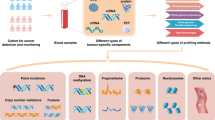
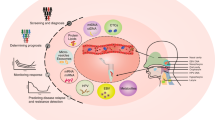
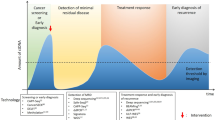
Abstract
Cancer screening is an essential public health intervention for diagnosing cancers at an early stage that can enable earlier treatment — ideally with curative intent — and thus lead to improved outcomes. Over the past decade, liquid biopsy-based tests have emerged as a promising, minimally invasive and broadly applicable screening approach by combining multi-cancer early detection (MCED) with tumour tissue-of-origin identification. Large-scale randomized clinical trials evaluating liquid biopsy-based MCED approaches are now under way, although whether the diagnostic performance of this first generation of MCED tests is sufficient to translate into clinical benefits remains to be determined. In this Review, we discuss the promises and pitfalls of current MCED tests and highlight possible trajectories for the field of early cancer detection.
Key points
-
Cancer screening programmes have led to clinical benefits across a number of tumour types, although a universal cancer screening test does not yet exist. Multi-cancer early detection (MCED) via a single test requires sampling or imaging of all sites in the body.
-
Liquid biopsies enable minimally invasive detection of circulating nucleic acids, proteins and/or cancer cells. As these technologies have matured and technical performance improved, investigators are exploring the potential for liquid biopsy-based screening.
-
The analytical sensitivity of liquid biopsy-based MCED tests is a function of both the volume of plasma sampled and the number of markers targeted. This concept is well-established in the field of personalized monitoring of cancer using liquid biopsy assays based on mutations; here, we appraise the performance of MCED tests through this same lens.
-
Methylation-based MCED tests have rapidly gained traction and are being tested in randomized controlled trials, which reflects their high sensitivity and specificity. Competing technologies might emerge over time, particularly as sequencing costs decrease further.
-
Ongoing clinical trials should not only assess whether screening with liquid biopsy-based MCED tests detects cancers earlier but also determine whether this translates into real clinical benefit on a population level.
-
Outcomes from upcoming randomized controlled trials will be crucial for guiding the development of the next generation of MCED assays, bringing the field a step closer to the implementation of blood test-based cancer screening.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
27,99 € / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
209,00 € per year
only 17,42 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Ferlay, J. et al. Cancer statistics for the year 2020: an overview. Int. J. Cancer https://doi.org/10.1002/ijc.33588 (2021).
World Health Organization. Global health estimates 2020: deaths by cause, age, sex, by country and by region, 2000-2019. World Health Organization http://who.int/data/gho/data/themes/mortality-and-global-health-estimates/ghe-leading-causes-of-death (2020).
Independent UK Panel on Breast Cancer Screening.The benefits and harms of breast cancer screening: an independent review. Lancet 380, 1778–1786 (2012).
Field, J. K. et al. Lung cancer mortality reduction by LDCT screening: UKLS randomised trial results and international meta-analysis. Lancet Reg. Health Eur. 10, 100179 (2021).
Lin, J. S., Perdue, L. A., Henrikson, N. B., Bean, S. I. & Blasi, P. R. Screening for colorectal cancer: updated evidence report and systematic review for the US preventive services task force. JAMA 325, 1978–1998 (2021).
World Health Organization. WHO guidelines for screening and treatment of precancerous lesions for cervical cancer prevention. World Health Organization https://iris.who.int/bitstream/handle/10665/94830/9789241548694_eng.pdf (2013).
NORC at the University of Chicago. Percent of Cancers Detected by Screening in the U.S. https://cancerdetection.norc.org/ (2022).
White, T. & Algeri, S. Estimating the lifetime risk of a false positive screening test result. PLoS ONE 18, e0281153 (2023).
Wan, J. C. M. et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat. Rev. Cancer 17, 223–238 (2017).
Dudley, J. C. et al. Detection and surveillance of bladder cancer using urine tumor DNA. Cancer Discov. 9, 500–509 (2019).
Mouliere, F. et al. Fragmentation patterns and personalized sequencing of cell‐free DNA in urine and plasma of glioma patients. EMBO Mol. Med. 13, e12881 (2021).
Wang, Y. et al. Detection of somatic mutations and HPV in the saliva and plasma of patients with head and neck squamous cell carcinomas. Sci. Transl. Med. 7, 293ra104 (2015).
Markar, S. R. et al. Assessment of a noninvasive exhaled breath test for the diagnosis of oesophagogastric cancer. JAMA Oncol. 4, 970–976 (2018).
Leon, S. A., Shapiro, B., Sklaroff, D. M. & Yaros, M. J. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 37, 646–650 (1977).
Kopreski, M. S., Benko, F. A., Kwak, L. W. & Gocke, C. D. Detection of tumor messenger RNA in the serum of patients with malignant melanoma. Clin. Cancer Res. 5, 1961–1965 (1999).
Papsidero, L. D., Wang, M. C., Valenzuela, L. A., Murphy, G. P. & Chu, T. M. A prostate antigen in sera of prostatic cancer patients. Cancer Res. 40, 2428–2432 (1980).
