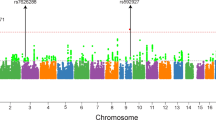
Abstract
Genome-wide association studies (GWASs) have reported multiple risk loci for schizophrenia (SCZ). However, the majority of the associations were from populations of European ancestry. Here we conducted a large-scale GWAS in Eastern Asian populations (29,519 cases and 44,392 controls) and identified ten Eastern Asian-specific risk loci, two of which have not been previously reported. A further cross-ancestry GWAS meta-analysis (96,806 cases and 492,818 controls) including populations from diverse ancestries identified 61 previously unreported risk loci. Systematic variant-level analysis, including fine mapping, functional genomics and expression quantitative trait loci, prioritized potential causal variants. Gene-level analyses, including transcriptome-wide association study, proteome-wide association study and Mendelian randomization, nominated the potential causal genes. By integrating evidence from layers of different analyses, we prioritized the most plausible causal genes for SCZ, such as ACE, CNNM2, SNAP91, ABCB9 and GATAD2A. Finally, drug repurposing showed that ACE, CA14, MAPK3 and MAPT are potential therapeutic targets for SCZ. Our study not only showed the power of cross-ancestry GWAS in deciphering the genetic aetiology of SCZ, but also uncovered new genetic risk loci, potential causal variants and genes and therapeutic targets for SCZ.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
27,99 € / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
118,99 € per year
only 9,92 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The genome-wide summary statistics of the SCZ (PGC3) are available via Figshare at https://doi.org/10.6084/m9.figshare.19426775.v6 (ref. 81). The genome-wide summary statistics of the EAS are available via Figshare at https://doi.org/10.6084/m9.figshare.19193084.v1 (ref. 82). The genome-wide summary statistics of the FinnGen are available at https://storage.googleapis.com/finngen-public-data-r9/summary_stats/. The SNP expression weights of PsychENCODE used in this study are available at http://resource.psychencode.org/. The processed protein weight files are available via the Synapse portal (Synapse IDs syn9884314 and syn23245237). The PsychENCODE cis-eQTL data are available via the SMR website at https://yanglab.westlake.edu.cn/data/SMR/PsychENCODE_cis_eqtl_HCP100_summary.tar.gz. The gene expression data used for MAGMA were from the Genotype-Tissue Expression (GTEx) consortium, GTEx_Analysis_2017-06-05_v8_RNASeQCv1.1.9. All Gene Ontology terms (including cellular components, biological processes and molecular functions) and Kyoto Encyclopedia of Genes and Genomes pathway gene sets are available via the MSigDB database at https://www.gsea-msigdb.org/gsea/msigdb/human/collections.jsp#C5. The BrainMeta V2 eQTL data are available via the BrainMeta portal at https://yanglab.westlake.edu.cn/data/SMR/BrainMeta_cis_eqtl_summary.tar.gz. Genome-wide summary statistics for EUR-ancestry meta-analysis of SCZ (59,901 cases and 441,418 controls) and GWAS summary statistics for cross-ancestry meta-analysis of SCZ (excluding the Chinese population, a total of 90,065 cases and 483,788 controls) are available via Figshare at https://doi.org/10.6084/m9.figshare.27313227 (ref. 83). The GWAS dataset for the Chinese population can be requested from the China National Genomics Data Center (https://ngdc.cncb.ac.cn/), with the data accession number OMIX005075.
References
Saha, S., Chant, D., Welham, J. & McGrath, J. A systematic review of the prevalence of schizophrenia. PLoS Med. 2, e141 (2005).
Owen, M. J., Sawa, A. & Mortensen, P. B. Schizophrenia. Lancet 388, 86–97 (2016).
Sullivan, P. F., Kendler, K. S. & Neale, M. C. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch. Gen. Psychiatry 60, 1187–1192 (2003).
Trubetskoy, V. et al. Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature 604, 502–508 (2022).
Lam, M. et al. Comparative genetic architectures of schizophrenia in East Asian and European populations. Nat. Genet. 51, 1670–1678 (2019).
Pardiñas, A. F. et al. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat. Genet. 50, 381–389 (2018).
