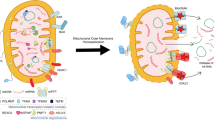
Abstract
Stress-induced oxidized mitochondrial DNA (Ox-mtDNA) fragments enter the cytoplasm, activating the NLRP3 inflammasome and caspase-1 and enabling gasdermin-D-mediated circulatory release of mtDNA. Elevated amounts of circulating mtDNA, presumably oxidized, have been detected in older individuals and patients with metabolic or autoimmune disorders. Here we show that sustained Ox-mtDNA release, triggered by a prototypical NLRP3 inflammasome activator, induces autoantibody production and glomerulonephritis in mice. Similar autoimmune responses, dependent on plasmacytoid dendritic cells (pDCs) and follicular helper T (TFH) cells, are elicited by in vitro-generated Ox-mtDNA, but not by non-oxidized mtDNA. Although both mtDNA forms are internalized by pDCs and induce interferon-α, only Ox-mtDNA stimulates autocrine interleukin (IL)-1β signaling that induces co-stimulatory molecules and IL-21, which enable mouse and human pDCs to induce functional TFH differentiation, supportive of autoantibody production. These findings underscore the role of pDC-generated IL-1β in autoantibody production and highlight Ox-mtDNA as an important autoimmune trigger, suggesting potential therapeutic opportunities.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
27,99 € / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
209,00 € per year
only 17,42 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
RNA-seq and scRNA-seq data have been deposited in the GEO under accession codes GSE264619 and GSE264620. GRCm39 (https://www.ncbi.nlm.nih.gov/datasets/genome/GCF_000001635.27/) and published scRNA-seq data (GSE196720) were used as a reference genome. Source data are provided with this paper.
References
Shimada, K. et al. Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity 36, 401–414 (2012).
Zhong, Z. et al. New mitochondrial DNA synthesis enables NLRP3 inflammasome activation. Nature 560, 198–203 (2018).
Xian, H. et al. Oxidized DNA fragments exit mitochondria via mPTP- and VDAC-dependent channels to activate NLRP3 inflammasome and interferon signaling. Immunity 55, 1370–1385 (2022).
Lehmann, J., Giaglis, S., Kyburz, D., Daoudlarian, D. & Walker, U. A. Plasma mtDNA as a possible contributor to and biomarker of inflammation in rheumatoid arthritis. Arthritis Res. Ther. 26, 97 (2024).
Xian, H. & Karin, M. Oxidized mitochondrial DNA: a protective signal gone awry. Trends Immunol. 44, 188–200 (2023).
Caielli, S. et al. Oxidized mitochondrial nucleoids released by neutrophils drive type I interferon production in human lupus. J. Exp. Med. 213, 697–713 (2016).
Miao, N. et al. Oxidized mitochondrial DNA induces gasdermin D oligomerization in systemic lupus erythematosus. Nat. Commun. 14, 872 (2023).
Wei, R. et al. Mitochondrial DNA content is linked to cardiovascular disease patient phenotypes. J. Am. Heart Assoc. 10, e018776 (2021).
Yuzefovych, L. V. et al. Plasma mitochondrial DNA is elevated in obese type 2 diabetes mellitus patients and correlates positively with insulin resistance. PLoS ONE 14, e0222278 (2019).
Varhaug, K. N. et al. Increased levels of cell-free mitochondrial DNA in the cerebrospinal fluid of patients with multiple sclerosis. Mitochondrion 34, 32–35 (2017).
Mahmoud, E. H., Fawzy, A., Ahmad, O. K. & Ali, A. M. Plasma circulating cell-free nuclear and mitochondrial DNA as potential biomarkers in the peripheral blood of breast cancer patients. Asian Pac. J. Cancer Prev. 16, 8299–8305 (2015).
Eisenbarth, S. C., Colegio, O. R., O’Connor, W., Sutterwala, F. S. & Flavell, R. A. Crucial role for the Nalp3 inflammasome in the immunostimulatory properties of aluminium adjuvants. Nature 453, 1122–1126 (2008).
Marichal, T. et al. DNA released from dying host cells mediates aluminum adjuvant activity. Nat. Med 17, 996–1002 (2011).