Bast, R. C. Jr. et al. Monitoring human ovarian carcinoma with a combination of CA 125, CA 19-9, and carcinoembryonic antigen. Am. J. Obstet. Gynecol. 149, 553–559 (1984).
Kahlert, C. & Kalluri, R. Exosomes in tumor microenvironment influence cancer progression and metastasis. J. Mol. Med. 91, 431–437 (2013).
Ashworth, T. R. A case of cancer in which cells similar to those in the tumours were seen in the blood after death. Med. J. Aust. 14, 146–147 (1869).
Bronkhorst, A. J. et al. Towards systematic nomenclature for cell-free DNA. Hum. Genet. 140, 565–578 (2021).
Cohen, J. D. et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 359, 926–930 (2018).
Shen, S. Y. et al. Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature 563, 579–583 (2018).
Neal, R. D. et al. Cell-free DNA-based multi-cancer early detection test in an asymptomatic screening population (NHS-Galleri): design of a pragmatic, prospective randomised controlled trial. Cancers 14, 4818 (2022).
Lennon, A. M. et al. Outcomes following a false positive multi-cancer early detection (MCED) test: results from DETECT-A, the first large, prospective, interventional MCED study. Cancer Prev. Res. 17, 355–359 (2024).
Rampinelli, C. et al. Exposure to low dose computed tomography for lung cancer screening and risk of cancer: secondary analysis of trial data and risk-benefit analysis. BMJ 356, j347 (2017).
Berrington de González, A. et al. Projected cancer risks from computed tomographic scans performed in the United States in 2007. Arch. Intern. Med. 169, 2071–2077 (2009).
Morin, S. H. X. et al. Incidental findings in healthy control research subjects using whole-body MRI. Eur. J. Radiol. 72, 529–533 (2009).
Park, J. H. et al. Systematic review and meta-analysis of the accuracy and applicability of blood-based multi-cancer early detection (MCED) in the general population. J. Clin. Oncol. 41, 3069 (2023).
LeeVan, E. & Pinsky, P. Predictive performance of cell-free nucleic acid-based multi-cancer early detection tests: a systematic review. Clin. Chem. 70, 90–101 (2024).
Wade, R. et al. Multi-cancer early detection tests for general population screening: a systematic literature review. Health Technol. Assess. 29, 1–105 (2025).
Sorenson, G. D. et al. Soluble normal and mutated DNA sequences from single-copy genes in human blood. Cancer Epidemiol. Biomarkers Prev. 3, 67–67 (1994).
Diehl, F. et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc. Natl Acad. Sci. USA 102, 16368–16373 (2005).
Stroun, M., Anker, P., Lyautey, J., Lederrey, C. & Maurice, P. A. Isolation and characterization of DNA from the plasma of cancer patients. Eur. J. Cancer Clin. Oncol. 23, 707–712 (1987).
Forshew, T. et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci. Transl. Med. 4, 136ra68 (2012).
Newman, A. M. et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med. 20, 548–554 (2014).
Wan, J. C. M. et al. Liquid biopsies for residual disease and recurrence. Med 2, 1292–1313 (2021).
Wan, J. C. M. et al. ctDNA monitoring using patient-specific sequencing and integration of variant reads. Sci. Transl. Med. 12, eaaz8084 (2020).
Heider, K. et al. Abstract 736: ctDNA detection in early stage non-small cell lung cancer. Cancer Res. 80, 736 (2020).
Newman, A. M. et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat. Biotechnol. 34, 547–555 (2016).
Avanzini, S. et al. A mathematical model of ctDNA shedding predicts tumor detection size. Sci. Adv. 6, eabc4308 (2020).
Phallen, J. et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci. Transl. Med. 9, eaan2415 (2017).
Bettegowda, C. et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 6, 224ra24 (2014).
Lennon, A. M. et al. Feasibility of blood testing combined with PET-CT to screen for cancer and guide intervention. Science 369, eabb9601 (2020).
Huang, B., Law, M. W. M. & Khong, P. L. Whole-body PET/CT scanning: estimation of radiation dose and cancer risk. Radiology 251, 166–174 (2009).
Mouliere, F. et al. High fragmentation characterizes tumour-derived circulating DNA. PLoS ONE 6, e23418 (2011).
Mouliere, F., El Messaoudi, S., Pang, D., Dritschilo, A. & Thierry, A. R. Multi-marker analysis of circulating cell-free DNA toward personalized medicine for colorectal cancer. Mol. Oncol. 8, 927–941 (2014).
Leary, R. J. et al. Detection of chromosomal alterations in the circulation of cancer patients with whole-genome sequencing. Sci. Transl. Med. 4, 162ra154 (2012).
Murtaza, M. et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 497, 108–112 (2013).
Cheng, A. P. et al. Error-corrected flow-based sequencing at whole-genome scale and its application to circulating cell-free DNA profiling. Nat. Methods 22, 973–981 (2025).
