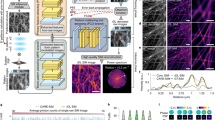
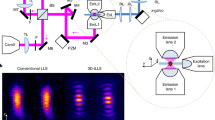
Abstract
Lattice light-sheet microscopy provides a crucial observation window into intra- and intercellular physiology of living specimens but at the diffraction-limited resolution or anisotropic super-resolution with structured illumination. Here we present meta-learning-empowered reflective lattice light-sheet virtual structured illumination microscopy (Meta-rLLS-VSIM), which upgrades lattice light-sheet microscopy to a near-isotropic super resolution of ~120 nm laterally and ~160 nm axially without modifications of the core optical system or loss of other live-cell imaging metrics. Moreover, we devised an adaptive online training approach by synergizing the front-end imaging system and back-end meta-learning framework, which alleviated the demand for training data by tenfold and reduced the total time for data acquisition and model training down to tens of seconds. We demonstrate the versatile functionalities of Meta-rLLS-VSIM by imaging a variety of bioprocesses with ultrahigh spatiotemporal resolution for hundreds of multicolor volumes, delineating the nanoscale distributions, dynamics and interaction patterns of multiple organelles in embryos and eukaryotic cells.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
27,99 € / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
269,00 € per year
only 22,42 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The datasets for training and testing the Meta-VSI-SR and RL-DFN models have been uploaded onto the publicly available Zenodo repository at https://doi.org/10.5281/zenodo.14322456 (ref. 57). Other long-term live-cell datasets that support the biological findings of this study in Figs. 3 and 5, Extended Data Fig. 10, Supplementary Figs. 12–15 and Supplementary Videos 2–10, which were not uploaded onto public repositories because of the excessive file size, are available upon request. Source data are provided with this paper.
Code availability
All source codes of Meta-VSI-SR, RL-DFN, and data processing pipeline of Meta-rLLS-VSIM are open-source on GitHub at https://github.com/Intelligent-SR-Imaging/Meta-rLLS-VSIM. The training datasets, representative testing data, as well as the pre-trained meta-model and several representative finetuned models are available at the Zenodo repository at https://doi.org/10.5281/zenodo.14322456 (ref. 57).
References
Stelzer, E. H. K. et al. Light sheet fluorescence microscopy. Nat. Rev. Methods Primers 1, 73 (2021).
Huisken, J., Swoger, J., Del Bene, F., Wittbrodt, J. & Stelzer, E. H. K. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science 305, 1007–1009 (2004).
Murphy, D. B. & Davidson, M. W. Fundamentals of Light Microscopy and Electronic Imaging (John Wiley & Sons, 2012).
Stephens, D. J. & Allan, V. J. Light microscopy techniques for live cell imaging. Science 300, 82–86 (2003).
Hu, Y. S., Zimmerley, M., Li, Y., Watters, R. & Cang, H. Single‐molecule super‐resolution light‐sheet microscopy. ChemPhysChem 15, 577–586 (2014).
Legant, W. R. et al. High-density three-dimensional localization microscopy across large volumes. Nat. Methods 13, 359–365 (2016).
Gustavsson, A.-K., Petrov, P. N., Lee, M. Y., Shechtman, Y. & Moerner, W. E. 3D single-molecule super-resolution microscopy with a tilted light sheet. Nat. Commun. 9, 123 (2018).
Chen, B. C. et al. Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science 346, 1257998 (2014).
Li, D. et al. Extended-resolution structured illumination imaging of endocytic and cytoskeletal dynamics. Science 349, aab3500 (2015).
Chen, B. et al. Resolution doubling in light-sheet microscopy via oblique plane structured illumination. Nat. Methods 19, 1419–1426 (2022).
Temma, K. et al. Selective-plane-activation structured illumination microscopy. Nat. Methods 21, 889–896 (2024).
Shi, Y. et al. Smart lattice light-sheet microscopy for imaging rare and complex cellular events. Nat. Methods 21, 301–310 (2024).
Weigert, M. et al. Content-aware image restoration: pushing the limits of fluorescence microscopy. Nat. Methods 15, 1090–1097 (2018).
Qiao, C. et al. Evaluation and development of deep neural networks for image super-resolution in optical microscopy. Nat. Methods 18, 194–202 (2021).
Wang, H. et al. Deep learning enables cross-modality super-resolution in fluorescence microscopy. Nat. Methods 16, 103–110 (2019).
Chen, J. et al. Three-dimensional residual channel attention networks denoise and sharpen fluorescence microscopy image volumes. Nat. Methods 18, 678–687 (2021).
Qiao, C. et al. 3D structured illumination microscopy via channel attention generative adversarial network. IEEE J. Sel. Top. Quantum Electron. 27, 1–11 (2021).
Zhao, Y. et al. Isotropic super-resolution light-sheet microscopy of dynamic intracellular structures at subsecond timescales. Nat. Methods 19, 359–369 (2022).