McKee, A. S. et al. Host DNA released in response to aluminum adjuvant enhances MHC class II-mediated antigen presentation and prolongs CD4 T-cell interactions with dendritic cells. Proc. Natl Acad. Sci. USA 110, E1122–E1131 (2013).
Caielli, S. et al. A CD4+ T cell population expanded in lupus blood provides B cell help through interleukin-10 and succinate. Nat. Med. 25, 75–81 (2019).
Ghosn, E. E. et al. Two physically, functionally, and developmentally distinct peritoneal macrophage subsets. Proc. Natl Acad. Sci. USA 107, 2568–2573 (2010).
Xian, H. et al. Metformin inhibition of mitochondrial ATP and DNA synthesis abrogates NLRP3 inflammasome activation and pulmonary inflammation. Immunity 54, 1463–1477 (2021).
Swiecki, M., Gilfillan, S., Vermi, W., Wang, Y. & Colonna, M. Plasmacytoid dendritic cell ablation impacts early interferon responses and antiviral NK and CD8+ T cell accrual. Immunity 33, 955–966 (2010).
Cho, Y. M. & Furie, R. The development of litifilimab (BIIB 059) for cutaneous and systemic lupus erythematosus. Immunotherapy 16, 15–20 (2024).
Hollister, K. et al. Insights into the role of Bcl6 in follicular Th cells using a new conditional mutant mouse model. J. Immunol. 191, 3705–3711 (2013).
Weber, J. P. et al. ICOS maintains the T follicular helper cell phenotype by down-regulating Krüppel-like factor 2. J. Exp. Med. 212, 217–233 (2015).
Stone, E. L. et al. ICOS coreceptor signaling inactivates the transcription factor FOXO1 to promote TFH cell differentiation. Immunity 42, 239–251 (2015).
Ballesteros-Tato, A. & Randall, T. D. Priming of T follicular helper cells by dendritic cells. Immunol. Cell Biol. 92, 22–27 (2014).
Ritvo, P. G. et al. T fr cells lack IL-2Rα but express decoy IL-1R2 and IL-1Ra and suppress the IL-1-dependent activation of TFH cells. Sci. Immunol. 2, eaan0368 (2017).
Barbet, G. et al. Sensing microbial viability through bacterial RNA augments T follicular helper cell and antibody responses. Immunity 48, 584–598 (2018).
Stetson, D. B. & Medzhitov, R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity 24, 93–103 (2006).
Liberzon, A. et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015).
Lou, H. & Pickering, M. C. Extracellular DNA and autoimmune diseases. Cell Mol. Immunol. 15, 746–755 (2018).
Krieg, A. M. TLR9 and DNA ‘feel’ RAGE. Nat. Immunol. 8, 475–477 (2007).
Newman, L. E. et al. Mitochondrial DNA replication stress triggers a pro-inflammatory endosomal pathway of nucleoid disposal. Nat. Cell Biol. 26, 194–206 (2024).
Day, R. A. & Sletten, E. M. Experimental perspectives on direct visualization of endosomal rupture. Chembiochem 22, 3277–3282 (2021).
Pinti, M. et al. Circulating mitochondrial DNA increases with age and is a familiar trait: Implications for ‘inflamm-aging’. Eur. J. Immunol. 44, 1552–1562 (2014).
Ye, W., Wen, C., Zeng, A. & Hu, X. Increased levels of circulating oxidized mitochondrial DNA contribute to chronic inflammation in metabolic syndrome, and MitoQ-based antioxidant therapy alleviates this DNA-induced inflammation. Mol. Cell Endocrinol. 560, 111812 (2023).
Zheng, Y., Liu, Q., Goronzy, J. J. & Weyand, C. M. Immune aging - a mechanism in autoimmune disease. Semin. Immunol. 69, 101814 (2023).
Caielli, S. et al. Type I IFN drives unconventional IL-1β secretion in lupus monocytes. Immunity 57, 2497–2513 (2024).
Walker, L. S. K. The link between circulating follicular helper T cells and autoimmunity. Nat. Rev. Immunol. 22, 567–575 (2022).
Fitzgerald-Bocarsly, P., Dai, J. & Singh, S. Plasmacytoid dendritic cells and type I IFN: 50 years of convergent history. Cytokine Growth Factor Rev. 19, 3–19 (2008).