Abbosh, C. et al. Phylogenetic ctDNA analysis depicts early stage lung cancer evolution. Nature 545, 446–451 (2017).
Heider, K. et al. ctDNA detection by personalised assays in early-stage NSCLC. Preprint at medRxiv https://doi.org/10.1101/2021.06.01.21258171 (2021).
Goldstraw, P. et al. The IASLC lung cancer staging project: proposals for revision of the TNM stage groupings in the forthcoming (eighth) edition of the TNM classification for lung cancer. J. Thorac. Oncol. 11, 39–51 (2016).
Black, J. R. M. et al. Ultrasensitive ctDNA detection for preoperative disease stratification in early-stage lung adenocarcinoma. Nat. Med. 31, 70–76 (2025).
Aigrain, L., Gu, Y. & Quail, M. A. Quantitation of next generation sequencing library preparation protocol efficiencies using droplet digital PCR assays — a systematic comparison of DNA library preparation kits for Illumina sequencing. BMC Genomics 17, 458 (2016).
Terp, S. K., Pedersen, I. S. & Stoico, M. P. Extraction of cell-free DNA: evaluation of efficiency, quantity, and quality. J. Mol. Diagn. 26, 310–319 (2024).
Husain, H. et al. Tumor fraction correlates with detection of actionable variants across >23,000 circulating tumor DNA samples. JCO Precis. Oncol. 6, e2200261 (2022).
Martin-Alonso, C. et al. Priming agents transiently reduce the clearance of cell-free DNA to improve liquid biopsies. Science 383, eadf2341 (2024).
Bae, J. H. et al. Single duplex DNA sequencing with CODEC detects mutations with high sensitivity. Nat. Genet. 55, 871–879 (2023).
Schmitt, M. W. et al. Detection of ultra-rare mutations by next-generation sequencing. Proc. Natl Acad. Sci. 109, 14508–14513 (2012).
Jaiswal, S. & Ebert, B. L. Clonal hematopoiesis in human aging and disease. Science 366, eaan4673 (2019).
Genovese, G. et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl. J. Med. 371, 2477–2487 (2014).
Hu, Y. et al. False-positive plasma genotyping due to clonal hematopoiesis. Clin. Cancer Res. 24, 4437–4443 (2018).
Lecomte, T. et al. Detection of free-circulating tumor-associated DNA in plasma of colorectal cancer patients and its association with prognosis. Int. J. Cancer 100, 542–548 (2002).
Jensen, S. Ø. et al. Novel DNA methylation biomarkers show high sensitivity and specificity for blood-based detection of colorectal cancer — a clinical biomarker discovery and validation study. Clin. Epigenetics 11, 158 (2019).
Lehmann-Werman, R. et al. Identification of tissue-specific cell death using methylation patterns of circulating DNA. Proc. Natl Acad. Sci. USA 113, E1826–E1834 (2016).
Alexander, G. E. et al. Analytical validation of a multi-cancer early detection test with cancer signal origin using a cell-free DNA-based targeted methylation assay. PLoS ONE 18, e0283001 (2023).
Liu, L. et al. Targeted methylation sequencing of plasma cell-free DNA for cancer detection and classification. Ann. Oncol. 29, 1445–1453 (2018).
Loyfer, N. et al. A DNA methylation atlas of normal human cell types. Nature 613, 355–364 (2023).
Sun, K. et al. Plasma DNA tissue mapping by genome-wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc. Natl Acad. Sci. 112, E5503–E5512 (2015).
Moss, J. et al. Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nat. Commun. 9, 5068 (2018).
Liu, M. C. et al. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann. Oncol. 31, 745–759 (2020).
Klein, E. A. et al. Clinical validation of a targeted methylation-based multi-cancer early detection test using an independent validation set. Ann. Oncol. 32, 1167–1177 (2021).
Liang, N. et al. Ultrasensitive detection of circulating tumour DNA via deep methylation sequencing aided by machine learning. Nat. Biomed. Eng. 5, 586–599 (2021).
Gao, Q. et al. Unintrusive multi-cancer detection by circulating cell-free DNA methylation sequencing (THUNDER): development and independent validation studies. Ann. Oncol. 34, 486–495 (2023).
Chen, X. et al. Non-invasive early detection of cancer four years before conventional diagnosis using a blood test. Nat. Commun. 11, 3475 (2020).
Schrag, D. et al. Blood-based tests for multicancer early detection (PATHFINDER): a prospective cohort study. Lancet 402, 1251–1260 (2023).
Sasieni, P. D. Effect of an invitation or the effect of participation: what should randomized controlled trials of cancer screening examine? J. Clin. Oncol. 42, 3266–3269 (2024).
Wood, M. et al. The Vanguard study: an NCI cancer screening research network multi-cancer detection feasibility study. J. Clin. Oncol. 43, e22522–e22522 (2025).
Hubbell, E., Clarke, C. A., Aravanis, A. M. & Berg, C. D. Modeled reductions in late-stage cancer with a multi-cancer early detection test. Cancer Epidemiol. Biomarkers Prev. 30, 460–468 (2021).
