Abstract
In the classical twin design, researchers compare trait resemblance in cohorts of identical and non-identical twins to understand how genetic and environmental factors correlate with resemblance in behaviour and other phenotypes. The twin design is also a valuable tool for studying causality, intergenerational transmission, and gene–environment correlation and interaction. Here we review recent developments in twin studies, recent results from twin studies of new phenotypes and recent insights into twinning. We ask whether the results of existing twin studies are representative of the general population and of global diversity, and we conclude that stronger efforts to increase representativeness are needed. We provide an updated overview of twin concordance and discordance for major diseases and mental disorders, which conveys a crucial message: genetic influences are not as deterministic as many believe. This has important implications for public understanding of genetic risk prediction tools, as the accuracy of genetic predictions can never exceed identical twin concordance rates.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
27,99 € / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
118,99 € per year
only 9,92 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Code availability
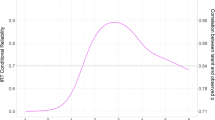
The code to reproduce Fig. 2b is available on GitHub: https://github.com/KuznetsovDima/NATHUMBEHAV-22123354/.
References
Boomsma, D., Busjahn, A. & Peltonen, L. Classical twin studies and beyond. Nat. Rev. Genet. 3, 872–882 (2002).
Craig, J. M., Calais-Ferreira, L., Umstad, M. P. & Buchwald, D. The value of twins for health and medical research: a third of a century of progress. Twin Res. Hum. Genet. 23, 8–15 (2020).
Falconer, D. S. & Mackay, T. F. Introduction to Quantitative Genetics (Prentice Hall, 1996).
McGue, M. When assessing twin concordance, use the probandwise not the pairwise rate. Schizophr. Bull. 18, 171–176 (1992).
Visscher, P. M. & Wray, N. R. Concepts and misconceptions about the polygenic additive model applied to disease. Hum. Hered. 80, 165–170 (2015).
Falconer, D. S. The inheritance of liability to certain diseases, estimated from the incidence among relatives. Ann. Hum. Genet 29, 51–76 (1965).
Smith, C. Heritability of liability and concordance in monozygous twins. Ann. Hum. Genet. 34, 85–91 (1970).
Owen, M. J., Sawa, A. & Mortensen, P. B. Schizophrenia. Lancet 388, 86–97 (2016).
Marion, M. C. et al. Nucleic acid-sensing and interferon-inducible pathways show differential methylation in MZ twins discordant for lupus and overexpression in independent lupus samples: implications for pathogenic mechanism and drug targeting. Genes 12, 1898 (2021).
Castellani, C. A. et al. DNA methylation differences in monozygotic twin pairs discordant for schizophrenia identifies psychosis related genes and networks. BMC Med. Genomics 8, 17 (2015).
Konki, M. et al. Peripheral blood DNA methylation differences in twin pairs discordant for Alzheimer’s disease. Clin. Epigenet. 11, 130 (2019).
Kazuno, A. et al. Proteomic analysis of lymphoblastoid cells derived from monozygotic twins discordant for bipolar disorder: a preliminary study. PLoS ONE 8, e53855 (2013).
O’Hanlon, T. P. et al. Plasma proteomic profiles from disease-discordant monozygotic twins suggest that molecular pathways are shared in multiple systemic autoimmune diseases. Arthritis Res. Ther. 13, R181 (2011).
Muniandy, M. et al. Plasma metabolites reveal distinct profiles associating with different metabolic risk factors in monozygotic twin pairs. Int J. Obes. 43, 487–502 (2019).
Tsang, T. M., Huang, J. T.-J., Holmes, E. & Bahn, S. Metabolic profiling of plasma from discordant schizophrenia twins: correlation between lipid signals and global functioning in female schizophrenia patients. J. Proteome Res. 5, 756–760 (2006).
Bondia‐Pons, I. et al. Metabolome and fecal microbiota in monozygotic twin pairs discordant for weight: a Big Mac challenge. FASEB J. 28, 4169–4179 (2014).
Zhu, Y. et al. Genome-wide profiling of DNA methylome and transcriptome in peripheral blood monocytes for major depression: a monozygotic discordant twin study. Transl. Psychiatry 9, 215 (2019).