Moseman, E. A. et al. Human plasmacytoid dendritic cells activated by CpG oligodeoxynucleotides induce the generation of CD4+CD25+ regulatory T cells. J. Immunol. 173, 4433–4442 (2004).
Jego, G. et al. Plasmacytoid dendritic cells induce plasma cell differentiation through type I interferon and interleukin 6. Immunity 19, 225–234 (2003).
Ogata, M. et al. Plasmacytoid dendritic cells have a cytokine-producing capacity to enhance ICOS ligand-mediated IL-10 production during T-cell priming. Int. Immunol. 25, 171–182 (2013).
Eto, D. et al. IL-21 and IL-6 are critical for different aspects of B cell immunity and redundantly induce optimal follicular helper CD4 T cell (TFH) differentiation. PLoS ONE 6, e17739 (2011).
Karnowski, A. et al. B and T cells collaborate in antiviral responses via IL-6, IL-21, and transcriptional activator and coactivator, Oct2 and OBF-1. J. Exp. Med 209, 2049–2064 (2012).
Arroyo Hornero, R. & Idoyaga, J. Plasmacytoid dendritic cells: a dendritic cell in disguise. Mol. Immunol. 159, 38–45 (2023).
Cornélie, S. et al. Direct evidence that toll-like receptor 9 (TLR9) functionally binds plasmid DNA by specific cytosine-phosphate-guanine motif recognition. J. Biol. Chem. 279, 15124–15129 (2004).
Longhi, M. P. et al. Dendritic cells require a systemic type I interferon response to mature and induce CD4+ Th1 immunity with poly IC as adjuvant. J. Exp. Med 206, 1589–1602 (2009).
Hornung, V. et al. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat. Immunol. 9, 847–856 (2008).
Xian, H., Yang, Q., Xiao, L., Shen, H. M. & Liou, Y. C. STX17 dynamically regulated by Fis1 induces mitophagy via hierarchical macroautophagic mechanism. Nat. Commun. 10, 2059 (2019).
Gehrke, N. et al. Oxidative damage of DNA confers resistance to cytosolic nuclease TREX1 degradation and potentiates STING-dependent immune sensing. Immunity 39, 482–495 (2013).
Lood, C. et al. Neutrophil extracellular traps enriched in oxidized mitochondrial DNA are interferogenic and contribute to lupus-like disease. Nat. Med. 22, 146–153 (2016).
Valente, M. et al. Novel mouse models based on intersectional genetics to identify and characterize plasmacytoid dendritic cells. Nat. Immunol. 24, 714–728 (2023).
Acknowledgements
We thank J. Idoyaga, L.-F. Lu, V. Pascual, S. Caielli and T. Greene for help and advice. We acknowledge eBioscience, Cell Signaling Technologies, Santa Cruz Technologies, Thermo Fisher, BioLegend, BD Biosciences and STEMCELL Technologies for gifts of reagents and the UCSD Tissue Technology Shared Resource supported by an NCI Cancer Center Support Grant (CCSG P30CA23100). We are grateful to A. Dent at Indiana University for providing us with Bcl6fl/fl/Cd4cre mice, J. Ravetch at the Rockefeller University, W. Leonard and R. Spolski at the NIH and Y.-G. Chen at the Medical College of Wisconsin, and K. King at UCSD, for Fcgr1−/−, Il21−/− and Ifnar1−/− mouse bones, respectively. J. Chung at the NIH kindly provided MEFs. We thank the UCSD Nikon Imaging Center and especially P. Guo for assistance with AXR Confocal and NSPARC Super Resolution imaging. Cartoons were prepared with BioRender.com. This publication includes data generated at the UCSD IGM Genomics Center utilizing an Illumina NovaSeq 6000 that was purchased through an NIH SIG grant (S10 OD026929). H.X. was supported by the Arthritis National Research Foundation (1291101). This work was supported by NIH grants R01 DK100640 and R37 AI043477 to M.K., who is an American Cancer Research Society Professor and holds the Ben and Wanda Hildyard Chair for Mitochondrial and Metabolic Diseases, NIH R01 grant AI145314 to E.I.Z. and NIH grants R01 AI155869, R01 DK113592 and P01 HL152958 to H.M.H.
