Abstract
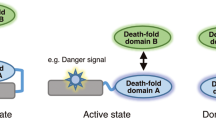
Caspase recruitment domains (CARDs) and pyrin domains are important facilitators of inflammasome activity and pyroptosis1. Following pathogen recognition by nucleotide binding-___domain, leucine-rich, repeat-containing (NLR) proteins, CARDs recruit and activate caspases, which, in turn, activate gasdermin pore-forming proteins to induce pyroptotic cell death2. Here we show that CARD domains are present in defence systems that protect bacteria against phage. The bacterial CARD ___domain is essential for protease-mediated activation of certain bacterial gasdermins, which promote cell death once phage infection is recognized. We further show that multiple anti-phage defence systems use CARD domains to activate a variety of cell death effectors, and that CARD domains mediate protein–protein interactions in these systems. We find that these systems are triggered by a conserved immune-evasion protein used by phages to overcome the bacterial defence system RexAB3, demonstrating that phage proteins inhibiting one defence system can activate another. Our results suggest that CARD domains represent an ancient component of innate immune systems conserved from bacteria to humans, and that CARD-dependent activation of gasdermins is shared in organisms across the tree of life.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
27,99 € / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
199,00 € per year
only 3,90 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
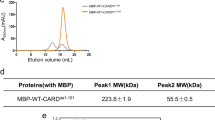
Data that support the findings of this study are available within the article and its Supplementary Tables. IMG accessions appear in Supplementary Tables 1 and 2, the sequences of toxic phage genes appear in Supplementary Table 3 and INPHARED accessions appear in Supplementary Table 4. Atomic coordinates and structure factors are available in the PDB database under accession number 9FRR for the Lysobacter CARD ___domain, and under accession numbers 9CS7 and 9CS8 for the Azospirillum CARD ___domain. Uncropped images of gels and blots from all figures are presented in Supplementary Fig. 1. Source Data are provided with this paper.
References
Broz, P. & Dixit, V. M. Inflammasomes: mechanism of assembly, regulation and signalling. Nat. Rev. Immunol. 16, 407–420 (2016).
Man, S. M. & Kanneganti, T.-D. Converging roles of caspases in inflammasome activation, cell death and innate immunity. Nat. Rev. Immunol. 16, 7–21 (2016).
Wong, S., Alattas, H. & Slavcev, R. A. A snapshot of the λ T4rII exclusion (Rex) phenotype in Escherichia coli. Curr. Genet. 67, 739–745 (2021).
Jorgensen, I., Rayamajhi, M. & Miao, E. A. Programmed cell death as a defence against infection. Nat. Rev. Immunol. 17, 151–164 (2017).
Broz, P., Pelegrín, P. & Shao, F. The gasdermins, a protein family executing cell death and inflammation. Nat. Rev. Immunol. 20, 143–157 (2020).
Jones, J. D. G., Vance, R. E. & Dangl, J. L. Intracellular innate immune surveillance devices in plants and animals. Science 354, aaf6395 (2016).
Ma, S. et al. Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme. Science 370, eabe3069 (2020).
Liu, X. et al. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature 535, 153–158 (2016).
Ruan, J., Xia, S., Liu, X., Lieberman, J. & Wu, H. Cryo-EM structure of the gasdermin A3 membrane pore. Nature 557, 62–67 (2018).
Lieberman, J., Wu, H. & Kagan, J. C. Gasdermin D activity in inflammation and host defense. Sci. Immunol. 4, eaav1447 (2019).
Johnson, A. G. et al. Bacterial gasdermins reveal an ancient mechanism of cell death. Science 375, 221–225 (2022).
Lopatina, A., Tal, N. & Sorek, R. Abortive infection: bacterial suicide as anantiviral immune strategy. Annu. Rev. Virol. 7, 371–384 (2020).
Kibby, E. M. et al. Bacterial NLR-related proteins protect against phage. Cell 186, 2410–2424 (2023).
Leipe, D. D., Koonin, E. V. & Aravind, L. STAND, a class of P-loop NTPases including animal and plant regulators of programmed cell death: multiple, complex ___domain architectures, unusual phyletic patterns, and evolution by horizontal gene transfer. J. Mol. Biol. 343, 1–28 (2004).
