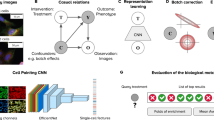
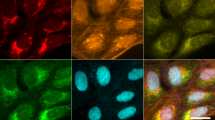
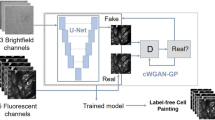
Abstract
Modern quantitative image analysis techniques have enabled high-throughput, high-content imaging experiments. Image-based profiling leverages the rich information in images to identify similarities or differences among biological samples, rather than measuring a few features, as in high-content screening. Here, we review a decade of advancements and applications of Cell Painting, a microscopy-based cell-labeling assay aiming to capture a cell’s state, introduced in 2013 to optimize and standardize image-based profiling. Cell Painting’s ability to capture cellular responses to various perturbations has expanded owing to improvements in the protocol, adaptations for different perturbations, and enhanced methodologies for feature extraction, quality control, and batch-effect correction. Cell Painting is a versatile tool that has been used in various applications, alone or with other -omics data, to decipher the mechanism of action of a compound, its toxicity profile, and other biological effects. Future advances will likely involve computational and experimental techniques, new publicly available datasets, and integration with other high-content data types.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
27,99 € / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
269,00 € per year
only 22,42 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
12 December 2024
A Correction to this paper has been published: https://doi.org/10.1038/s41592-024-02578-y
References
Swinney, D. C. & Anthony, J. How were new medicines discovered? Nat. Rev. Drug Discov. 10, 507–519 (2011).
Lin, A. et al. Off-target toxicity is a common mechanism of action of cancer drugs undergoing clinical trials. Sci. Transl. Med. 11, eaaw8412 (2019).
Moffat, J. G., Vincent, F., Lee, J. A., Eder, J. & Prunotto, M. Opportunities and challenges in phenotypic drug discovery: an industry perspective. Nat. Rev. Drug Discov. 16, 531–543 (2017).
Perlman, Z. E. et al. Multidimensional drug profiling by automated microscopy. Science 306, 1194–1198 (2004).
Schulze, C. J. et al. ‘Function-first’ lead discovery: mode of action profiling of natural product libraries using image-based screening. Chem. Biol. 20, 285 (2013).
Feng, Y., Mitchison, T. J., Bender, A., Young, D. W. & Tallarico, J. A. Multi-parameter phenotypic profiling: using cellular effects to characterize small-molecule compounds. Nat. Rev. Drug Discov. 8, 567–578 (2009).
Woehrmann, M. H. et al. Large-scale cytological profiling for functional analysis of bioactive compounds. Mol. Biosyst. 9, 2604–2617 (2013).
Gustafsdottir, S. M. et al. Multiplex cytological profiling assay to measure diverse cellular states. PLoS ONE 8, e80999 (2013).
Stirling, D. R. et al. CellProfiler 4: improvements in speed, utility and usability. BMC Bioinf. 22, 1–11 (2021).
Bray, M. A. et al. Cell Painting, a high-content image-based assay for morphological profiling using multiplexed fluorescent dyes. Nat. Protoc. 11, 1757–1774 (2016).
Cimini, B. A. et al. Optimizing the Cell Painting assay for image-based profiling. Nat. Protoc. 18, 1981–2013 (2023).
Tromans-Coia, C. et al. Assessing the performance of the Cell Painting assay across different imaging systems. Cytom. Part A 103, 915–926 (2023).
Chandrasekaran, S. N. et al. Three million images and morphological profiles of cells treated with matched chemical and genetic perturbations. Nat. Methods 21, 1114–1121 (2024).
Chandrasekaran, S. N. et al. JUMP Cell Painting dataset: morphological impact of 136,000 chemical and genetic perturbations. Preprint at bioRxiv https://doi.org/10.1101/2023.03.23.534023 (2023).
Heinrich, L., Kumbier, K., Li, L., Altschuler, S. J. & Wu, L. F. Selection of optimal cell lines for high-content phenotypic screening. ACS Chem. Biol. 18, 679–685 (2023).
Willis, C., Nyffeler, J. & Harrill, J. Phenotypic profiling of reference chemicals across biologically diverse cell types using the Cell Painting assay. SLAS Discov. 25, 755–769 (2020).
Laber, S. et al. Discovering cellular programs of intrinsic and extrinsic drivers of metabolic traits using LipocyteProfiler. Cell Genomics 3, 100346 (2023).