Author information
Authors and Affiliations
Contributions
Conceptualization: H.X. and M.K. Methodology: H.X. and K.W. Investigation: H.X., K.W., M.O., J.S.B. and P.H. Resources: J.O., H.M.H., E.I.Z. and M.K. Formal analysis: H.X. and M.O. Supervision: M.K. Funding acquisition: E.I.Z., H.M.H. and M.K. Writing—original draft: H.X. and M.K. Writing—review and editing: E.I.Z., H.M.H. and M.K.
Corresponding author
Ethics declarations
Competing interests
M.K. is a founder of Elgia Pharmaceuticals and received research support from Gossamer Bio and Jansen Pharmaceuticals. M.K. holds an interest in PF-06835375, a TFH cell-depleting CXCR5 antibody. H.M.H. is a consultant for SOBI and Akros and received research funds from Takeda and Inapill. The other authors declare no competing interests.
Peer review
Peer review information
Nature Immunology thanks Moshe Arditi, Timothy Crother and Michelle Linterman for their contribution to the peer review of this work. Peer reviewer reports are available. Primary Handling Editor: Nick Bernard, in collaboration with the Nature Immunology team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Alum injections induce pathological autoantibody production.
a, Treatment and analysis scheme. 6–8-week-old female B6 mice were i.p. injected with PBS or alum (1 mg/mouse) and analyzed as indicated. b,c, ELISA of serum IL-1β (b) and anti-dsDNA IgG (c). d-f, FC percentages of splenic nucleosome- or DNA-reactive antibody-forming cells (AFCs) (d), IgM–IgG1+CD19+ B, plasma cells (PC, B220intCD138+CD3−) and IgM–IgG2b+CD19+ B cells (e), TFH cells (CXCR5+PD1+CD44+CD4+) and GC B cells (GL7+CD38-B220+CD3-CD11b-CD11c−) (f). g, Treatment and analysis scheme of female B6 mice used in h,i. h, ELISA of serum anti-dsDNA IgG. i, FC percentages of splenic Tfh cells (CXCR5+ICOS+Foxp3-CD4+), GC B and MZ B cells (CD21hiCD23−B220+). j, ELISA of serum anti-dsDNA IgG 100 days after the 1st alum injection as in g (male: n = 6/PBS, n = 9/Alum; Female: n = 8/PBS, n = 14/Alum). Results in (b-f and h-j) are mean ± SD. n = 5/group (b-f). n = 10/0 d, n = 13/100 d, n = 10/130 d (h, i). Kruskal–Wallis test (b-f, h and i) and two-way ANOVA with Tukey multiple-comparison test (j).
Extended Data Fig. 2 DNase I injections blunt alum-induced autoimmunity.
a, Treatment and analysis scheme of male B6 mice in three independent biological experiments. b, Spleen gross morphology, mass to body weight ratio (n = 9/PBS, n = 10/alum, n = 12/alum+DNase I) and absolute splenocyte numbers (n = 5/PBS, n = 7 alum, n = 5/alum+DNase I). Scale bar, 1 cm. c, CLIFT assay of serum anti-dsDNA IgG. Scale bar, 20 μm. d, FC plots of anti-mtDNA antibody forming cells (AFC) in spleens. e, GC B cell number (n = 5/PBS, n = 7/alum, n = 5/alum+DNase I) and frequencies of PC (n = 7/PBS, n = 9/alum, n = 7/alum+DNase I) and MZ B cells (n = 9/PBS, n = 9/alum, n = 12/alum+DNase I) in the spleens. f, Percentages of splenic proliferating Ki67+ B220+cells and CD4+ T cells. g, ELISA of serum IL-21, IL-6, IL-1β and IFN-α. h, Relative amounts of circulating mtDNA. i, 8-Oxo-dG containing DNA amounts (n = 5/PBS, n = 7/alum, n = 5/alum+DNase I) and ratio of Fpg-treated (+) to nontreated (−) circulating mtDNA, indicating the fraction of non-Ox-mtDNA (Fpg-resistant) in mtDNA (D-loop, Cox1 and Non-Numt, n = 9/group). j, Ratio of serum nDNA (Tert or B2m) to mtDNA (D-loop). Results in (b and e-j) are mean ± SD. n = 9/PBS, n = 9/alum, n = 12/alum+DNase I (f and g). n = 7/PBS, n = 9/alum, n = 7/alum+DNase I (h and j). Kruskal–Wallis test. Micrographs (b and c) are representative of at least three independent experiments.