Gao, L. A. et al. Prokaryotic innate immunity through pattern recognition of conserved viral proteins. Science 377, eabm4096 (2022).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Park, H. H. et al. The Death ___domain superfamily in intracellular signaling of apoptosis and inflammation. Annu. Rev. Immunol. 25, 561–586 (2007).
Werts, C., Girardin, S. E. & Philpott, D. J. TIR, CARD and PYRIN: three domains for an antimicrobial triad. Cell Death Differ. 13, 798–815 (2006).
Wu, H. & Lo, Y.-C. Structures, domains and functions in cell death (DD, DED, CARD, PYD). eLS https://doi.org/10.1002/9780470015902.a0021579 (2009).
Davis, B. K., Wen, H. & Ting, J. P.-Y. The inflammasome NLRs in immunity, inflammation, and associated diseases. Annu. Rev. Immunol. 29, 707–735 (2011).
Holm, L. Dali server: structural unification of protein families. Nucleic Acids Res. 50, W210–W215 (2022).
Matyszewski, M. et al. Cryo-EM structure of the NLRC4CARD filament provides insights into how symmetric and asymmetric supramolecular structures drive inflammasome assembly. J. Biol. Chem. 293, 20240–20248 (2018).
Humke, E. W., Shriver, S. K., Starovasnik, M. A., Fairbrother, W. J. & Dixit, V. M. ICEBERG: a novel inhibitor of interleukin-1β generation. Cell 103, 99–111 (2000).
Pinheiro, A. S. et al. Three-dimensional structure of the NLRP7 pyrin ___domain: Insight into pyrin-pyrin-mediated effector ___domain signaling in innate immunity. J. Biol. Chem. 285, 27402–27410 (2010).
Eibl, C. et al. Structural and functional analysis of the NLRP4 pyrin ___domain. Biochemistry 51, 7330–7341 (2012).
Hou, X. & Niu, X. The NMR solution structure of AIM2 PYD ___domain from Mus musculus reveals a distinct α2–α3 helix conformation from its human homologues. Biochem. Biophys. Res. Commun. 461, 396–400 (2015).
Rousset, F. & Sorek, R. The evolutionary success of regulated cell death in bacterial immunity. Curr. Opin. Microbiol. 74, 102312 (2023).
van Kempen, M. et al. Fast and accurate protein structure search with Foldseek. Nat. Biotechnol. 42, 243–246 (2024).
Suzek, B. E. et al. UniRef clusters: a comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics 31, 926–932 (2015).
Stokar-Avihail, A. et al. Discovery of phage determinants that confer sensitivity to bacterial immune systems. Cell 186, 1863–1876 (2023).
Cook, R. et al. INfrastructure for a PHAge REference Database: identification of large-scale biases in the current collection of cultured phage genomes. Phage 2, 214–223 (2021).
Millman, A., Melamed, S., Amitai, G. & Sorek, R. Diversity and classification of cyclic-oligonucleotide-based anti-phage signalling systems. Nat. Microbiol. 5, 1608–1615 (2020).
Miller, E. S. et al. Bacteriophage T4 genome. Microbiol. Mol. Biol. Rev. 67, 86–156 (2003).
Shinedling, S., Parma, D. & Gold, L. Wild-type bacteriophage T4 is restricted by the lambda rex genes. J. Virol. 61, 3790–3794 (1987).
Landsmann, J., Kroger, M. & Hobom, G. The rex region of bacteriophage lambda: two genes under three-way control. Gene 20, 11–24 (1982).
Rousset, F. et al. Phages and their satellites encode hotspots of antiviral systems. Cell Host Microbe 30, 740–753 (2022).
Isaev, A. et al. Phage T7 DNA mimic protein Ocr is a potent inhibitor of BREX defence. Nucleic Acids Res. 48, 5397–5406 (2020).
Bedoui, S., Herold, M. J. & Strasser, A. Emerging connectivity of programmed cell death pathways and its physiological implications. Nat. Rev. Mol. Cell Biol. 21, 678–695 (2020).
