging data on the web. Preprint at OSF https://doi.org/10.31219/osf.io/wd2gu (2020).
Balhoff, J. & Curtis, C. K. Ubergraph (2021); https://github.com/INCATools/ubergraph
Li, W., Germain, R. N. & Gerner, M. Y. Multiplex, quantitative cellular analysis in large tissue volumes with clearing-enhanced 3D microscopy (Ce3D). Proc. Natl Acad. Sci. USA 114, E7321–E7330 (2017).
Fedorov, A. et al. 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn. Reson. Imaging 30, 1323–1341 (2012).
Slicer Community. 3D Slicer image computing platform (2021); https://www.slicer.org/
ZBrush, the all-in-one digital sculpting solution (Pixologic, 2021); https://pixologic.com/
Maya: 3D computer animation, modeling, simulation, and rendering software (Autodesk, 2021); https://www.autodesk.com/products/maya/overview
Acknowledgements
We thank B. B. Lake from University of California, San Diego, for assistance with annotations and analysing the single-nucleus RNA-seq HUBMAP data for several of the markers in the kidney ASCT+B tables; B. Steck and R. Dull from the University of Michigan for assistance with the nomenclature and curation of kidney partonomy; S. Winfree, IUPUI, for discussions regarding the kidney ASCT+B table; and staff at the KPMP, especially the Tissue Interrogation Sites and the Controlled Cell Vocabulary working group, for guidance and development of the initial sets of ASCT+B kidney tables. We acknowledge L. Yao for segmenting and optimizing the mouse popliteal lymph node model from high-resolution microscopy data. The work was funded, in part, by NIH Awards OT2OD026671, U54DK120058, 1UH3CA246594, 1U54AI142766, 1UG3CA256960, 1UG3HL145609, U54HL145608, U54HL145611, UH3DK114933, DK110814 and DK107350; National Institute of Allergy and Infectious Diseases (NIAID), Department of Health and Human Services under BCBB Support Services Contract HHSN316201300006W/HHSN27200002; the Intramural Research Program of the NIH at NIAID; and Helmsley Charitable Trust 2018PG-T1D071.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
In the past 3 years, S.A.T. has received remuneration for consulting and Scientific Advisory Board membership from Genentech, Roche, Biogen, GlaxoSmithKline, Foresite labs, Qiagen and Transition Bio and she is a co-founder and equity holder of Transition Bio. R.M. receives research funding from Bayer and Amgen and serves as a consultant for Myokardia/BMS and Third Pole; he is a co-founder of Patch Inc; he is a co-inventor for a patent no. PCT/US2O12/O22119 on pharmacologic BMP inhibitors (along with Mass General Brigham) for which he receives royalties from Keros Therapeutics, Inc.; he also receives royalties from UpToDate for scientific content authorship. The other authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
ASCT+B Tables
Rights and permissions
About this article
Cite this article
Börner, K., Teichmann, S.A., Quardokus, E.M. et al. Anatomical structures, cell types and biomarkers of the Human Reference Atlas. Nat Cell Biol 23, 1117–1128 (2021). https://doi.org/10.1038/s41556-021-00788-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-021-00788-6
This article is cited by
-
Spatial top-down proteomics for the functional characterization of human kidney
Clinical Proteomics (2025)
-
Construction, Deployment, and Usage of the Human Reference Atlas Knowledge Graph
Scientific Data (2025)
-
Functional tissue units in the Human Reference Atlas
Nature Communications (2025)
-
MicroRNA in intestinal tight junction regulation
npj Gut and Liver (2025)
-
Human BioMolecular Atlas Program (HuBMAP): 3D Human Reference Atlas construction and usage
Nature Methods (2025)