Extended Data Fig. 3 Alum-induced autoimmunity depends on NLRP3 and mtDNA oxidation.
a, Treatment and analysis scheme of WT & Nlrp3−/− (b-d) and WT & mt-Ogg1Tg (e-n) females. b-d, Relative amounts of circulating ccf-mtDNA (b), ratio of Fpg-treated (+) to nontreated (−) circulating ccf-mtDNA (c), and nDNA (Tert or B2m) to mtDNA (D-loop) ratio (d) (n = 8/WT PBS; n = 14/WT alum; n = 9/Nlrp3−/− alum). e, IF of IgG (green), GL7 (blue) and B220 (red) stained spleen sections. Scale bar, 50 μm. f,g, Percentages of splenic GC B and Tfh cells (f) and splenic CXCR5−CXCR3+PD-1+CD4+ and IFNγ+IL10+CXCR5−CD4+ memory T cells (g) (n = 7/WT PBS, n = 10/WT alum, n = 7/mt-Ogg1Tg alum). h, Percentages of blood CXCR5−CXCR3+PD-1+CD4+ T helper and IFNγ+IL10+CXCR5−CD4+ T memory (n = 6/WT PBS, n = 4/WT Alum, n = 4/mt-Ogg1Tg Alum). i, Percentages of blood Tfh cells (n = 7/WT PBS, n = 10/WT Alum, n = 7/mt-Ogg1Tg Alum) and CD138+CD19+CD3−CD4− cells (n = 6/WT PBS, n = 4/WT Alum, n = 4/mt-Ogg1Tg Alum). j, Glomeruli sizes in H&E-stained kidney sections (n = 165/WT PBS, n = 147/WT Alum, n = 158/mt-Ogg1Tg Alum). k, MFI of IgG (n = 158/WT PBS, n = 166/WT Alum, n = 123/mt-Ogg1Tg Alum), C3 deposits (n = 115/WT PBS, n = 119/WT Alum, n = 101/mt-Ogg1Tg Alum) and area (in %) occupied by CD45+ cells (n = 74/WT PBS, n = 78/WT Alum, n = 72/mt-Ogg1Tg Alum) in kidney sections. l, Urinary protein ELISA (n = 4/WT PBS, n = 7/WT Alum, n = 5/mt-Ogg1Tg Alum). m,n, ELISA of serum anti-dsDNA and anti-nucleosome IgGs (m) and IL-1β (n) (n = 5/WT PBS or Alum, n = 6/mt-Ogg1Tg PBS or Alum). Results in (b-d and f-n) are mean ± SD. Two-way ANOVA with Tukey multiple-comparison test. Micrographs (e) are representative of at least three independent experiments.
Extended Data Fig. 4 mt-Ogg1 prevents pristane-induced B6 autoimmunity.
8-week-old wild-type (WT) and mt-Ogg1Tg females (B6 background) were given a single i.p. injection of 0.5 ml PBS or pristane and analyzed 12 months later. a, Serum titers of anti-dsDNA IgG. b, Serum IL-1β amounts. c, Spleen mass to body weight ratio. d, Percentages of splenic GC B, Tfh and IgG1+IgM-CD19+ B cells. e, Sizes of glomeruli (n = 112/WT PBS, n = 91/WT Pristane, n = 107/mt-Ogg1Tg PBS, n = 100/mt-Ogg1Tg Pristane), MFI quantitation of IgG (n = 81/WT PBS or Pristane, n = 102/mt-Ogg1Tg PBS, n = 85/mt-Ogg1Tg Pristane) and C3 deposits (n = 81/WT PBS, n = 89/WT Pristane, n = 95/mt-Ogg1Tg PBS, n = 88/mt-Ogg1Tg Pristane) and CD45+ cell areas % (n = 31/WT PBS, n = 28/WT Pristane, n = 41/mt-Ogg1Tg PBS, n = 40/mt-Ogg1Tg Pristane) in kidney sections. f, Relative amounts of serum ccf-mtDNA. g, Serum non-Ox-mtDNA (Fpg-resistant). h, Serum nDNA (Tert or B2m) to mtDNA (D-loop) ratio. Results are mean ± SD. n = 4/group (a-d and f-h). Two-way ANOVA with Tukey multiple-comparison test.