Vitaro, F., Brendgen, M. & Arseneault, L. The discordant MZ-twin method: one step closer to the holy grail of causality. Int. J. Behav. Dev. 33, 376–382 (2009).
Vink, J. M. et al. Differential gene expression patterns between smokers and non-smokers: cause or consequence? Addict. Biol. 22, 550–560 (2017).
Goldman, S. M. et al. Concordance for Parkinson’s disease in twins: a 20‐year update. Ann. Neurol. 85, 600–605 (2019).
Tanner, C. M. et al. Parkinson disease in twins: an etiologic study. JAMA 281, 341–346 (1999).
Plomin, R., DeFries, J. C., Knopik, V. S. & Neiderhiser, J. M. Behavioral Genetics (Worth, 2012).
Willoughby, E. A., Polderman, T. J. C. & Boutwell, B. B. Behavioural genetics methods. Nat. Rev. Methods Prim. 3, 11 (2023).
Christensen, K. & McGue, M. in Twin Research for Everyone (eds Tarnoki, A. et al.) 439–456 (Academic Press, 2022).
Willemsen, G., Odintsova, V., de Geus, E. & Boomsma, D. I. in Twin and Higher-Order Pregnancies (eds Khalil, A. et al.) 51–71 (Springer International, 2021).
de Geus, E. J. C., Posthuma, D., IJzerman, R. G. & Boomsma, D. I. Comparing blood pressure of twins and their singleton siblings: being a twin does not affect adult blood pressure. Twin Res. 4, 385–391 (2001).
Christensen, K. et al. Comparison of academic performance of twins and singletons in adolescence: follow-up study. Br. Med. J. 333, 1095 (2006).
Barnes, J. C. & Boutwell, B. B. A demonstration of the generalizability of twin-based research on antisocial behavior. Behav. Genet. 43, 120–131 (2013).
Johnson, W., Krueger, R. F., Bouchard, T. J. & McGue, M. The personalities of twins: just ordinary folks. Twin Res. Hum. Genet. 5, 125–131 (2002).
Kendler, K. S., Ohlsson, H., Lichtenstein, P., Sundquist, J. & Sundquist, K. The genetic epidemiology of treated major depression in Sweden. Am. J. Psychiatry 175, 1137–1144 (2018).
Johnson, B. N. et al. Male microchimerism in females: a quantitative study of twin pedigrees to investigate mechanisms. Hum. Reprod. 36, 2529–2537 (2021).
Mook-Kanamori, D. O. et al. Heritability estimates of body size in fetal life and early childhood. PLoS ONE 7, e39901 (2012).
Silventoinen, K., Magnusson, P. K. E., Tynelius, P., Kaprio, J. & Rasmussen, F. Heritability of body size and muscle strength in young adulthood: a study of one million Swedish men. Genet. Epidemiol. 32, 341–349 (2008).
Estourgie-van Burk, G. F., Bartels, M., Boomsma, D. I. & Delemarre-van de Waal, H. A. Body size of twins compared with siblings and the general population: from birth to late adolescence. J. Pediatr. 156, 586–591 (2010).
Beck, J. J. et al. Genetic meta-analysis of twin birth weight shows high genetic correlation with singleton birth weight. Hum. Mol. Genet. 30, 1894–1905 (2021).
Lykken, D. T. Research with twins: the concept of emergenesis. Psychophysiology 19, 361–372 (1982).
Jonsson, H. et al. Differences between germline genomes of monozygotic twins. Nat. Genet. 53, 27–34 (2021).
Kong, A. et al. Rate of de novo mutations and the importance of father’s age to disease risk. Nature 488, 471–475 (2012).
Veltman, J. A. & Brunner, H. G. De novo mutations in human genetic disease. Nat. Rev. Genet. 13, 565–575 (2012).
Francioli, L. C. et al. Genome-wide patterns and properties of de novo mutations in humans. Nat. Genet. 47, 822–826 (2015).
Dolan, C. V., Huijskens, R. C. A., Minică, C. C., Neale, M. C. & Boomsma, D. I. Incorporating polygenic risk scores in the ACE twin model to estimate A–C covariance. Behav. Genet. 51, 237–249 (2021).
