Abstract
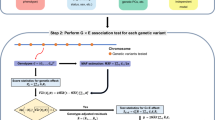
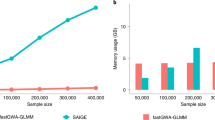
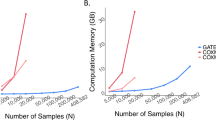
In genome-wide association studies (GWAS) for thousands of phenotypes in large biobanks, most binary traits have substantially fewer cases than controls. Both of the widely used approaches, the linear mixed model and the recently proposed logistic mixed model, perform poorly; they produce large type I error rates when used to analyze unbalanced case-control phenotypes. Here we propose a scalable and accurate generalized mixed model association test that uses the saddlepoint approximation to calibrate the distribution of score test statistics. This method, SAIGE (Scalable and Accurate Implementation of GEneralized mixed model), provides accurate P values even when case-control ratios are extremely unbalanced. SAIGE uses state-of-art optimization strategies to reduce computational costs; hence, it is applicable to GWAS for thousands of phenotypes by large biobanks. Through the analysis of UK Biobank data of 408,961 samples from white British participants with European ancestry for > 1,400 binary phenotypes, we show that SAIGE can efficiently analyze large sample data, controlling for unbalanced case-control ratios and sample relatedness.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
27,99 € / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
209,00 € per year
only 17,42 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Bush, W. S., Oetjens, M. T. & Crawford, D. C. Unravelling the human genome-phenome relationship using phenome-wide association studies. Nat. Rev. Genet. 17, 129–145 (2016).
Denny, J. C. et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat. Biotechnol. 31, 1102–1110 (2013).
Kang, H. M. et al. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 42, 348–354 (2010).
Zhang, Z. et al. Mixed linear model approach adapted for genome-wide association studies. Nat. Genet. 42, 355–360 (2010).
Yang, J., Lee, S. H., Goddard, M. E. & Visscher, P. M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88, 76–82 (2011).
Lippert, C. et al. FaST linear mixed models for genome-wide association studies. Nat. Methods 8, 833–835 (2011).
Zhou, X. & Stephens, M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 44, 821–824 (2012).
Loh, P. R. et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat. Genet. 47, 284–290 (2015).
Chen, H. et al. Control for population structure and relatedness for binary traits in genetic association studies via logistic mixed models. Am. J. Hum. Genet. 98, 653–666 (2016).
Ma, C., Blackwell, T., Boehnke, M. & Scott, L. J., GoT2D investigators. Recommended joint and meta-analysis strategies for case-control association testing of single low-count variants. Genet. Epidemiol 37, 539–550 (2013).
Kuonen, D. Saddlepoint approximations for distributions of quadratic forms in normal variables. Biometrika 86, 4 (1999).
Dey, R., Schmidt, E. M., Abecasis, G. R. & Lee, S. A fast and accurate algorithm to test for binary phenotypes and its application to PheWAS. Am. J. Hum. Genet. 101, 37–49 (2017).
Kaasschieter, E. F. Preconditioned conjugate gradients for solving singular systems. J. Comput. Appl. Math. 24, 265–275 (1988).
Sudlow, C. et al. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 12, e1001779 (2015).
Bycroft, C. et al. Genome-wide genetic data on ~500,000 UK Biobank participants. Preprint at bioRxiv, https://doi.org/10.1101/166298 (2017).
Gilmour, A. R., Thompson, R. & Cullis, B. R. Average information REML: an efficient algorithm for variance parameter estimation in linear mixed models. Biometrics 51, 1440–1450 (1995).
Aulchenko, Y. S., Ripke, S., Isaacs, A. & van Duijn, C. M. GenABEL: an R library for genome-wide association analysis. Bioinformatics 23, 1294–1296 (2007).
Svishcheva, G. R., Axenovich, T. I., Belonogova, N. M., van Duijn, C. M. & Aulchenko, Y. S. Rapid variance components-based method for whole-genome association analysis. Nat. Genet. 44, 1166–1170 (2012).