Extended Data Fig. 5 Alum induced ccf-Ox-mtDNA is sensed by functionally important peritoneal pDC.
a-f, Relative amounts of peritoneal mtDNA (a,c and e), and non-Ox-mtDNA (Fpg-resistant) (b,d and f) 24 h after PBS or alum injection into the indicated mice in Fig. 3f, g and i. (n = 7/WT PBS, n = 5/WT alum, n = 7/Nlrp3−/− alum (a); n = 7/WT PBS, n = 4/WT alum, n = 9/Gsdmd−/− alum (c); n = 8/WT PBS, n = 8/Cmpk2ff alum, n = 9/Cmpk2ΔMye alum). g, Treatment and analysis scheme (short-term). BDCA2-DTR− and BDCA2-DTR+ male mice were i.p. injected with DT (100 ng/mice), followed by PBS (n = 6/BDCA2-DTR− and n = 4/BDCA2-DTR+) or alum (n = 6/genotype) injections and FC analyses of splenic and peritoneal pDC after 24 h. h, Treatment and analysis scheme (long term) for i-n. Alum-treated BDCA2-DTR− and BDCA2-DTR+ female mice were repetitively injected with PBS (n = 5/BDCA2-DTR- and n = 4/BDCA2-DTR+) or alum (n = 8/genotype) and analyzed. i, Spleen mass to body weight ratio. j, k, ELISA of serum anti-dsDNA and anti-nucleosome IgGs (j), IL-21 and IFN-α (k). l, IF of GL7 (blue) and B220 (red) stained spleen sections. Scale bar, 50 μm. m, FC frequencies of splenic GC B, Tfh, IgG1+IgM-CD19+ and IgG2b+IgM-CD19+ cells. n, Sizes of glomeruli (n = 130/PBS/genotype, n = 125/BDCA2-DTR− alum, n = 122/BDCA2-DTR+ alum) in H&E-stained kidney sections, MFI of kidney IgG deposits (n = 112/PBS/genotype, n = 118/BDCA2-DTR− alum, n = 103/BDCA2-DTR+ alum) and urinary protein ELISA (n = 5/BDCA2-DTR− PBS, n = 4/BDCA2-DTR+ PBS, n = 8/BDCA2-DTR− or BDCA2-DTR+ Alum). Results in (a-g, i-k, m and n) are mean ± SD. n = 5/BDCA2-DTR− PBS, n = 4/BDCA2-DTR+ PBS, n = 8/BDCA2-DTR− or BDCA2-DTR+ Alum (i-m). Two-way ANOVA with Tukey multiple-comparison test. Micrographs (l) are representative of at least three independent experiments.
Extended Data Fig. 6 Ox-mtDNA treatment of mouse and human pDC enables ex vivo induction of Tfh differentiation.
a, Naïve splenic CD4+ T cells (see gating strategy in Supplementary Fig. 4a) were co-cultured with purified peritoneal or splenic pDC collected 24 h after two alum injections (72 h apart). Tfh generation was analyzed 72 h after (n = 5). Created with BioRender.com. b, BM Flt3L-induced BMDC labeled with CellTracker Green CMFDA were i.p. injected into B6 males. After 24 h, the presence of peritoneal CMFDA+ pDC in spleen was examined (n = 3 biological replicates). c, Naïve CD4+ T cells were co-cultured with FACS-sorted BM Flt3L-induced pDC (see gating strategy in Supplementary Fig. 4b), −/+ mtDNA or Ox-mtDNA (50 μg) for 72 h. Tfh percentages were analyzed (n = 5). d,e, Tfh percentages after naïve CD4+ T cells co-culture with either pDC, cDC1, or cDC2, −/+ Ox-mtDNA (d) or with pDC incubated with different types of DNA (e) (n = 3). f, Naïve CD4+ T cells were cultured in transwell plates separately from or together with pDC incubated with Ox-mtDNA for 72 h. Tfh differentiation in either the upper (left) or lower (right) wells was analyzed (n = 4). g, Treatment and analysis scheme for h,i. Created with BioRender.com. CD4+ T after naïve CD4+ T cells co-cultured with pDC, −/+ mtDNA or Ox-mtDNA were isolated and co-cultured with naïve splenic B cells for 72 h, after which B cell proliferation, differentiation and IgG secretion were analyzed. h,i, Percentages of different B cell types (h) and ELISA of secreted IgG (i) (n = 4). j,k, Secreted IgG (j) and BCL6+ B cells and plasmablasts (PB; CD138hiB220loIgD-CD3-CD19+, k) 72 h after co-culture of naïve splenic B cells with CD4+ T cells from PBS or alum injected mice (n = 4). l,m, Freshly isolated pDC and CD4+ T cells purified from healthy human buffy coats were co-cultured as in c. Tfh percentages (l) and intracellular BCL6 (m) were analyzed (n = 3 biological replicates). Results are mean ± SD. Kruskal–Wallis test (c, f and h-l) and two-way ANOVA with Tukey multiple-comparison test (a, d and e).