Dolan, C. V., de Kort, J. M., van Beijsterveldt, T. C. E. M., Bartels, M. & Boomsma, D. I. GE covariance through phenotype to environment transmission: an assessment in longitudinal twin data and application to childhood anxiety. Behav. Genet. 44, 240–253 (2014).
Wang, B. et al. Robust genetic nurture effects on education: a systematic review and meta-analysis based on 38,654 families across 8 cohorts. Am. J. Hum. Genet. 108, 1780–1791 (2021).
D’Onofrio, B. M. et al. The role of the children of twins design in elucidating causal relations between parent characteristics and child outcomes. J. Child Psychol. Psychiatry 44, 1130–1144 (2003).
McAdams, T. A. et al. Revisiting the children-of-twins design: improving existing models for the exploration of intergenerational associations. Behav. Genet. 48, 397–412 (2018).
McAdams, T. A. et al. Accounting for genetic and environmental confounds in associations between parent and child characteristics: a systematic review of children-of-twins studies. Psychol. Bull. 140, 1138–1173 (2014).
Balbona, J. V., Kim, Y. & Keller, M. C. Estimation of parental effects using polygenic scores. Behav. Genet. 51, 264–278 (2021).
Balbona, J. V., Kim, Y. & Keller, M. C. The estimation of environmental and genetic parental influences. Dev. Psychopathol. 34, 1876–1886 (2022).
Conley, D. et al. Is the effect of parental education on offspring biased or moderated by genotype? Sociol. Sci. 2, 82–105 (2015).
Hart, S. A., Little, C. & van Bergen, E. Nurture might be nature: cautionary tales and proposed solutions. NPJ Sci. Learn. 6, 2 (2021).
Willoughby, E. A., McGue, M., Iacono, W. G., Rustichini, A. & Lee, J. J. The role of parental genotype in predicting offspring years of education: evidence for genetic nurture. Mol. Psychiatry 26, 3896–3904 (2021).
de Vries, L. P. et al. Gene-by-crisis interaction for optimism and meaning in life: the effects of the COVID-19 pandemic. Behav. Genet. 52, 13–25 (2022).
Purcell, S. Variance components models for gene–environment interaction in twin analysis. Twin Res. 5, 554–571 (2002).
van der Sluis, S., Posthuma, D. & Dolan, C. V. A note on false positives and power in G × E modelling of twin data. Behav. Genet. 42, 170–186 (2012).
Molenaar, D., van der Sluis, S., Boomsma, D. I. & Dolan, C. V. Detecting specific genotype by environment interactions using marginal maximum likelihood estimation in the classical twin design. Behav. Genet 42, 483–499 (2012).
Jinks, J. L. & Fulker, D. W. Comparison of the biometrical genetical, MAVA, and classical approaches to the analysis of human behavior. Psychol. Bull. 73, 311–349 (1970).
van der Sluis, S., Dolan, C. V., Neale, M. C., Boomsma, D. I. & Posthuma, D. Detecting genotype–environment interaction in monozygotic twin data: comparing the Jinks and Fulker test and a new test based on marginal maximum likelihood estimation. Twin Res. Hum. Genet. 9, 377–392 (2006).
Plomin, R., DeFries, J. C. & Loehlin, J. C. Genotype–environment interaction and correlation in the analysis of human behavior. Psychol. Bull. 84, 309–322 (1977).
Eaves, L. A model for sibling effects in man. Heredity 36, 205–214 (1976).
Hunter, M. D. Multilevel modeling in classical twin and modern molecular behavior genetics. Behav. Genet. 51, 301–318 (2021).
Tamimy, Z. et al. Multilevel twin models: geographical region as a third level variable. Behav. Genet. 51, 319–330 (2021).
Davey Smith, G. & Hemani, G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum. Mol. Genet. 23, R89–R98 (2014).
Heath, A. C. et al. Testing hypotheses about direction of causation using cross-sectional family data. Behav. Genet. 23, 29–50 (1993).
Castro-de-Araujo, L. F. S. et al. MR-DoC2: bidirectional causal modeling with instrumental variables and data from relatives. Behav. Genet. 53, 63–73 (2022).