Rietdijk, J. et al. A phenomics approach for antiviral drug discovery. BMC Biol. 19, 156 (2021).
Singh, S. et al. Morphological profiles of RNAi-induced gene knockdown are highly reproducible but dominated by seed effects. PLoS One 10, e0131370 (2015).
Caicedo, J. C. et al. Cell Painting predicts impact of lung cancer variants. Mol. Biol. Cell 33, ar49 (2022).
Way, G. P. et al. Predicting cell health phenotypes using image-based morphology profiling. Mol. Biol. Cell 32, 995–1005 (2021).
Dahlin, J. L. et al. Reference compounds for characterizing cellular injury in high-content cellular morphology assays. Nat. Commun. 14, 1364 (2023).
Smith, K. et al. Phenotypic image analysis software tools for exploring and understanding big image data from cell-based assays. Cell Syst. 6, 636–653 (2018).
Moshkov, N. et al. Learning representations for image-based profiling of perturbations. Nat. Commun. 15, 1594 (2024).
Caron, M. et al. Emerging Properties in Self-Supervised Vision Transformers. In Proc.IEEE/CVF International Conference on Computer Vision 9630–9640 (IEEE, 2021)
He, K. et al. Masked autoencoders are scalable vision learners. in 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 15979–15988 (IEEE, 2022).
Kim, V., Adaloglou, N., Osterland, M., Morelli, F. M. & Zapata, P. A. M. Self-supervision advances morphological profiling by unlocking powerful image representations. Preprint at bioRxiv https://doi.org/10.1101/2023.04.28.538691 (2023).
Cross-Zamirski, J. O. et al. Label-free prediction of cell painting from brightfield images. Sci. Rep. 12, 10001 (2022).
Harrison, P. J. et al. Evaluating the utility of brightfield image data for mechanism of action prediction. PLoS Comput. Biol. 19, e1011323 (2023).
Belli, B. Brightfield is back: a 17th century cell imaging technique is making a comeback thanks to machine learning. https://www.recursion.com/news/brightfield-is-back-a-17th-century-cell-imaging-technique-is-making-a-comeback-thanks-to-machine-learningRecursion (2024).
Serrano, E. et al. Reproducible image-based profiling with Pycytominer. Preprint at arXiv https://doi.org/10.48550/arXiv.2311.13417 (2024).
Siegismund, D., Fassler, M., Heyse, S. & Steigele, S. Benchmarking feature selection methods for compressing image information in high-content screening. SLAS Technol. 27, 85–93 (2022).
Janosch, A., Kaffka, C. & Bickle, M. Unbiased phenotype detection using negative controls. SLAS Discov. 24, 234–241 (2019).
Way, G. P. et al. Morphology and gene expression profiling provide complementary information for mapping cell state. Cell Syst. 13, 911–923 (2022).
Caicedo, J. C. et al. Data-analysis strategies for image-based cell profiling. Nat. Methods 14, 849–863 (2017).
Altschuler, S. J. & Wu, L. F. Cellular heterogeneity: do differences make a difference? Cell 141, 559–563 (2010).
van Dijk, R., Arevalo, J., Babadi, M., Carpenter, A. E. & Singh, S. Capturing cell heterogeneity in representations of cell populations for image-based profiling using contrastive learning. PLOS Comput. Biol. 20, e1012547 (2024).
Arevalo, J. et al. Evaluating batch correction methods for image-based cell profiling. Nat. Commun. 15, 6516 (2024).
Yang, S. et al. DeepNoise: signal and noise disentanglement based on classifying fluorescent microscopy images via deep learning. Genom. Proteom. Bioinform. 20, 989–1001 (2022).
Weisbart, E. et al. Cell Painting Gallery: an open resource for image-based profiling. Nat. Methods 21, 1775–1777 (2024).
Trapotsi, M. A., Hosseini-Gerami, L. & Bender, A. Computational analyses of mechanism of action (MoA): data, methods and integration. RSC Chem. Biol. 3, 170–200 (2022).
Akbarzadeh, M. et al. Morphological profiling by means of the Cell Painting assay enables identification of tubulin-targeting compounds. Cell Chem. Biol. 29, 1053–1064 (2022).