Extended Data Fig. 7 Alum-induced Ox-mtDNA-dependent autoimmunity relies on Tfh cells.
a, Tfh frequencies after co-culture of murine Bcl6ff and Bcl6ΔCD4 naïve splenic CD4+ T cells with FACS-sorted B6 BM-Flt3L-induced pDC, −/+ Ox-mtDNA for 3 days (n = 3 biological replicates). b-j, Alum-induced autoimmunity in 6–8-week-old Bcl6ff and Bcl6ΔCD4 males treated as in Extended Data Fig. 3a. Intracellular BCL6 staining of splenic CD4+ T cells (b), serum anti-dsDNA IgG (c), FC analyses of splenic GC B frequencies (d), IF of GL7 (blue) and B220 (red) stained spleen sections (Scale bar, 50 μm, e), serum IL-21 and IL-1β (f), urinary protein ELISA (n = 4/group, g), sizes of glomeruli in kidney sections (n = 123/group, h), MFI of renal IgG deposits (n = 120/group) and C3 (n = 120/group) and renal area (in %) occupied by CD45+ cells (n = 29/Bcl6ff PBS; n = 38/Bcl6ff Alum; n = 33/Bcl6ΔCD4 PBS; n = 32/Bcl6ΔCD4 Alum) (i) and spleen mass to body weight ratio (j). k, Numbers and percentages of peritoneal pDC 24 h after PBS or alum injection into Bcl6ff and Bcl6ΔCD4 males (n = 5/Bcl6ff PBS; n = 9/Bcl6ff Alum; n = 5/Bcl6ΔCD4 PBS; n = 8/Bcl6ΔCD4 Alum). l, Active Casp1 (FLICAhi) in pre-gated peritoneal pDC 24 h after alum injections into Bcl6ff and Bcl6ΔCD4 males (representative of 3 independent experiments). FMO, fluorescence minus one. Results in (a, c, d and f-k) are mean ± SD. n = 8/Bcl6ff PBS; n = 7/Bcl6ff Alum; n = 5/Bcl6ΔCD4 PBS; n = 7/Bcl6ΔCD4 Alum (b-d, f and j). Two-way ANOVA with Tukey multiple-comparison test. Micrographs (e) are representative of at least three independent experiments.
Extended Data Fig. 8 Ox-mtDNA activates Casp1 in primary pDC.
a,b, FC analyses of active Casp1 (FLICAhi) in pre-gated Flt3L-induced DC subsets (n = 4 biological replicates, a) or BMDM (n = 3 biological replicates, b) 4 h after incubation with different types of DNA. Results in (a) are mean ± SD. Two-way ANOVA with Tukey multiple-comparison test.