Minică, C. C., Dolan, C. V., Boomsma, D. I., de Geus, E. & Neale, M. C. Extending causality tests with genetic instruments: an integration of Mendelian randomization with the classical twin design. Behav. Genet. 48, 337–349 (2018).
Evans, L. M. et al. Comparison of methods that use whole genome data to estimate the heritability and genetic architecture of complex traits. Nat. Genet. 50, 737–745 (2018).
Zaitlen, N. et al. Using extended genealogy to estimate components of heritability for 23 quantitative and dichotomous traits. PLoS Genet. 9, e1003520 (2013).
Ouwens, K. G. et al. A characterization of cis- and trans-heritability of RNA-seq-based gene expression. Eur. J. Hum. Genet. 28, 253–263 (2020).
Hagenbeek, F. A. et al. Heritability estimates for 361 blood metabolites across 40 genome-wide association studies. Nat. Commun. 11, 39 (2020).
Hall, J. G. Twinning. Lancet 362, 735–743 (2003).
Bulmer, M. G. The Biology of Twinning in Man (Clarendon, 1970).
Lewis, C. M., Healey, S. C. & Martin, N. G. Genetic contribution to DZ twinning. Am. J. Med. Genet. 61, 237–246 (1996).
Meulemans, W. J. et al. Genetic modelling of dizygotic twinning in pedigrees of spontaneous dizygotic twins. Am. J. Med. Genet. 61, 258–263 (1996).
Duffy, D. L. & Martin, N. G. The heritability of twinning in seven large historic pedigrees. Twin Res. Hum. Genet. 25, 63–66 (2022).
Mbarek, H. et al. Identification of common genetic variants influencing spontaneous dizygotic twinning and female fertility. Am. J. Hum. Genet. 98, 898–908 (2016).
van Dongen, J. et al. Identical twins carry a persistent epigenetic signature of early genome programming. Nat. Commun. 12, 5618 (2021).
Levi, S. Ultrasonic assessment of the high rate of human multiple pregnancy in the first trimester. J. Clin. Ultrasound 4, 3–5 (1976).
Hall, J. G. The mystery of monozygotic twinning I: what can amyoplasia tell us about monozygotic twinning and the possible role of twin–twin transfusion? Am. J. Med. Genet. A 185, 1816–1821 (2021).
Eaves, L. J. & Eysenck, H. J. Genetics and the development of social attitudes. Nature 249, 288–289 (1974).
Taubman, P. Earnings, education, genetics, and environment. J. Hum. Resour. 11, 447–461 (1976).
Cesarini, D. et al. Heritability of cooperative behavior in the trust game. Proc. Natl Acad. Sci. USA 105, 3721–3726 (2008).
Hatemi, P. K. et al. Genetic influences on political ideologies: twin analyses of 19 measures of political ideologies from five democracies and genome-wide findings from three populations. Behav. Genet. 44, 282–294 (2014).
Williams, F. M. K. et al. Self-reported symptoms of COVID-19, including symptoms most predictive of SARS-CoV-2 infection, are heritable. Twin Res. Hum. Genet. 23, 316–321 (2020).
Baird, P. N. & Hysi, P. Twin registries moving forward and meeting the future: a review. Twin Res. Hum. Genet. 22, 201–209 (2019).
Geserick, M. et al. Acceleration of BMI in early childhood and risk of sustained obesity. N. Engl. J. Med. 379, 1303–1312 (2018).
Elks, C. et al. Variability in the heritability of body mass index: a systematic review and meta-regression. Front. Endocrinol. 3, 29 (2012).
Huston, A. C. From research to policy and back. Child Dev. 79, 1–12 (2008).
Bird, S., Segall, I. & Lopatka, M. Replication: why we still can’t browse in peace—on the uniqueness and reidentifiability of web browsing histories. in Sixteenth Symposium on Usable Privacy and Security (SOUPS 2020), 489–503 (USENIX Association, 2020).
Olejnik, L., Castelluccia, C. & Janc, A. Why Johnny can’t browse in peace: on the uniqueness of web browsing history patterns. In 5th Workshop on Hot Topics in Privacy Enhancing Technologies (HotPETs 2012), hal-00747841 (2012).