Yu, H. et al. Common variants on 2p16.1, 6p22.1 and 10q24.32 are associated with schizophrenia in Han Chinese population. Mol. Psychiatry 22, 954–960 (2017).
Li, Z. et al. Genome-wide association analysis identifies 30 new susceptibility loci for schizophrenia. Nat. Genet. 49, 1576–1583 (2017).
Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427 (2014).
Hamshere, M. L. et al. Genome-wide significant associations in schizophrenia to ITIH3/4, CACNA1C and SDCCAG8, and extensive replication of associations reported by the Schizophrenia PGC. Mol. Psychiatry 18, 708–712 (2013).
Shi, Y. et al. Common variants on 8p12 and 1q24.2 confer risk of schizophrenia. Nat. Genet. 43, 1224–1227 (2011).
Schizophrenia Psychiatric Genome-Wide Association Study Consortium. Genome-wide association study identifies five new schizophrenia loci. Nat. Genet. 43, 969–976 (2011).
Stefansson, H. et al. Common variants conferring risk of schizophrenia. Nature 460, 744–747 (2009).
Purcell, S. M. et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 460, 748–752 (2009).
O’Donovan, M. C. et al. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat. Genet. 40, 1053–1055 (2008).
Liu, J. et al. Genome-wide association study followed by trans-ancestry meta-analysis identify 17 new risk loci for schizophrenia. BMC Med. 19, 177 (2021).
Ikeda, M. et al. Genome-wide association study detected novel susceptibility genes for schizophrenia and shared trans-populations/diseases genetic effect. Schizophr. Bull. 45, 824–834 (2019).
Hamdan, F. F. et al. Mutations in SYNGAP1 in autosomal non-syndromic mental retardation. N. Engl. J. Med. 360, 599–605 (2009).
Clement, J. P. et al. Pathogenic SYNGAP1 mutations impair cognitive development by disrupting maturation of dendritic spine synapses. Cell 151, 709–723 (2012).
Aceti, M. et al. Syngap1 haploinsufficiency damages a postnatal critical period of pyramidal cell structural maturation linked to cortical circuit assembly. Biol. Psychiatry 77, 805–815 (2015).
Berryer, M. H. et al. Decrease of SYNGAP1 in GABAergic cells impairs inhibitory synapse connectivity, synaptic inhibition and cognitive function. Nat. Commun. 7, 13340 (2016).
Carreno-Munoz, M. I. et al. Sensory processing dysregulations as reliable translational biomarkers in SYNGAP1 haploinsufficiency. Brain 145, 754–769 (2022).
Deciphering Developmental Disorders Study. Prevalence and architecture of de novo mutations in developmental disorders. Nature 542, 433–438 (2017).
Lim, Y. S. & Tang, B. L. The Evi5 family in cellular physiology and pathology. FEBS Lett. 587, 1703–1710 (2013).
Itoh, T., Satoh, M., Kanno, E. & Fukuda, M. Screening for target Rabs of TBC (Tre-2/Bub2/Cdc16) ___domain-containing proteins based on their Rab-binding activity. Genes Cells 11, 1023–1037 (2006).
Bulik-Sullivan, B. K. et al. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015).
Zheng, J. et al. LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics 33, 272–279 (2017).
Forde, A., Hemani, G. & Ferguson, J. Review and further developments in statistical corrections for winner’s curse in genetic association studies. PLoS Genet. 19, e1010546 (2023).
Song, J. et al. The impact of educational attainment, intelligence and intellectual disability on schizophrenia: a Swedish population-based register and genetic study. Mol. Psychiatry 27, 2439–2447 (2022).
Speed, D., Holmes, J. & Balding, D. J. Evaluating and improving heritability models using summary statistics. Nat. Genet. 52, 458–462 (2020).
Huo, Y., Li, S., Liu, J., Li, X. & Luo, X. J. Functional genomics reveal gene regulatory mechanisms underlying schizophrenia risk. Nat. Commun. 10, 670 (2019).
Nishizaki, S. S. et al. Predicting the effects of SNPs on transcription factor binding affinity. Bioinformatics 36, 364–372 (2020).
Yang, Z. et al. CARMA is a new Bayesian model for fine-mapping in genome-wide association meta-analyses. Nat. Genet. 55, 1057–1065 (2023).