Sasieni, P. et al. Modelled mortality benefits of multi-cancer early detection screening in England. Br. J. Cancer 129, 72–80 (2023).
Tabár, L. et al. Insights from the breast cancer screening trials: how screening affects the natural history of breast cancer and implications for evaluating service screening programs. Breast J. 21, 13–20 (2015).
Hu, P. et al. Design considerations and challenges in the CHinA National CancEr Screening (CHANCES) trial and Tomosynthesis Mammographic Imaging Screening Trial (TMIST). J. Natl Cancer Inst. Monogr. 68, 42–48 (2025).
Dai, J. Y. et al. Strong association between reduction of late-stage cancers and reduction of cancer-specific mortality in meta-regression of randomized screening trials across multiple cancer types. J. Med. Screen. https://doi.org/10.1177/09691413241256744 (2024).
Feng, X. et al. Cancer stage compared with mortality as end points in randomized clinical trials of cancer screening: a systematic review and meta-analysis. JAMA 331, 1910–1917 (2024).
Webb, A. B. et al. Considerations for using potential surrogate endpoints in cancer screening trials. Lancet Oncol. 25, e183–e192 (2024).
Sasieni, P., Swanton, C. & Neal, R. Advanced cancer: a robust surrogate of cancer mortality in early detection trials? Ann. Oncol. https://doi.org/10.1016/j.annonc.2025.03.001 (2025).
Melton, C. A. et al. A novel tissue-free method to estimate tumor-derived cell-free DNA quantity using tumor methylation patterns. Cancers 16, 82 (2023).
Uddin, Md. M. et al. Clonal hematopoiesis of indeterminate potential, DNA methylation, and risk for coronary artery disease. Nat. Commun. 13, 5350 (2022).
Song, C. X. et al. 5-Hydroxymethylcytosine signatures in cell-free DNA provide information about tumor types and stages. Cell Res. 27, 1231–1242 (2017).
Park, M. K., Lee, J. C., Lee, J. W. & Hwang, S. J. Alu cell-free DNA concentration, Alu index, and LINE-1 hypomethylation as a cancer predictor. Clin. Biochem. 94, 67–73 (2021).
Zhang, X. et al. Evaluating methylation of human ribosomal DNA at each CpG site reveals its utility for cancer detection using cell-free DNA. Brief. Bioinform. 23, bbac278 (2022).
Snyder, M. W., Kircher, M., Hill, A. J., Daza, R. M. & Shendure, J. Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell 164, 57–68 (2016).
Bai, J. et al. Histone modifications of circulating nucleosomes are associated with changes in cell-free DNA fragmentation patterns. Proc. Natl Acad. Sci. USA 121, e2404058121 (2024).
Zhou, X. et al. CRAG: de novo characterization of cell-free DNA fragmentation hotspots in plasma whole-genome sequencing. Genome Med. 14, 138 (2022).
Esfahani, M. S. et al. Inferring gene expression from cell-free DNA fragmentation profiles. Nat. Biotechnol. 40, 585–597 (2022).
Underhill, H. R. et al. Fragment length of circulating tumor DNA. PLoS Genet. 12, 426–437 (2016).
Jiang, P. et al. Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc. Natl Acad. Sci. USA 112, E1317–E1325 (2015).
Mouliere, F. et al. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci. Transl. Med. 10, eaat4921 (2018).
Jiang, P. et al. Plasma DNA end-motif profiling as a fragmentomic marker in cancer, pregnancy, and transplantation. Cancer Discov. 10, 664–673 (2020).
Zhou, Z. et al. Fragmentation landscape of cell-free DNA revealed by deconvolutional analysis of end motifs. Proc. Natl Acad. Sci. USA 120, e2220982120 (2023).
Ulz, P. et al. Inferring expressed genes by whole-genome sequencing of plasma DNA. Nat. Genet. 48, 1273–1278 (2016).
Ulz, P. et al. Inference of transcription factor binding from cell-free DNA enables tumor subtype prediction and early detection. Nat. Commun. 10, 4666 (2019).
Jiang, P. et al. Preferred end coordinates and somatic variants as signatures of circulating tumor DNA associated with hepatocellular carcinoma. Proc. Natl Acad. Sci. USA 115, E10925–E10933 (2018).
Wan, N. et al. Machine learning enables detection of early-stage colorectal cancer by whole-genome sequencing of plasma cell-free DNA. BMC Cancer 19, 832 (2019).
Sun, K. et al. Orientation-aware plasma cell-free DNA fragmentation analysis in open chromatin regions informs tissue of origin. Genome Res. 29, 418–427 (2019).
Jiang, P. & Lo, Y. M. D. The long and short of circulating cell-free DNA and the ins and outs of molecular diagnostics. Trends Genet. 32, 360–371 (2016).
Cristiano, S. et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature 570, 385–389 (2019).
Helzer, K. T. et al. Fragmentomic analysis of circulating tumor DNA-targeted cancer panels. Ann. Oncol. 34, 813–825 (2023).