Hofmann, K. The evolutionary origins of programmed cell death signaling. Cold Spring Harb. Perspect. Biol. 12, a036442 (2020).
Wein, T. & Sorek, R. Bacterial origins of human cell-autonomous innate immune mechanisms. Nat. Rev. Immunol. 22, 629–638 (2022).
Kaur, G., Iyer, L. M., Burroughs, A. M. & Aravind, L. Bacterial death and TRADD-N domains help define novel apoptosis and immunity mechanisms shared by prokaryotes and metazoans. eLife 10, e70394 (2021).
Kaur, G., Burroughs, A. M., Iyer, L. M. & Aravind, L. Highly regulated, diversifying NTP-dependent biological conflict systems with implications for the emergence of multicellularity. eLife 9, e52696 (2020).
Mitchell, P. S., Sandstrom, A. & Vance, R. E. The NLRP1 inflammasome: new mechanistic insights and unresolved mysteries. Curr. Opin. Immunol. 60, 37–45 (2019).
Shi, J. et al. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature 514, 187–192 (2014).
Tesson, F. et al. Systematic and quantitative view of the antiviral arsenal of prokaryotes. Nat. Commun. 13, 2561 (2022).
Jones, J. D. G. & Dangl, J. L. The plant immune system. Nature 444, 323–329 (2006).
Adachi, H., Derevnina, L. & Kamoun, S. NLR singletons, pairs, and networks: evolution, assembly, and regulation of the intracellular immunoreceptor circuitry of plants. Curr. Opin. Plant Biol. 50, 121–131 (2019).
Remick, B. C., Gaidt, M. M. & Vance, R. E. Effector-triggered immunity. Annu. Rev. Immunol. 41, 453–481 (2023).
Sandstrom, A. et al. Functional degradation: a mechanism of NLRP1 inflammasome activation by diverse pathogen enzymes. Science 364, eaau1330 (2019).
Robinson, K. S. et al. Enteroviral 3C protease activates the human NLRP1 inflammasome in airway epithelia. Science 370, eaay2002 (2020).
Johnston, J. B. et al. A poxvirus-encoded pyrin ___domain protein interacts with ASC-1 to inhibit host inflammatory and apoptotic responses to infection. Immunity 23, 587–598 (2005).
Doron, S. et al. Systematic discovery of antiphage defense systems in the microbial pangenome. Science 359, eaar4120 (2018).
Millman, A. et al. Bacterial retrons function in anti-phage defense. Cell 183, 1551–1561 (2020).
Bernheim, A. et al. Prokaryotic viperins produce diverse antiviral molecules. Nature 589, 120–124 (2021).
Lee, T. S. et al. BglBrick vectors and datasheets: a synthetic biology platform for gene expression. J. Biol. Eng. 5, 12 (2011).
Mazzocco, A., Waddell, T. E., Lingohr, E. & Johnson, R. P. In Bacteriophages: Methods and Protocols (eds Kropinski, A. M. & Cloike, M. R. J.) 81–85 (Humana Press, 2009).
Kropinski, A. M., Mazzocco, A., Waddell, T. E., Lingohr, E. & Johnson, R. P. In Bacteriophages: Methods and Protocols (eds Kropinski, A. M. & Cloike, M. R. J.) 69–76 (Humana Press, 2009).
Baym, M. et al. Inexpensive multiplexed library preparation for megabase-sized genomes. PLoS ONE 10, e0128036 (2015).
Deatherage, D. E. & Barrick, J. E. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol. Biol. (Clifton, NJ) 1151, 165–188 (2014).
Chung, C. T. & Miller, R. H. In Methods in Enzymology Vol. 218 (ed. Wu, R.) 621–627 (Academic Press, 1993).
Frey, S. & Görlich, D. A new set of highly efficient, tag-cleaving proteases for purifying recombinant proteins. J. Chromatogr. A 1337, 95–105 (2014).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Williams, C. J. et al. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018).
Zhou, W. et al. Structure of the human cGAS–DNA complex reveals enhanced control of immune surveillance. Cell 174, 300–311 (2018).
Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. Sect. Struct. Biol. 75, 861–877 (2019).