McCarthy, S. et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 48, 1279–1283 (2016).
Nelis, M. et al. Genetic structure of Europeans: a view from the North-East. PLoS One 4, e5472 (2009).
Shameer, K. et al. A genome- and phenome-wide association study to identify genetic variants influencing platelet count and volume and their pleiotropic effects. Hum. Genet. 133, 95–109 (2014).
Yang, J., Zaitlen, N. A., Goddard, M. E., Visscher, P. M. & Price, A. L. Advantages and pitfalls in the application of mixed-model association methods. Nat. Genet. 46, 100–106 (2014).
Listgarten, J. et al. Improved linear mixed models for genome-wide association studies. Nat. Methods 9, 525–526 (2012).
Breslow, N. E. & Clayton, D. G. Approximate inference in generalized linear mixed models. J. Am. Stat. Assoc 88, 9–25 (1993).
Hestenes, M. R. & Stiefel, E. Methods of conjugate gradients for solving linear systems. J. Res. Natl Bur. Stand. 49, 409–436 (1952).
Imhof, J. P. Computing the distribution of quadratic forms in normal variables. Biometrika 48, 419–426 (1961).
Abecasis, G. R., Cherny, S. S., Cookson, W. O. & Cardon, L. R. Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet. 30, 97–101 (2002).
de Villemereuil, P., Schielzeth, H., Nakagawa, S. & Morrissey, M. General methods for evolutionary quantitative genetic inference from generalized mixed models. Genetics 204, 1281–1294 (2016).
Bulik-Sullivan, B. K. et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015).
Acknowledgements
This research has been conducted using the UK Biobank Resource under application number 24460. S.L. and R.D. were supported by NIH R01 HG008773. C.J.W. was supported by NIH R35 HL135824. W.Z. was supported by the University of Michigan Rackham Predoctoral Fellowship. J.B.N. was supported by the Danish Heart Foundation and the Lundbeck Foundation. J.C.D., A.G., L.A.B., and W.-Q.W. were supported by NIH R01 LM010685 and U2C OD023196.
Author information
Authors and Affiliations
Contributions
W.Z., C.J.W., and S.L. designed the experiments. W.Z. and S.L. performed the experiments. J.B.N., L.G.F., A.G., L.A.B., W.-Q.W., and J.C.D. constructed the phenotypes for the UK Biobank data. W.Z., J.L., S.A.G., B.N.W., M.L., H.M.K., C.J.W., S.L., and G.R.A. analyzed the UK Biobank data. P.V. created the PheWeb. M.E.G. and K.H. provided the data. W.Z., J.B.N., A.G., J.C.D., R.D., C.J.W., and S.L. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–18, Supplementary Tables 1–8 and Supplementary Note
Rights and permissions
About this article
Cite this article
Zhou, W., Nielsen, J.B., Fritsche, L.G. et al. Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic association studies. Nat Genet 50, 1335–1341 (2018). https://doi.org/10.1038/s41588-018-0184-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-018-0184-y
This article is cited by
-
Associations between RetNet gene polymorphisms and the efficacy of orthokeratology for myopia control: a retrospective clinical study
Eye and Vision (2025)
-
Dynamic clustering of genomics cohorts beyond race, ethnicity—and ancestry
BMC Medical Genomics (2025)
-
Genetic insights into idiopathic pulmonary fibrosis: a multi-omics approach to identify potential therapeutic targets
Journal of Translational Medicine (2025)
-
Sequence-based GWAS in 180,000 German Holstein cattle reveals new candidate variants for milk production traits
Genetics Selection Evolution (2025)
-
Dose-dependent role of AMH and AMHR2 signaling in male differentiation and regulation of sex determination in Spotted knifejaw (Oplegnathus punctatus) with X1X1X2X2/X1X2Y chromosome system
Cell Communication and Signaling (2025)