Eichstaedt, J. C. et al. Facebook language predicts depression in medical records. Proc. Natl Acad. Sci. USA 115, 11203–11208 (2018).
Long, E. C. et al. The genetic and environmental contributions to internet use and associations with psychopathology: a twin study. Twin Res. Hum. Genet 19, 1–9 (2016).
Langner, I., Garbe, E., Banaschewski, T. & Mikolajczyk, R. T. Twin and sibling studies using health insurance data: the example of attention deficit/hyperactivity disorder (ADHD). PLoS ONE 8, e62177 (2013).
Bullmore, E. & Sporns, O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat. Rev. Neurosci. 10, 186–198 (2009).
Borsboom, D. & Cramer, A. O. J. Network analysis: an integrative approach to the structure of psychopathology. Annu. Rev. Clin. Psychol. 9, 91–121 (2013).
Koenis, M. M. G. et al. Association between structural brain network efficiency and intelligence increases during adolescence. Hum. Brain Mapp. 39, 822–836 (2018).
Koenis, M. M. G. et al. Development of the brain’s structural network efficiency in early adolescence: a longitudinal DTI twin study. Hum. Brain Mapp. 36, 4938–4953 (2015).
Bohlken, M. M. et al. Structural brain connectivity as a genetic marker for schizophrenia. JAMA Psychiatry 73, 11–19 (2016).
Olatunji, B. O., Christian, C., Strachan, E. & Levinson, C. A. Central and peripheral symptoms in network analysis are differentially heritable: a twin study of anxious misery. J. Affect. Disord. 274, 986–994 (2020).
Forbes, M. K., Wright, A. G. C., Markon, K. E. & Krueger, R. F. Quantifying the reliability and replicability of psychopathology network characteristics. Multivar. Behav. Res. 56, 224–242 (2021).
Fried, E. I. & Cramer, A. O. J. Moving forward: challenges and directions for psychopathological network theory and methodology. Perspect. Psychol. Sci. 12, 999–1020 (2017).
Zhang, H. The review of transcriptome sequencing: principles, history and advances. IOP Conf. Ser. Earth Environ. Sci. 332, 042003 (2019).
Grundberg, E. et al. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat. Genet. 44, 1084–1089 (2012).
Wright, F. A. et al. Heritability and genomics of gene expression in peripheral blood. Nat. Genet. 46, 430–437 (2014).
Aristizabal, M. J. et al. Biological embedding of experience: a primer on epigenetics. Proc. Natl Acad. Sci. USA 117, 23261–23269 (2020).
van Dongen, J. et al. Genetic and environmental influences interact with age and sex in shaping the human methylome. Nat. Commun. 7, 11115 (2016).
Li, S. et al. Early life affects late-life health through determining DNA methylation across the lifespan: a twin study. eBioMedicine 77, 103927 (2022).
Liu, Y. et al. Quantitative variability of 342 plasma proteins in a human twin population. Mol. Syst. Biol. 11, 786 (2015).
Menni, C. et al. Glycosylation of immunoglobulin G: role of genetic and epigenetic influences. PLoS ONE 8, e82558 (2013).
Zaytseva, O. O. et al. Heritability of human plasma N-glycome. J. Proteome Res. 19, 85–91 (2020).
Yang, Q. et al. Metabolomics biotechnology, applications, and future trends: a systematic review. RSC Adv. 9, 37245–37257 (2019).
Pool, R. et al. Genetics and not shared environment explains familial resemblance in adult metabolomics data. Twin Res. Hum. Genet. 23, 145–155 (2020).
Shin, S.-Y. et al. An atlas of genetic influences on human blood metabolites. Nat. Genet. 46, 543–550 (2014).
Bermingham, K. M. et al. Genetic and environmental contributions to variation in the stable urinary NMR metabolome over time: a classic twin study. J. Proteome Res. 20, 3992–4000 (2021).
Hagenbeek, F. A. et al. Heritability of urinary amines, organic acids, and steroid hormones in children. Metabolites 12, 474 (2022).
Hasin, Y., Seldin, M. & Lusis, A. Multi-omics approaches to disease. Genome Biol. 18, 83 (2017).