Chen, I.-M. A. et al. IMG/M v.5.0: an integrated data management and comparative analysis system for microbial genomes and microbiomes. Nucleic Acids Res. 47, D666–D677 (2019).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Waterhouse, A. M., Procter, J. B., Martin, D. M. A., Clamp, M. & Barton, G. J. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 (2009).
Cretin, G. et al. SWORD2: hierarchical analysis of protein 3D structures. Nucleic Acids Res. 50, W732–W738 (2022).
Steinegger, M. & Söding, J. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat. Biotechnol. 35, 1026–1028 (2017).
Millman, A. et al. An expanded arsenal of immune systems that protect bacteria from phages. Cell Host Microbe 30, 1556–1569 (2022).
Mirdita, M. et al. ColabFold: making protein folding accessible to all. Nat. Methods 19, 679–682 (2022).
Nguyen, L.-T., Schmidt, H. A., von Haeseler, A. & Minh, B. Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015).
Letunic, I. & Bork, P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296 (2021).
Acknowledgements
We thank the Sorek and Kranzusch laboratory members for comments on the manuscript and fruitful discussions. R.S. was supported, in part, by the European Research Council (grant no. ERC-AdG GA 101018520), Israel Science Foundation (MAPATS grant no. 2720/22), Deutsche Forschungsgemeinschaft (SPP 2330, grant no. 464312965), the Ernest and Bonnie Beutler Research Program of Excellence in Genomic Medicine and the Knell Family Center for Microbiology. P.J.K. was supported, in part, by the Pew Biomedical Scholars programme, The G. Harold and Leila Y. Mathers Charitable Foundation, the Parker Institute for Cancer Immunotherapy and the National Institutes of Health (no. 1DP2GM146250). T.W. was supported by a Minerva Foundation postdoctoral fellowship and by a European Molecular Biology Organization postdoctoral fellowship (no. ALTF 946-2020). A.M. was supported by a fellowship from the Ariane de Rothschild Women Doctoral Program and, in part, by the Israeli Council for Higher Education by the Weizmann Data Science Research Center. E.Y. was supported, in part, by the Israeli Council for Higher Education by the Weizmann Data Science Research Center. X-ray data were collected at Northeastern Collaborative Access Team beamlines 24-ID-C and 24-ID-E (P30 GM124165), including the use of a Pilatus detector (S10RR029205), an Eiger detector (S10OD021527) and the Argonne National Laboratory Advanced Photon Source (DE-AC02-06CH11357).
Author information
Authors and Affiliations
Contributions
T.W. performed genomic analyses, biochemical pulldown experiments, cellular imaging, phage infection experiments and gasdermin cleavage assays. J.G. and R.H. performed phage infection experiments. A.M., K.L. and E.Y. performed genomic analyses of bacterial defence systems and phage CARD proteins. S.M. cloned RIIB homologues and toxic genes. G.A., O.D., F.S., A.B.H. and P.J.K. performed protein purification, crystallography, model building and structural analyses. T.W., P.J.K. and R.S. wrote the manuscript. R.S. and P.J.K. supervised the study.
Corresponding authors
Ethics declarations
Competing interests
R.S. is a scientific cofounder and advisor of BiomX and Ecophage. The other authors declare no competing interests.
Peer review
Peer review information
Nature thanks Pu Gao, Eugene Koonin, Russell Vance and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Anti-phage defense of tagged Lysobacter gasdermin system.
Efficiency of plating of phages infecting E. coli cells that express the WT Lysobacter gasdermin system, as well as systems in which gasdermin was N-terminally fused to an HA-tag or to GFP. Negative control is a strain in which GFP is expressed instead of the gasdermin system. Data represent plaque-forming units (PFU) per ml. Average of three independent biological replicates, with individual data points overlaid.
Extended Data Fig. 2 Multiple sequence alignment of the Nucleotide binding oligomerization ___domain (NOD) module of STAND ATPases.
Shown are NOD modules from human proteins (NLRC4, NCBI accession: AAH31555; NLRP1, AAG30288; NLRP3, AAL33911; NAIP, A55478; NOD1, AAD29125) and bacterial proteins (bNACHT01, NCBI accession: WP_015632533.1; bNACHT25, WP_001702659.1; ECAvs2, WP_063118745.1; SeAvs3, WP_126523998.1), including the ATPase protein from the Lysobacter gasdermin system (IMG gene ID 2841794910). The intensity of shading indicates degree of residue conservation.