Seal, S. et al. Integrating cell morphology with gene expression and chemical structure to aid mitochondrial toxicity detection. Commun. Biol. 5, 858 (2022).
Herman, D. et al. Leveraging Cell Painting images to expand the applicability ___domain and actively improve deep learning quantitative structure–activity relationship models. Chem. Res. Toxicol. 36, 1028–1036 (2023).
Garcia de Lomana, M., Marin Zapata, P. A. & Montanari, F. Predicting the mitochondrial toxicity of small molecules: insights from mechanistic assays and cell painting data. Chem. Res. Toxicol. 36, 1107–1120 (2023).
Laraia, L., Robke, L. & Waldmann, H. BioactiVe compound collections: from design to target identification. Chem 4, 705–730 (2018).
Cox, M. J. et al. Tales of 1,008 small molecules: phenomic profiling through live-cell imaging in a panel of reporter cell lines. Sci. Rep. 10, 13262 (2020).
C. Herbert Waldmann—celebrating more than three decades in academia. J. Med. Chem. 66, 15055–15060 (2023).
Laraia, L. et al. Image-based morphological profiling identifies a lysosomotropic, iron-sequestering autophagy inhibitor. Angew. Chem. Int. Ed. 59, 5721–5729 (2020).
Svenningsen, E. B. & Poulsen, T. B. Establishing cell painting in a smaller chemical biology lab—a report from the frontier. Bioorg. Med. Chem. 27, 2609–2615 (2019).
Schölermann, B. et al. Identification of dihydroorotate dehydrogenase inhibitors using the Cell Painting assay. ChemBioChem 23, e202200475 (2022).
Wilke, J. et al. Discovery of a σ1 receptor antagonist by combination of unbiased cell painting and thermal proteome profiling. Cell Chem. Biol. 28, 848–854 (2021).
Wassermann, A. M. et al. Dark chemical matter as a promising starting point for drug lead discovery. Nat. Chem. Biol. 11, 958–966 (2015).
Pahl, A. et al. Illuminating dark chemical matter using the Cell Painting assay. J. Med. Chem. 67, 8862–8876 (2024).
Dürr, O. & Sick, B. Single-cell phenotype classification using deep convolutional neural networks. J. Biomol. Screen 21, 998–1003 (2016).
Kensert, A., Harrison, P. J. & Spjuth, O. Transfer learning with deep convolutional neural networks for classifying cellular morphological changes. SLAS Discov. 24, 466–475 (2019).
Lafarge, M. W. et al. Capturing single-cell phenotypic variation via unsupervised representation learning. Proc. Mach. Learn Res. 102, 315–325 (2019).
Wong, D. R. et al. Deep representation learning determines drug mechanism of action from cell painting images. Digital Discov. 2, 1354–1367 (2023).
Liu, G., Seal, S., Arevalo, J. & Liang, Z. Learning molecular representation in a cell. Preprint at arXiv https://doi.org/10.48550/arXiv.2406.12056 (2024).
Simm, J. et al. Repurposing high-throughput image assays enables biological activity prediction for drug discovery. Cell Chem. Biol. 25, 611–618 (2018).
Hofmarcher, M., Rumetshofer, E., Clevert, D. A., Hochreiter, S. & Klambauer, G. Accurate prediction of biological assays with high-throughput microscopy images and convolutional networks. J. Chem. Inf. Model. 59, 1163–1171 (2019).
Nyffeler, J. et al. Comparison of approaches for determining bioactivity hits from high-dimensional profiling data. SLAS Discov. 26, 292–308 (2021).
Trapotsi, M. A. et al. Comparison of chemical structure and cell morphology information for multitask bioactivity predictions. J. Chem. Inf. Model. 61, 1444–1456 (2021).
Seal, S. et al. Merging bioactivity predictions from cell morphology and chemical fingerprint models using similarity to training data. J. Cheminform. 15, 56 (2023).
Sanchez-Fernandez, A., Rumetshofer, E., Hochreiter, S. & Klambauer, G. CLOOME: contrastive learning unlocks bioimaging databases for queries with chemical structures. Nat. Commun. 14, 1–14 (2023).
Tian, G., Harrison, P. J., Sreenivasan, A. P., Carreras-Puigvert, J. & Spjuth, O. Combining molecular and cell painting image data for mechanism of action prediction. Artif. Intell. Life Sci. 3, 100060 (2023).