Duruflé, H. et al. A powerful framework for an integrative study with heterogeneous omics data: from univariate statistics to multi-block analysis. Brief. Bioinform. 22, bbaa166 (2021).
Hur, Y.-M., Odintsova, V. V., Ordoñana, J. R., Silventoinen, K. & Willemsen, G. in Twin Research for Everyone (eds Tarnoki, A. et al.) 23–50 (Academic Press, 2022).
Silventoinen, K. et al. The CODATwins Project: the current status and recent findings of COllaborative Project of Development of Anthropometrical Measures in Twins. Twin Res. Hum. Genet. 22, 800–808 (2019).
Odintsova, V. V. et al. Establishing a twin register: an invaluable resource for (behavior) genetic, epidemiological, biomarker, and ‘omics’ studies. Twin Res. Hum. Genet. 21, 239–252 (2018).
Boutwell, B. B., Narvey, C. S., Helton, J. J. & Piquero, A. R. Why twin studies are important for health span science research: the case of maltreatment of aging adults. BMC Geriatr. 22, 943 (2022).
Austerberry, C., Mateen, M., Fearon, P. & Ronald, A. Heritability of psychological traits and developmental milestones in infancy: a systematic review and meta-analysis. JAMA Netw. Open 5, e2227887 (2022).
Polderman, T. J. C. et al. Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nat. Genet. 47, 702–709 (2015).
Hur, Y.-M. et al. A comparison of twin birthweight data from Australia, the Netherlands, the United States, Japan, and South Korea: are genetic and environmental variations in birthweight similar in Caucasians and East Asians? Twin Res. Hum. Genet. 8, 638–648 (2005).
Laursen, M. et al. Genetic influence on prolonged gestation: a population-based Danish twin study. Am. J. Obstet. Gynecol. 190, 489–494 (2004).
Garrett-Bakelman, F. E. et al. The NASA Twins Study: a multidimensional analysis of a year-long human spaceflight. Science 364, eaau8650 (2019).
Vinkhuyzen, A. A. E., Van Der Sluis, S., De Geus, E. J. C., Boomsma, D. I. & Posthuma, D. Genetic influences on ‘environmental’ factors. Genes Brain Behav. 9, 276–287 (2010).
Friedman, N. P., Banich, M. T. & Keller, M. C. Twin studies to GWAS: there and back again. Trends Cogn. Sci. 25, 855–869 (2021).
Hatemi, P. K. The intersection of behavioral genetics and political science: introduction to the special issue. Twin Res. Hum. Genet. 15, 1–5 (2012).
Cesarini, D. & Visscher, P. M. Genetics and educational attainment. NPJ Sci. Learn. 2, 4 (2017).
Plomin, R. The next 10 years of behavioural genomic research. JCPP Adv. 2, e12112 (2022).
Acknowledgements
We thank N. G. Martin, C. V. Dolan, J. Couvée and J. van Dongen for their valuable contributions to the draft versions of this manuscript. The current work is supported by the Royal Netherlands Academy of Science Professor Award (no. PAH/6635) to D.I.B. The funder had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
Conceptualization: D.I.B. Funding acquisition: D.I.B. Project administration: F.A.H. and D.I.B. Supervision: D.I.B. Visualization: F.A.H., S. Bruins, D.V.K. and V.V.O. Writing—original draft: all authors. Writing—review and editing: all authors. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Human Behaviour thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Tables 1–3 and references.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hagenbeek, F.A., Hirzinger, J.S., Breunig, S. et al. Maximizing the value of twin studies in health and behaviour. Nat Hum Behav 7, 849–860 (2023). https://doi.org/10.1038/s41562-023-01609-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41562-023-01609-6
This article is cited by
-
Genetic and environmental contributions to epigenetic aging across adolescence and young adulthood
Clinical Epigenetics (2025)
-
Twin modelling reveals partly distinct genetic pathways to music enjoyment
Nature Communications (2025)
-
Genetic and environment influences on childhood victimization: a systematic review and meta-analysis
Molecular Psychiatry (2025)
-
Mental health-related genes that are sensitive to the environment
Nature Human Behaviour (2025)
-
Therapeutic use of plant parts as food or medicine at succulent state: an alternative healthcare system for the prevention and treatment of diseases
Discover Plants (2025)