Extended Data Fig. 3 Predicted structures of CARD domains.
AlphaFold2 prediction of N-terminal CARD domains from the proteases homologous to the protease of the Lysobacter gasdermin system (Supplementary Table 1).
Extended Data Fig. 4 Systems homologous to the Lysobacter gasdermin system.
a. Efficiency of plating of phages infecting E. coli cells that express the Lysobacter or Pedobacter defense systems. Negative control is a strain in which GFP is expressed instead of the gasdermin system. Data represent plaque-forming units (PFU) per milliliter. Average of three independent biological replicates, with individual data points overlaid. b. Representative instances of homologous systems in their genomic neighborhood. Genes known to be involved in anti-phage defense are shown in yellow (RM, restriction-modification; Pycsar, pyrimidine cyclase system for anti-phage resistance; AIPR, abortive infection phage resistance; REase, restriction endonuclease). c. A plot of DALI Z-scores of protein structures similar to the Azospirillum CARD ___domain structure.
Extended Data Fig. 5 Lysobacter NLR-like protein encodes a CARD ___domain at the C-terminus.
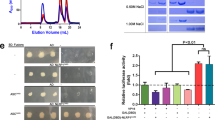
a. An AlphaFold2 model of the Lysobacter NLR-like ATPase protein. b. An AlphaFold2 model of the C-terminal ___domain of the NLR-like ATPase. The human ICEBERG CARD ___domain (PDB ID 1DGN) is shown for comparison. c. A plot of DALI Z-scores of protein structures similar to the Lysobacter NLR-like CARD ___domain structure predicted by AlphaFold2. d. Deletion of the C-terminus of the NLR-like gene in the Lysobacter gasdermin system leads to toxicity. Bacteria expressing the WT or CARD-deleted Lysobacter gasdermin system were plated in 10-fold serial dilution on LB-agar plates in conditions that repress expression (1% glucose) or induce expression (0.2% arabinose).
Extended Data Fig. 6 Bacterial CARD domains mediate protein-protein interactions in multiprotein anti-phage defense complexes.
a, b, AlphaFold2-Multimer model of the trypsin-like protease and the NLR-like protein from the Lysobacter gasdermin system. The model confidence score for protein-protein interactions is depicted below. c, Predicted Aligned Error (PAE) of the AlphaFold2-Multimer predicted interactions between the trypsin-like protease and the NLR-like protein. d, AlphaFold2-Multimer models of the trypsin-like protease and the NLR-like protein from systems homologous to the Lysobacter gasdermin system. The model confidence score for protein-protein interactions is depicted below each model. Percent sequence identity between the Lysobacter NLR/protease and the respective protein is presented for each model. e, Western blot analyses of the experiments presented in Fig. 3h. The FLAG-tagged NLR-like protein was expressed together with either the full-length HA-tagged protease, the tagged protease in which the CARD ___domain was deleted or only with the HA-tagged CARD ___domain. Left panel, anti-FLAG beads were used to immunoprecipitate the FLAG-tagged NLR-like protein, and anti-HA antibody was used for western blotting. Right panel, anti-HA beads were used to immunoprecipitate the HA-tagged proteins, and anti-FLAG antibody used for western blotting. Representative of three replicates. f, Control experiments showing specificity of pulldown. Shown is SDS-PAGE with Coomassie stain analysis, with the following immunoprecipitation results: FLAG-tagged NLR-like protein from Lysobacter that was co-expressed with RFP; HA-tagged trypsin-like protease; HA-tagged CARD-deleted protease; and HA-tagged CARD ___domain from Lysobacter that was co-expressed with GFP g, Cell lysates of the samples used for immunoprecipitations shown in Fig. 3h, presented here as control. h, Cell lysates of the samples used for immunoprecipitations shown in panel f, presented here as control.