Mazzone, P. J. et al. Clinical validation of a cell-free DNA fragmentome assay for augmentation of lung cancer early detection. Cancer Discov. 14, 2224–2242 (2024).
Yang, S. et al. 1263 — Interim results from a large-scale, prospective cohort study (JINLING) for multi-cancer early detection test in average-risk asymptomatic patients. Cancer Res. 84, 1263 (2024).
Bao, H. et al. Letter to the Editor: An ultra-sensitive assay using cell-free DNA fragmentomics for multi-cancer early detection. Mol. Cancer 21, 129 (2022).
Nguyen, T. H. H. et al. Clinical validation of a ctDNA-based assay for multi-cancer detection: an interim report from a Vietnamese longitudinal prospective cohort study of 2795 participants. Cancer Invest. 41, 232–248 (2023).
Liu, Y. et al. FinaleMe: predicting DNA methylation by the fragmentation patterns of plasma cell-free DNA. Nat. Commun. 15, 2790 (2024).
Bruhm, D. C. et al. Single-molecule genome-wide mutation profiles of cell-free DNA for non-invasive detection of cancer. Nat. Genet. 55, 1301–1310 (2023).
Adalsteinsson, V. A. et al. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat. Commun. 8, 1324 (2017).
Wan, J. C. M. et al. Genome-wide mutational signatures in low-coverage whole genome sequencing of cell-free DNA. Nat. Commun. 13, 4953 (2022).
Larson, M. H. et al. A comprehensive characterization of the cell-free transcriptome reveals tissue- and subtype-specific biomarkers for cancer detection. Nat. Commun. 12, 2357 (2021).
Roskams-Hieter, B. et al. Plasma cell-free RNA profiling distinguishes cancers from pre-malignant conditions in solid and hematologic malignancies. NPJ Precis. Oncol. 6, 28 (2022).
Reggiardo, R. E. et al. Profiling of repetitive RNA sequences in the blood plasma of patients with cancer. Nat. Biomed. Eng. 7, 1627–1635 (2023).
Pan, H. et al. Serum long non-coding RNA LOC553103 as non-specific diagnostic and prognostic biomarker for common types of human cancer. Clin. Chim. Acta 508, 69–76 (2020).
Chen, X. et al. Evaluation on the diagnostic and prognostic values of long non-coding RNA BLACAT1 in common types of human cancer. Mol. Cancer 16, 160 (2017).
Mitchell, P. S. et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl Acad. Sci. USA 105, 10513–10518 (2008).
Hashimoto, K., Inada, M., Yamamoto, Y. & Ochiya, T. Preliminary evaluation of miR-1307-3p in human serum for detection of 13 types of solid cancer using microRNA chip. Heliyon 7, e07919 (2021).
Zhang, A. & Hu, H. A novel blood-based microRNA diagnostic model with high accuracy for multi-cancer early detection. Cancers 14, 1450 (2022).
Best, M. G. et al. RNA-seq of tumor-educated platelets enables blood-based pan-cancer, multiclass, and molecular pathway cancer diagnostics. Cancer Cell 28, 666–676 (2015).
In’T Veld, S. G. J. G. et al. Detection and localization of early- and late-stage cancers using platelet RNA. Cancer Cell 40, 999–1009.e6 (2022).
Nilsson, R. J. A. et al. Blood platelets contain tumor-derived RNA biomarkers. Blood 118, 3680–3683 (2011).
Yan, F. et al. Deep neural network based tissue deconvolution of circulating tumor cell RNA. J. Transl. Med. 21, 783 (2023).
Nesselbush, M. C. et al. An ultrasensitive method for detection of cell-free RNA. Nature 641, 759–768 (2025).
Wen, Y. H. et al. Cancer screening through a multi-analyte serum biomarker panel during health check-up examinations: results from a 12-year experience. Clin. Chim. Acta 450, 273–276 (2015).
Tang, C. et al. 1268 — Immune activation characterization via amino acid concentration signatures for multi-cancer early detection and CDKi treatment response prediction. Cancer Res. 84, 1268 (2024).
Budnik, B., Amirkhani, H., Forouzanfar, M. H. & Afshin, A. Novel proteomics-based plasma test for early detection of multiple cancers in the general population. BMJ Oncol. 3, e000073 (2024).
Wik, L. et al. Proximity extension assay in combination with next-generation sequencing for high-throughput proteome-wide analysis. Mol. Cell. Proteom. 20, 100168 (2021).
Allard, W. J. et al. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with Nonmalignant diseases. Clin. Cancer Res. 10, 6897–6904 (2004).
Kagan, M. et al. A sample preparation and analysis system for identification of circulating tumor cells. J. Clin. Ligand Assay. 25, 104–110 (2002).
Coumans, F. & Terstappen, L. Detection and characterization of circulating tumor cells by the CellSearch approach. Methods Mol. Biol. 1347, 263–278 (2015).
Akolkar, D. et al. Circulating ensembles of tumor‐associated cells: a redoubtable new systemic hallmark of cancer. Int. J. Cancer 146, 3485–3494 (2020).