Extended Data Fig. 9 Ox-mtDNA enables primary pDC to induce Tfh differentiation by activating NLRP3.
a, Naïve B6 splenic CD4+ T cells were co-cultured with purified peritoneal pDC collected 24 h after 4 repetitive mtDNA or Ox-mtDNA injections (72 h apart). No peritoneal pDC were present after PBS injection. After 3 days of co-culture, Tfh (CXCR5+BCL6+Foxp3-CD44+CD4+) generation was analyzed (n = 3 independent experiments). b, BM Flt3L-induced BMDC (CMFDA+) were i.p. injected into 8-week-old B6 males pre-treated with 4 repetitive PBS or mtDNA injections. 24 h later, splenic DC were analyzed for presence of peritoneal CMFDA+ pDC (n = 3 independent experiments). c, Naïve B6 splenic CD4+ T cells were co-cultured with purified splenic DC collected 24 h after 4 repetitive PBS or Ox-mtDNA injections into DT treated BDCA2-DTR- and BDCA2-DTR+ mice. After 3 days, Tfh generation was analyzed (n = 5 independent experiments). d, Experimental strategy for generating mixed BM chimeras by transplanting lethally irradiated females with 50:50% mixtures of Nlrp3−/− and BDCA2-DTR− (bearing Nlrp3+/+) or BDCA2-DTR+ (bearing Nlrp3+/+) BM, followed by i.p. injections of DT and Ox-mtDNA, as indicated. e-h, Serum titers of anti-dsDNA and anti-nucleosome IgGs (e), percentages of splenic GC, IgG1+IgM-CD19+, IgG2b+IgM-CD19+ B cells and Tfh (f). serum IL-21 amounts (g) and spleen mass to body weight ratio (h) (n = 4/group). Results (a-c and e-h) are mean ± SD. Kruskal–Wallis test (a, b) and two-way ANOVA with Tukey multiple-comparison test (c and e-h).
Extended Data Fig. 10 FcγR1 mediates mtDNA uptake followed by delivery to TLR9 and endosomal rupture.
a, Treatment and analysis scheme. 8-week-old B6 males were i.p. injected with 2 μg Biotin-Ox-mtDNA and 24 h later, BM, splenic and peritoneal pDC were purified and FC analyzed for intracellular biotin-Ox-mtDNA uptake (n = 3 independent experiments). b, IF analysis of splenic pDC that were incubated with 2 μg Biotin-mtDNA or Biotin-Ox-mtDNA for 1 h and stained for biotin (red) and TLR9 (green). Scale bars, 5 μm and 1 μm (n = 3). c, Relative fluorescence intensities of the biotin and TLR9 signals along the broken white lines in b, demonstrating (Ox-)mtDNA-TLR9 co-localization. d, FC analyses of TLR9hi cells in WT and Tlr9−/− Flt3L-induced BMDC cultures (including pDC, cDC1 and cDC2) 4 h after incubation with Ox-mtDNA (n = 3 independent experiments). e, GSEA pathway analyses of bulk RNA-seq data (n = 3). f, IF analysis of splenic pDC that were incubated with 2 μg biotin-labelled non-oxidized or oxidized mtDNA (red) for 1 h and stained for ruptured endosomes with galectin 8 (green) and TLR9 (cyan) antibodies. Scale bar, 5 μm. g, A working model illustrating how (Ox-) mtDNAs are internalized by pDC and activate TLR9-dependent NF-κB and NLRP3 inflammasome signaling (created with BioRender). Although both mtDNA forms activate TLR9 and access the cytoplasm via ruptured endosomes, only Ox-mtDNA binds NLRP3 and activates the inflammasome to produce IL-1β that potentiates NF-κB activation via IL-1R. Results in (a and d) are mean ± SD. Kruskal–Wallis test (a and d). Two-sided adaptive multilevel splitting Monte Carlo approach with Benjamini-Hochberg procedure (e). Micrographs (b, c and f) are representative of at least three independent experiments.
Supplementary information
Supplementary Information
Supplementary Figs. 1–4 and Supplementary Tables 1 and 2.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 4
Unprocessed western blots.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 7
Statistical source data.
Source Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 9
Statistical source data.
Source Data Extended Data Fig. 10
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xian, H., Watari, K., Ohira, M. et al. Mitochondrial DNA oxidation propagates autoimmunity by enabling plasmacytoid dendritic cells to induce TFH differentiation. Nat Immunol 26, 1168–1181 (2025). https://doi.org/10.1038/s41590-025-02179-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-025-02179-7
This article is cited by
-
pDCs drive immunopathology by sensing oxidized mitochondrial DNA
Nature Immunology (2025)