Extended Data Fig. 7 Expression of RIIB proteins activate gasdermin-mediated defense.
a Bacteria expressing RIIB homologs together with a negative control or the Lysobacter gasdermin system were plated in 10-fold serial dilution on LB-agar plates in conditions that repress expression (1% glucose) or induce expression (0.2% arabinose and 0.1 mM IPTG). b. Transformation efficiency assays of plasmids encoding rIIB or RFP into cells that contain the Lysobacter or Pedobacter systems on a pBAD plasmid. Bars represent the average of three biological replicates with individual data points overlaid. c. Phylogenetic tree of phage RIIB homologs (Supplementary Table 4). Names of phages from which RIIB homologs were experimentally tested are indicated. d. Bacteria co-expressing RIIB homologs together with the Lysobacter gasdermin system or a negative control were plated in 10-fold serial dilution on LB-agar plates in conditions that repress expression (1% glucose) or induce expression (0.2% arabinose and 1 mM IPTG). Numbering of RIIB homologs corresponds to the numbering on the tree in panel c. e. Western blot analyses of N-terminally HA-tagged gasdermin co-expressed with RIIB homologs, before (0 min) and 80 min after RIIB expression was induced. GroEL was used as a loading control. f. Genomic neighborhoods of two homologous phage CARD-only proteins with 57.5% sequence identity. g. DALI Z-scores of protein structures similar to the AlphaFold2 model of the phage CARD-only protein from Acinetobacter phage Acj9.
Extended Data Fig. 8 Toxic genes of phage origin do not activate the Lysobacter gasdermin system.
a. Bacteria co-expressing the Lysobacter gasdermin system with toxic genes of phage origin that are unrelated to RIIB (Supplementary Table 3) were plated in 10-fold serial dilution on LB-agar plates in conditions that repress expression (1% glucose) or induce expression (0.2% arabinose and 1 mM IPTG). Negative control represents cells expressing GFP instead of the gasdermin system. b. Expression of toxic phage genes does not lead to gasdermin cleavage. Western blot analyses of N-terminally HA-tagged gasdermin co-expressed with toxic genes of phage origin, before (0 min) and 80 min after the expression of the toxic gene was induced. GroEL was used as a loading control.
Supplementary information
Supplentary Figure 1
Uncropped images of gels and blots from all figures.
Supplementary Table 1
Systems homologous to gasdermin-associated protease and ATPase proteins.
Supplementary Table 2
Systems with structural homology to gasdermin-associated protease.
Supplementary Table 3
Toxic phage genes used in this study.
Supplementary Table 4
RIIB homologues.
Supplementary Video 1
Time-lapse microscopy, corresponding to Fig. 1a, of live E. coli cells expressing GFP (green) mixed with cells expressing the Lysobacter gasdermin system (black). Cells were infected with phage T6 in the presence of propidium iodide (PI) and incubated at room temperature on an agar pad. Overlay images of phase contrast, green channel (GFP) and magenta channel (PI) are presented. Frames were taken every 3 min.
Supplementary Video 2
Time-lapse microscopy, corresponding to Fig. 1d, of live E. coli cells expressing the WT Lysobacter gasdermin system in which gasdermin was N-terminally fused to GFP. Cells were infected with phage T6 in the presence of propidium iodide (PI) and incubated at room temperature on an agar pad. Overlay images of phase contrast, green channel (GFP) and magenta channel (PI) are presented. Frames were taken every 3 min.
Supplementary Video 3
Time-lapse microscopy, corresponding to Fig. 4c, of live E. coli cells coexpressing RIIB with the WT Lysobacter gasdermin system, in which gasdermin was N-terminally fused to GFP. Cells were visualized at room temperature on an agar pad in the presence of propidium iodide (PI). Overlay images of phase contrast, green channel (GFP) and magenta channel (PI) are presented. Frames were taken every 3 min.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wein, T., Millman, A., Lange, K. et al. CARD domains mediate anti-phage defence in bacterial gasdermin systems. Nature 639, 727–734 (2025). https://doi.org/10.1038/s41586-024-08498-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-024-08498-3
This article is cited by
-
Modularity of Zorya defense systems during phage inhibition
Nature Communications (2025)
-
A DNA-gated molecular guard controls bacterial Hailong anti-phage defence
Nature (2025)