Lin, D. et al. Circulating tumor cells: biology and clinical significance. Signal. Transduct. Target. Ther. 6, 404 (2021).
Dawson, S.-J. et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N. Engl. J. Med. 368, 1199–1209 (2013).
Gorges, T. M. et al. Enumeration and molecular characterization of tumor cells in lung cancer patients using a novel in vivo device for capturing circulating tumor cells. Clin. Cancer Res. 22, 2197–2206 (2016).
Gambhir, M., Zerda, A. D. La & Knysh, B. D. Wearable ultrasonic device for circulating tumor cell detection. US Patent US20150126861A1 (2020).
Kim, S. Y. et al. Cancer signature ensemble integrating cfDNA methylation, copy number, and fragmentation facilitates multi-cancer early detection. Exp. Mol. Med. 55, 2445–2460 (2023).
Stackpole, M. L. et al. Cost-effective methylome sequencing of cell-free DNA for accurately detecting and locating cancer. Nat. Commun. 13, 5566 (2022).
Prime, P. Harnessing the power of CD3. Cancer Research UK https://news.cancerresearchuk.org/2023/05/30/harnessing-the-power-of-cd3/ (2023).
Hernström, V. et al. Screening performance and characteristics of breast cancer detected in the mammography screening with artificial intelligence trial (MASAI): a randomised, controlled, parallel-group, non-inferiority, single-blinded, screening accuracy study. Lancet Digit. Health 7, E175–E183 (2025).
Widman, A. J. et al. Ultrasensitive plasma-based monitoring of tumor burden using machine-learning-guided signal enrichment. Nat. Med. 30, 1655–1666 (2024).
Deng, Z. et al. Early detection of hepatocellular carcinoma via no end-repair enzymatic methylation sequencing of cell-free DNA and pre-trained neural network. Genome Med. 15, 93 (2023).
Devlin, J., Chang, M.-W., Lee, K. & Toutanova, K. BERT: pre-training of deep bidirectional transformers for language understanding. Proc. 2019 Conf. North Am. Chapter Assoc. Comput. Linguist.: Hum. Lang. Technol. 1, 4171–4186 (2019).
Minasian, L. & Castle, P. E. Cancer screening research network/multi-cancer early detection evaluation. National Cancer Institute https://prevention.cancer.gov/sites/default/files/2023-02/Cancer-Screening-Research-Network-MCED-20220615.pdf (2022).
Hu, P., Prorok, P. C. & Katki, H. A. Design of randomized controlled trials to estimate cancer-mortality reductions from multicancer detection screening. J. Natl Cancer Inst. 117, 303–311 (2025).
Catto, J. W. F. et al. Protocol for the YORKSURe prospective multistage study testing the feasibility for early detection of bladder cancer in populations with high disease-specific mortality risk. BMJ Open 13, e076612 (2023).
Hackshaw, A. & Berg, C. D. An efficient randomised trial design for multi-cancer screening blood tests: nested enhanced mortality outcomes of screening trial. Lancet Oncol. 22, 1360–1362 (2021).
Katki, H. A., Prorok, P. C., Castle, P. E., Minasian, L. M. & Pinsky, P. F. Increasing power in screening trials by testing control-arm specimens: application to multicancer detection screening. J. Natl Cancer Inst. 116, 1675–1682 (2024).
Sasieni, P. & Brentnall, A. R. More efficient, smaller multicancer screening trials. J. Natl Cancer Inst. 117, 450–455 (2025).
Sasieni, P., Clarke, C. A. & Hubbell, E. 1135P Impact of MCED screening interval on reduction in late- stage cancer diagnosis and mortality. Ann. Oncol. 32, S925 (2021).
White, E. et al. Biennial versus annual mammography and the risk of late-stage breast cancer. J. Natl Cancer Inst. 96, 1832–1839 (2004).
Mandelblatt, J. S. et al. Effects of mammography screening under different screening schedules: model estimates of potential benefits and harms. Ann. Intern. Med. 151, 738–747 (2009).
Canelo-Aybar, C. et al. Benefits and harms of annual, biennial, or triennial breast cancer mammography screening for women at average risk of breast cancer: a systematic review for the European Commission Initiative on Breast Cancer (ECIBC). Br. J. Cancer 126, 673–688 (2022).
Barzilai, O. et al. Survival, local control, and health-related quality of life in patients with oligometastatic and polymetastatic spinal tumors: a multicenter, international study. Cancer 125, 770–778 (2019).
Gomez, D. R., Niibe, Y. & Chang, J. Y. Oligometastatic disease at presentation or recurrence for nonsmall cell lung cancer. Pulm. Med. 2012, 396592 (2012).
Palma, D. A. et al. Stereotactic ablative radiotherapy for the comprehensive treatment of Oligometastatic Cancers: long-term results of the SABR-COMET phase II randomized trial. J. Clin. Oncol. 38, 2830–2838 (2020).
Lu, S. et al. Osimertinib after chemoradiotherapy in stage III EGFR-mutated NSCLC. N. Engl. J. Med. 391, 585–597 (2024).