Peterson, R. E. et al. Genome-wide association studies in ancestrally diverse populations: opportunities, methods, pitfalls, and recommendations. Cell 179, 589–603 (2019).
Gao, B. & Zhou, X. MESuSiE enables scalable and powerful multi-ancestry fine-mapping of causal variants in genome-wide association studies. Nat. Genet. 56, 170–179 (2024).
Qi, T. et al. Genetic control of RNA splicing and its distinct role in complex trait variation. Nat. Genet. 54, 1355–1363 (2022).
Gallagher, M. D. & Chen-Plotkin, A. S. The post-GWAS era: from association to function. Am. J. Hum. Genet 102, 717–730 (2018).
Gusev, A. et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet. 48, 245–252 (2016).
Gandal, M. J. et al. Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science 362, eaat8127 (2018).
Zhu, Z. et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. 48, 481–487 (2016).
Reay, W. R. & Cairns, M. J. Advancing the use of genome-wide association studies for drug repurposing. Nat. Rev. Genet. 22, 658–671 (2021).
Schmidt, A. F. et al. Genetic drug target validation using Mendelian randomisation. Nat. Commun. 11, 3255 (2020).
Gaziano, L. et al. Actionable druggable genome-wide Mendelian randomization identifies repurposing opportunities for COVID-19. Nat. Med. 27, 668–676 (2021).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010).
Weeks, E. M. et al. Leveraging polygenic enrichments of gene features to predict genes underlying complex traits and diseases. Nat. Genet. 55, 1267–1276 (2023).
Liu, J. Z. et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet. 47, 979–986 (2015).
Loh, P. R. et al. Contrasting genetic architectures of schizophrenia and other complex diseases using fast variance-components analysis. Nat. Genet. 47, 1385–1392 (2015).
Zuk, O. et al. Searching for missing heritability: designing rare variant association studies. Proc. Natl Acad. Sci. USA 111, E455–E464 (2014).
Singh, T. et al. Rare coding variants in ten genes confer substantial risk for schizophrenia. Nature 604, 509–516 (2022).
Marshall, C. R. et al. Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat. Genet. 49, 27–35 (2017).
Trieu, B. H. et al. Angiotensin-converting enzyme gates brain circuit-specific plasticity via an endogenous opioid. Science 375, 1177–1182 (2022).
Sa, M. et al. Hypothalamic GABRA5-positive neurons control obesity via astrocytic GABA. Nat. Metab. 5, 1506–1525 (2023).
Lichter, D. I. et al. Sequence analysis of beta-subunit genes of the 20S proteasome in patients with relapsed multiple myeloma treated with bortezomib or dexamethasone. Blood 120, 4513–4516 (2012).
Sebastian, R. et al. Schizophrenia-associated NRXN1 deletions induce developmental-timing- and cell-type-specific vulnerabilities in human brain organoids. Nat. Commun. 14, 3770 (2023).
Howes, O., McCutcheon, R. & Stone, J. Glutamate and dopamine in schizophrenia: an update for the 21st century. J. Psychopharmacol. 29, 97–115 (2015).
Pocklington, A. J. et al. Novel findings from CNVs implicate inhibitory and excitatory signaling complexes in schizophrenia. Neuron 86, 1203–1214 (2015).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet 81, 559–575 (2007).
Manichaikul, A. et al. Robust relationship inference in genome-wide association studies. Bioinformatics 26, 2867–2873 (2010).
Price, A. L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006).
Yang, J., Lee, S. H., Goddard, M. E. & Visscher, P. M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet 88, 76–82 (2011).
Das, S. et al. Next-generation genotype imputation service and methods. Nat. Genet. 48, 1284–1287 (2016).
Loh, P. R. et al. Reference-based phasing using the Haplotype Reference Consortium panel. Nat. Genet. 48, 1443–1448 (2016).
Yifan, L. et al. Cross-ancestry genome-wide association study and systems-level integrative analyses implicate new risk genes and therapeutic targets for depression. Preprint at medRxiv https://doi.org/10.1101/2023.02.24.23286411 (2023).
Li, W. et al. Genome-wide meta-analysis, functional genomics and integrative analyses implicate new risk genes and therapeutic targets for anxiety disorders. Nat. Hum. Behav. 8, 361–379 (2023).