Moshkov, N. et al. Predicting compound activity from phenotypic profiles and chemical structures. Nat. Commun. 14, 1967 (2023).
Fredin Haslum, J. et al. Cell Painting-based bioactivity prediction boosts high-throughput screening hit-rates and compound diversity. Nat. Commun. 15, 1–11 (2024).
Gerry, C. J. et al. Real-time biological annotation of synthetic compounds. J. Am. Chem. Soc. 138, 8920–8927 (2016).
Nelson, S. D., Wawer, M. J. & Schreiber, S. L. Divergent synthesis and real-time biological annotation of optically active tetrahydrocyclopenta[c]pyranone derivatives. Org. Lett. 18, 6280–6283 (2016).
Gerlach, E. M., Korkmaz, M. A., Pavlinov, I., Gao, Q. & Aldrich, L. N. Systematic diversity-oriented synthesis of reduced flavones from γ-pyrones to probe biological performance diversity. ACS Chem. Biol. 14, 1536–1545 (2019).
Melillo, B. et al. Synergistic effects of stereochemistry and appendages on the performance diversity of a collection of synthetic compounds. J. Am. Chem. Soc. 140, 11784–11790 (2018).
Christoforow, A. et al. Design, synthesis, and phenotypic profiling of pyrano-furo-pyridone pseudo natural products. Angew. Chem. Int. Ed. 58, 14715–14723 (2019).
Foley, D. J. et al. Phenotyping reveals targets of a pseudo-natural-product autophagy inhibitor. Angew. Chem. Int. Ed. 59, 12470–12476 (2020).
Hippman, R. S. et al. Multiple chemical features impact biological performance diversity of a highly active natural product-inspired library. ChemBioChem 21, 3137–3145 (2020).
Singh, M., Garza, N., Pearson, Z., Douglas, J. & Boskovic, Z. Broad assessment of bioactivity of a collection of spiroindane pyrrolidines through ‘cell painting’. Bioorg. Med. Chem. 28, 115547 (2020).
Liu, J. et al. Design, synthesis, and biological evaluation of chemically and biologically diverse pyrroquinoline pseudo natural products. Angew. Chem. Int. Ed. 60, 4648–4656 (2021).
Burhop, A. et al. Synthesis of indofulvin pseudo-natural products yields a new autophagy inhibitor chemotype. Adv. Sci. 8, e2102042 (2021).
Seal, S. et al. From pixels to phenotypes: integrating image-based profiling with cell health data as BioMorph features improves interpretability. Mol. Biol. Cell 35, mr2 (2024).
Seal, S., Yang, H., Vollmers, L. & Bender, A. Comparison of cellular morphological descriptors and molecular fingerprints for the prediction of cytotoxicity- and proliferation-related assays. Chem. Res. Toxicol. 34, 422–437 (2021).
Trapotsi, M. A. et al. Cell morphological profiling enables high-throughput screening for proteolysis targeting chimera (PROTAC) phenotypic signature. ACS Chem. Biol. 17, 1733–1744 (2022).
Seal, S. et al. Improved detection of drug-induced liver injury by integrating predicted in vivo and in vitro data. Chem. Res Toxicol. 37, 1290–1305 (2024).
Nyffeler, J. et al. Bioactivity screening of environmental chemicals using imaging-based high-throughput phenotypic profiling. Toxicol. Appl. Pharmacol. 389, 114876 (2020).
Nyffeler, J. et al. Application of Cell Painting for chemical hazard evaluation in support of screening-level chemical assessments. Toxicol. Appl. Pharmacol. 468, 116513 (2023).
Thomas, R. S. et al. The next generation blueprint of computational toxicology at the U.S. Environmental Protection Agency. Toxicol. Sci. 169, 317–332 (2019).
Chen, M. et al. DILIrank: the largest reference drug list ranked by the risk for developing drug-induced liver injury in humans. Drug Discov. Today 21, 648–653 (2016).
Seal, S. et al. Improved detection of drug-induced liver injury by integrating predicted in vivo and in vitro data. Chem. Res. Toxicol. 37, 1290–1305 (2024).
Seal, S. et al. Insights into drug cardiotoxicity from biological and chemical data: the first public classifiers for FDA drug-induced cardiotoxicity rank. J. Chem. Inf. Model. 64, 1172–1186 (2024).