Solomon, B. J. et al. Lorlatinib versus crizotinib in patients with advanced ALK-positive non-small cell lung cancer: 5-year outcomes from the phase III CROWN study. J. Clin. Oncol. 42, 3400–3409 (2024).
Cercek, A. et al. PD-1 blockade in mismatch repair-deficient, locally advanced rectal cancer. N. Engl. J. Med. 386, 2363–2376 (2022).
Hemminki, K., Bevier, M., Hemminki, A. & Sundquist, J. Survival in cancer of unknown primary site: population-based analysis by site and histology. Ann. Oncol. 23, 1854–1863 (2012).
Rosenfeld, N. et al. MicroRNAs accurately identify cancer tissue origin. Nat. Biotechnol. 26, 462–469 (2008).
Conway, A. M. et al. A cfDNA methylation-based tissue-of-origin classifier for cancers of unknown primary. Nat. Commun. 15, 3292 (2024).
Bochtler, T. & Krämer, A. Does cancer of unknown primary (CUP) truly exist as a distinct cancer entity? Front. Oncol. 9, 402 (2019).
Blom, J., Saraste, D., Törnberg, S. & Jonsson, H. Routine fecal occult blood screening and colorectal cancer mortality in Sweden. JAMA Netw. Open 7, e240516 (2024).
Libby, G. et al. The impact of population-based faecal occult blood test screening on colorectal cancer mortality: a matched cohort study. Br. J. Cancer 107, 255–259 (2012).
IARC Working Group on the Evaluation of Cancer-Preventive Interventions. Colorectal Cancer Screening vol. 17 (International Agency for Research on Cancer, 2019).
US Food & Drug Administration. FDA news release — FDA roundup: July 30, 2024. FDA https://www.fda.gov/news-events/press-announcements/fda-roundup-july-30-2024 (2024).
Chung, D. C. et al. A cell-free DNA blood-based test for colorectal cancer screening. N. Engl. J. Med. 390, 973–983 (2024).
Imperiale, T. et al. Next-generation multitarget stool DNA test for colorectal cancer screening. N. Engl. J. Med. 390, 984–993 (2024).
Amant, F. et al. Presymptomatic identification of cancers in pregnant women during noninvasive prenatal testing. JAMA Oncol. 1, 814–819 (2015).
Turiff, E. et al. Prenatal cfDNA sequencing and incidental detection of maternal cancer. N. Engl. J. Med. 391, 2123–2132 (2024).
Roschewski, M., Rossi, D., Kurtz, D. M., Alizadeh, A. A. & Wilson, W. H. Circulating tumor DNA in lymphoma: principles and future directions. Blood Cancer Discov. 3, 5–15 (2022).
Pantel, K., Cote, R. J. & Fodstad, Ø. Detection and clinical importance of micrometastatic disease. J. Natl Cancer Inst. 91, 1113–1124 (1999).
Draisma, G. et al. Lead time and overdiagnosis in prostate-specific antigen screening: Importance of methods and context. J. Natl Cancer Inst. 101, 374–383 (2009).
Cuzick, J. et al. Tamoxifen for prevention of breast cancer: extended long-term follow-up of the IBIS-I breast cancer prevention trial. Lancet Oncol. 16, 67–75 (2015).
Cuzick, J. et al. Use of anastrozole for breast cancer prevention (IBIS-II): long-term results of a randomised controlled trial. Lancet 395, 117–122 (2020).
Cuzick, J. et al. Selective oestrogen receptor modulators in prevention of breast cancer: an updated meta-analysis of individual participant data. Lancet 381, 1827–1834 (2013).
NHS England. Evaluation of the rapid diagnostic centres. NHS England https://www.england.nhs.uk/contact-us/privacy-notice/how-we-use-your-information/our-services/evaluation-of-the-rapid-diagnostic-centres/ (2024).
Marlow, L. A. V., Schmeising-Barnes, N., Warwick, J. & Waller, J. Psychological impact of the Galleri test (sIG(n)al): protocol for a longitudinal evaluation of the psychological impact of receiving a cancer signal in the NHS-Galleri trial. BMJ Open 13, e072657 (2023).
Kim, A., Chung, K. C., Keir, C. & Patrick, D. L. Patient-reported outcomes associated with cancer screening: a systematic review. BMC Cancer 22, 223 (2022).
Faupel-Badger, J. et al. Defining precancer: a grand challenge for the cancer community. Nat. Rev. Cancer 24, 792–809 (2024).
Koliopoulos, G. et al. Cytology versus HPV testing for cervical cancer screening in the general population. Cochrane Database Syst. Rev. 8, CD008587 (2017).
Fitzgerald, R. C. et al. Cytosponge-trefoil factor 3 versus usual care to identify Barrett’s oesophagus in a primary care setting: a multicentre, pragmatic, randomised controlled trial. Lancet 396, 333–344 (2020).
Bonde, J. et al. Methylation markers FAM19A4 and miR124-2 as triage strategy for primary human papillomavirus screen positive women: a large European multicenter study. Int. J. Cancer 148, 396–405 (2021).