Watanabe, K., Taskesen, E., van Bochoven, A. & Posthuma, D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 8, 1826 (2017).
Evans, L. M. et al. Comparison of methods that use whole genome data to estimate the heritability and genetic architecture of complex traits. Nat. Genet. 50, 737–745 (2018).
Bulik-Sullivan, B. et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 47, 1236–1241 (2015).
Speed, D. & Balding, D. J. SumHer better estimates the SNP heritability of complex traits from summary statistics. Nat. Genet. 51, 277–284 (2019).
Bryois, J. et al. Genetic identification of cell types underlying brain complex traits yields insights into the etiology of Parkinson’s disease. Nat. Genet. 52, 482–493 (2020).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Wallace, C. Statistical testing of shared genetic control for potentially related traits. Genet. Epidemiol. 37, 802–813 (2013).
Wingo, A. P. et al. Integrating human brain proteomes with genome-wide association data implicates new proteins in Alzheimer’s disease pathogenesis. Nat. Genet. 53, 143–146 (2021).
Chen, R. et al. Functional genomics elucidates regulatory mechanisms of Parkinson’s disease-associated variants. BMC Med. 20, 68 (2022).
ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Bailey, T. L. & Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int Conf. Intell. Syst. Mol. Biol. 2, 28–36 (1994).
Auton, A. et al. A global reference for human genetic variation. Nature 526, 68–74 (2015).
Grant, C. E., Bailey, T. L. & Noble, W. S. FIMO: scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018 (2011).
Zeng, B. et al. Multi-ancestry eQTL meta-analysis of human brain identifies candidate causal variants for brain-related traits. Nat. Genet. 54, 161–169 (2022).
Sullivan, P. et al. Genome-wide summary statistics of the SCZ (PGC3). figshare https://doi.org/10.6084/m9.figshare.19426775.v6 (2022).
Sullivan, P. et al. Genome-wide summary statistics of the EAS. figshare https://doi.org/10.6084/m9.figshare.19193084.v1 (2022).
Dang, X. et al. Summary statistics from the schizophrenia cross-ancestry GWAS meta-analysis. figshare https://doi.org/10.6084/m9.figshare.27313227 (2024).
Acknowledgements
This study was equally supported by the National Natural Science Foundation of China (U2102205 and U22A20304) and startup funds from Southeast University (RF1028623032). It was also supported by the Special Project for Social Development of Yunnan Province (no. 202203AC100007), Key Research and Development Projects of Henan Province (241111312800 and 235101610004 to W.L.), Intestinal Microbiota Transplantation Engineering Research Center, Yunnan Provincial Department of Education (to Z.T.), the National Natural Science Foundation of China (nos. 82171498, 82230044, 81920108018, U21A20364, 82260276, 82301690, 82371508 and 82371508), the Talent and Fundamental Research Project of Yunnan Province (nos. 202301AY070001-299, 202105AC160004 and 202301AT070037), Key R&D Program of Zhejiang Province (grant number 2022C03096 to T.L.), the Key R & D by Hangzhou Science and Technology Bureau (grant number 20241203A14 to T.L.) and the Construction Fund of Key Medical Disciplines of Hangzhou (OO20200065 to T.L.). We thank Q. Li and Z. Ding for their technical assistance. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
X.-J.L. conceived, designed and supervised the whole study. X.D. performed most of the analyses. Z.T., Y. Yang, W.L., J.L., L.H., D.Z., X.L., L.L., Y.Z., D.G., S.-S.D., Y.L., Y. Yuan, X.M., Z.L. and T.L. contributed to sample collection, genotyping and paper writing. All authors revised the paper critically and approved the final version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Human Behaviour thanks Stephanie Le Hellard, Derek Morris and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–40.
Supplementary Table 1
Supplementary Tables 1–19.
Source Data Table 1
Statistical source data for Supplementary Figs. 31–39.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Dang, X., Teng, Z., Yang, Y. et al. Gene-level analysis reveals the genetic aetiology and therapeutic targets of schizophrenia. Nat Hum Behav 9, 609–624 (2025). https://doi.org/10.1038/s41562-024-02091-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41562-024-02091-4