Horne, R. I. et al. Using generative modeling to endow with potency initially inert compounds with good bioavailability and low toxicity. J. Chem. Inf. Model. 64, 590–596 (2024).
Seal, S. et al. PKSmart: an open-source computational model to predict in vivo pharmacokinetics of small molecules. Preprint at bioRxiv https://doi.org/10.1101/2024.02.02.578658 (2024).
Pierozan, P., Kosnik, M. & Karlsson, O. High-content analysis shows synergistic effects of low perfluorooctanoic acid (PFOS) and perfluorooctane sulfonic acid (PFOA) mixture concentrations on human breast epithelial cell carcinogenesis. Environ. Int. 172, 107746 (2023).
Rietdijk, J. et al. Morphological profiling of environmental chemicals enables efficient and untargeted exploration of combination effects. Sci. Total Environ. 832, 155058 (2022).
Chow, Y. L., Singh, S., Carpenter, A. E. & Way, G. P. Predicting drug polypharmacology from cell morphology readouts using variational autoencoder latent space arithmetic. PLoS Comput. Biol. 18, e1009888 (2022).
Rohban, M. H. et al. Systematic morphological profiling of human gene and allele function via cell painting. eLife 6, e24060 (2017).
Rohban, M. H. et al. Virtual screening for small-molecule pathway regulators by image-profile matching. Cell Syst. 13, 724–736 (2022).
Hughes, R. E., Elliott, R. J. R., Dawson, J. C. & Carragher, N. O. High-content phenotypic and pathway profiling to advance drug discovery in diseases of unmet need. Cell Chem. Biol. 28, 338–355 (2021).
Hughes, R. E. et al. High-content phenotypic profiling in esophageal adenocarcinoma identifies selectively active pharmacological classes of drugs for repurposing and chemical starting points for novel drug discovery. SLAS Discov. 25, 770–782 (2020).
Cuccarese, M. F. et al. Functional immune mapping with deep-learning enabled phenomics applied to immunomodulatory and COVID-19 drug discovery. Preprint at bioRxiv https://doi.org/10.1101/2020.08.02.233064 (2020).
Heiser, K. et al. Identification of potential treatments for COVID-19 through artificial intelligence-enabled phenomic analysis of human cells infected with SARS-CoV-2. Preprint at bioRxiv https://doi.org/10.1101/2020.04.21.054387 (2020).
Carey, K. L. et al. TFEB transcriptional responses reveal negative feedback by BHLHE40 and BHLHE41. Cell Rep. 33, 108371 (2020).
Kelley, M. E. et al. High-content microscopy reveals a morphological signature of bortezomib resistance. eLife. 12, e91362 (2023).
Tegtmeyer, M. et al. High-dimensional phenotyping to define the genetic basis of cellular morphology. Nat. Commun. 15, 347 (2024).
McDiarmid, A. H. et al. Morphological profiling in human neural progenitor cells classifies hits in a pilot drug screen for Alzheimer’s disease. Brain Commun. 6, fcae101 (2024).
Schiff, L. et al. Integrating deep learning and unbiased automated high-content screening to identify complex disease signatures in human fibroblasts. Nat. Commun. 13, 1590 (2022).
Yang, S. J. et al. Applying deep neural network analysis to high-content image-based assays. SLAS Discov. 24, 829–841 (2019).
Liu, A., Seal, S., Yang, H. & Bender, A. Using chemical and biological data to predict drug toxicity. SLAS Discov. 28, 53–64 (2023).
Nassiri, I. & McCall, M. N. Systematic exploration of cell morphological phenotypes associated with a transcriptomic query. Nucleic Acids Res. 46, e116 (2018).
Haghighi, M., Caicedo, J. C., Cimini, B. A., Carpenter, A. E. & Singh, S. High-dimensional gene expression and morphology profiles of cells across 28,000 genetic and chemical perturbations. Nat. Methods 19, 1550–1557 (2022).
Nyffeler, J. et al. Combining phenotypic profiling and targeted RNA-Seq reveals linkages between transcriptional perturbations and chemical effects on cell morphology: retinoic acid as an example. Toxicol. Appl. Pharmacol. 444, 116032 (2022).
Cerisier, N., Dafniet, B., Badel, A. & Taboureau, O. Linking chemicals, genes and morphological perturbations to diseases. Toxicol. Appl. Pharmacol. 461, 116407 (2023).