Hu, X. et al. Evolution of DNA methylome from precancerous lesions to invasive lung adenocarcinomas. Nat. Commun. 12, 687 (2021).
Cassinotti, E. et al. DNA methylation patterns in blood of patients with colorectal cancer and adenomatous colorectal polyps. Int. J. Cancer 131, 1153–1157 (2012).
Campbell, T. W. et al. Abstract 1056: Development of a B-cell epitope classifier for early detection of renal cell carcinoma. Cancer Res. 84, 1056 (2024).
Papier, K. et al. Identifying proteomic risk factors for cancer using prospective and exome analyses of 1463 circulating proteins and risk of 19 cancers in the UK Biobank. Nat. Commun. 15, 4010 (2024).
Robbins, H. A. et al. Absolute risk of oropharyngeal cancer after an HPV16-E6 serology test and potential implications for screening: results from the human papillomavirus cancer cohort consortium. J. Clin. Oncol. 40, 3613–3622 (2022).
Placido, D. et al. A deep learning algorithm to predict risk of pancreatic cancer from disease trajectories. Nat. Med. 29, 1113–1122 (2023).
National Institute for Health and Care Excellence. Familial breast cancer: classification, care and managing breast cancer and related risks in people with a family history of breast cancer. NICE guideline CG164. NICE https://www.nice.org.uk/guidance/cg164 (2023).
Burn, J. et al. Cancer prevention with aspirin in hereditary colorectal cancer (Lynch syndrome), 10-year follow-up and registry-based 20-year data in the CAPP2 study: a double-blind, randomised, placebo-controlled trial. Lancet 395, 1855–1863 (2020).
McCarthy, R. L. et al. Risk-reducing surgery for individuals with cancer-predisposing germline pathogenic variants and no personal cancer history: a review of current UK guidelines. Br. J. Cancer 129, 383–392 (2023).
Vinayak, S. & Ford, J. M. PARP inhibitors for the treatment and prevention of breast cancer. Curr. Breast Cancer Rep. 2, 190–197 (2010).
To, C. et al. The PARP inhibitors, veliparib and olaparib, are effective chemopreventive agents for delaying mammary tumor development in BRCA1-deficient mice. Cancer Prev. Res. 7, 698–707 (2014).
Geneseeq. FDA Grants Breakthrough Device Designation for Geneseeq’s Multi-cancer Early Detection Solution https://na.geneseeq.com/fda-grants-breakthrough-designation-for-geneseeqs-multi-cancer-early-detection-solution/ (2024).
Nguyen, V. T. C. et al. Multimodal analysis of methylomics and fragmentomics in plasma cell-free DNA for multi-cancer early detection and localization. eLife 12, RP89083 (2023).
Oesterling, J. E. et al. Serum prostate-specific antigen in a community-based population of healthy men: establishment of age-specific reference ranges. JAMA 270, 860–864 (1993).
Moss, E. L., Hollingworth, J. & Reynolds, T. M. The role of CA125 in clinical practice. J. Clin. Pathol. 58, 308–312 (2005).
Acknowledgements
This Review was not explicitly funded from any grant. The authors acknowledge funding from Cancer Research UK (CTUQQR-Dec22/100005, awarded to P.S. for the Cancer Prevention Trials Unit, and C1287/A26886, EDDRPG-May24/100002, C36857/A27548, EDDCPGM\100001, A20240, C9545/A29580, SEBINT-2024/100003, C7893/A26233 and CTRQQR-2021\100004 to N.R.). Cancer Research UK did not have a role in the design of this Review, the collection, analysis, and interpretation of the data, the writing of the manuscript, and the decision to submit the manuscript for publication. The work of N.R. is supported by the European Union and the UK Research & Innovation (UKRI) under the UK government’s Horizon Europe funding guarantee (EU PANCAID 101096309, UKRI 10070284). Views and opinions expressed in this manuscript are, however, those of the authors only and do not necessarily reflect those of the European Union, the European Health and Digital Executive Agency (HADEA) or the UKRI; neither the European Union nor the granting authorities can be held responsible for them. The authors acknowledge feedback on figures from R. Lam.
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding authors
Ethics declarations
Competing interests
J.C.M.W. and N.R. are inventors on patents for methods of circulating tumour DNA detection. J.C.M.W. is a consultant for, co-founder and shareholder of Prima Mente and has been a consultant for Cleary Gottlieb, Delfi Diagnostics and Rostrum. P.S. is compensated by Grail for time spent on their advisory board and is also Director of the Cancer Prevention Trials Unit at Queen Mary University of London, which is contracted by GRAIL to act as the clinical trials unit for the NHS-Galleri trial.
Peer review
Peer review information
Nature Reviews Clinical Oncology thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wan, J.C.M., Sasieni, P. & Rosenfeld, N. Promises and pitfalls of multi-cancer early detection using liquid biopsy tests. Nat Rev Clin Oncol (2025). https://doi.org/10.1038/s41571-025-01033-x
Accepted:
Published:
DOI: https://doi.org/10.1038/s41571-025-01033-x