Camunas-Soler, J. Integrating single-cell transcriptomics with cellular phenotypes: cell morphology, Ca2+ imaging and electrophysiology. Biophys. Rev. 16, 89–107 (2023).
Dagher, M. et al. nELISA: a high-throughput, high-plex platform enables quantitative profiling of the secretome. Preprint at bioRxiv https://doi.org/10.1101/2023.04.17.535914 (2023).
Schneidewind, T. et al. Combined morphological and proteome profiling reveals target-independent impairment of cholesterol homeostasis. Cell Chem. Biol. 28, 1780–1794 (2021).
Way, G. P., Sailem, H., Shave, S., Kasprowicz, R. & Carragher, N. O. Evolution and impact of high content imaging. SLAS Discov. 28, 292–305 (2023).
Lukonin, I., Zinner, M. & Liberali, P. Organoids in image-based phenotypic chemical screens. Exp. Mol. Med. 53, 1495–1502 (2021).
Cottet, M. et al. Live cell painting: New nontoxic dye to probe cell physiology in high content screening. SLAS Discov. 29, 100121 (2023).
Bray, M.-A. et al. A dataset of images and morphological profiles of 30 000 small-molecule treatments using the Cell Painting assay. Gigascience 6, 1–5 (2017).
Fay, M. M. et al. RxRx3: phenomics map of biology. Preprint at bioRxiv https://doi.org/10.1101/2023.02.07.527350 (2023).
Ramezani, M. et al. A genome-wide atlas of human cell morphology. Preprint at bioRxiv https://doi.org/10.1101/2023.08.06.552164 (2023).
Schneidewind, T. et al. Morphological profiling identifies a common mode of action for small molecules with different targets. ChemBioChem 21, 3197–3207 (2020).
Pahl, A. et al. Morphological subprofile analysis for bioactivity annotation of small molecules. Cell Chem. Biol. 30, 839–853 (2023).
Lapins, M. & Spjuth, O. Evaluation of gene expression and phenotypic profiling data as quantitative descriptors for predicting drug targets and mechanisms of action. Preprint at bioRxiv https://doi.org/10.1101/580654 (2019).
Acknowledgements
S. Seal acknowledges funding from the Cambridge Centre for Data-Driven Discovery (C2D3) and Accelerate Programme for Scientific Discovery. A.E.C., S. Singh, and S. Seal acknowledge funding from the National Institutes of Health (R35 GM122547 to A.E.C.). O.S. acknowledges funding from the Swedish Research Council (Grants 2020-03731 and 2020-01865), FORMAS (Grant 2022-00940), Swedish Cancer Foundation (22 2412 Pj 03 H), and Horizon Europe (Grant Agreements 101057014 (PARC) and 101057442 (REMEDI4ALL)).
Author information
Authors and Affiliations
Contributions
S. Seal and M.-A.T. designed and performed the systematic review on studies using Cell Painting data. S. Seal, M.-A.T., and A.E.C. wrote the manuscript with extensive discussions with all authors. All of the authors reviewed, edited, and contributed to discussions on the manuscript and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
S. Singh and A.E.C. serve as scientific advisors for companies that use image-based profiling and Cell Painting (A.E.C.: Recursion, SyzOnc, Quiver Bioscience; S. Singh: Waypoint Bio, Dewpoint Therapeutics, DeepCell) and receive honoraria for occasional talks at pharmaceutical and biotechnology companies. J.C.P. and O.S. declare ownership in Phenaros Pharmaceuticals. M.-A.T. and N.G. were formerly employed at AstraZeneca. M.-A.T. and N.G. are currently employed at Recursion Pharmaceuticals. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Methods thanks Jeremy Jenkins and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary handling editor: Rita Strack, in collaboration with the Nature Methods team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Table 1
Definitions of essential terms in image-based profiling
Supplementary Table 2
90 studies included in this study
Supplementary Table 3
65 studies excluded in this study
Supplementary Table 4
Academic institutions, government agencies, pharmaceutical companies, non-profit organizations that led studies evaluated in this work and/or are members of the JUMPCP or OASIS consortiums.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Seal, S., Trapotsi, MA., Spjuth, O. et al. Cell Painting: a decade of discovery and innovation in cellular imaging. Nat Methods 22, 254–268 (2025). https://doi.org/10.1038/s41592-024-02528-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-024-02528-8