Park, H. et al. Deep learning enables reference-free isotropic super-resolution for volumetric fluorescence microscopy. Nat. Commun. 13, 3297 (2022).
Li, X. et al. Three-dimensional structured illumination microscopy with enhanced axial resolution. Nat. Biotechnol. 41, 1307–1319 (2023).
Ning, K. et al. Deep self-learning enables fast, high-fidelity isotropic resolution restoration for volumetric fluorescence microscopy. Light Sci. Appl. 12, 204 (2023).
Hoffman, D. P., Slavitt, I. & Fitzpatrick, C. A. The promise and peril of deep learning in microscopy. Nat. Methods 18, 131–132 (2021).
Laine, R. F., Arganda-Carreras, I., Henriques, R. & Jacquemet, G. Avoiding a replication crisis in deep-learning-based bioimage analysis. Nat. Methods 18, 1136–1144 (2021).
Finn, C., Abbeel, P. & Levine, S. Model-agnostic meta-learning for fast adaptation of deep networks. In Proc. 34th. International Conference on Machine Learning 1126–1135 (PMLR, 2017).
Kerepecky, T., Liu, J., Ng, X. W., Piston, D. W. & Kamilov, U. S. Dual-cycle: self-supervised dual-view fluorescence microscopy image reconstruction using cycleGAN. In ICASSP 2023 - 2023 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP) 1–5 (IEEE, 2023).
Wu, Y. et al. Reflective imaging improves spatiotemporal resolution and collection efficiency in light sheet microscopy. Nat. Commun. 8, 1452 (2017).
Preibisch, S. et al. Efficient Bayesian-based multiview deconvolution. Nat. Methods 11, 645–648 (2014).
Dong, L., Chang, Q., Ziwei, L. & Siwei, Z. Three-dimensional super-resolution light sheet microscopic imaging method and microscope. CN113917677B (2023).
Wu, Y. et al. Multiview confocal super-resolution microscopy. Nature 600, 279–284 (2021).
Hospedales, T., Antoniou, A., Micaelli, P. & Storkey, A. Meta-learning in neural networks: a survey. IEEE Trans. Pattern Anal. Mach. Intell. 44, 5149–5169 (2022).
Miller, A. L. The contractile ring. Curr. Biol. 21, R976–R978 (2011).
Kumar, A. et al. Using stage-and slit-scanning to improve contrast and optical sectioning in dual-view inverted light sheet microscopy (diSPIM). Biol. Bull. 231, 26–39 (2016).
Motta, P. M., Nottola, S. A., Makabe, S. & Heyn, R. Mitochondrial morphology in human fetal and adult female germ cells. Hum. Reprod. 15, 129–147 (2000).
Brukman, N. G., Uygur, B., Podbilewicz, B. & Chernomordik, L. V. How cells fuse. J. Cell Biol. 218, 1436–1451 (2019).
Guo, M. et al. Rapid image deconvolution and multiview fusion for optical microscopy. Nat. Biotechnol. 38, 1337–1346 (2020).
Valm, A. M. et al. Applying systems-level spectral imaging and analysis to reveal the organelle interactome. Nature 546, 162–167 (2017).
Ba, Q., Raghavan, G., Kiselyov, K. & Yang, G. Whole-cell scale dynamic organization of lysosomes revealed by spatial statistical analysis. Cell Rep. 23, 3591–3606 (2018).
Guo, Y. et al. Visualizing intracellular organelle and cytoskeletal interactions at nanoscale resolution on millisecond timescales. Cell 175, 1430–1442 e1417 (2018).
Wong, Y. C., Ysselstein, D. & Krainc, D. Mitochondria–lysosome contacts regulate mitochondrial fission via RAB7 GTP hydrolysis. Nature 554, 382–386 (2018).
Qiao, C. et al. Rationalized deep learning super-resolution microscopy for sustained live imaging of rapid subcellular processes. Nat. Biotechnol. 41, 367–377 (2023).
Dong, J. et al. Phase retrieval: from computational imaging to machine learning: a tutorial. IEEE Signal Process. Mag. 40, 45–57 (2023).
Bai, B. et al. Deep learning-enabled virtual histological staining of biological samples. Light Sci. Appl. 12, 57 (2023).
Rivenson, Y. et al. PhaseStain: the digital staining of label-free quantitative phase microscopy images using deep learning. Light Sci. Appl. 8, 1–11 (2019).
Mirza, M. & Osindero, S. Conditional generative adversarial nets. Preprint at https://arxiv.org/abs/1411.1784 (2014).
Ledig, C. et al. Photo-realistic single image super-resolution using a generative adversarial network. In Proc. 2017 IEEE Conference on Computer Vision and Pattern Recognition 4681–4690 (IEEE, 2017).
Zhang, Y. et al. Image super-resolution using very deep residual channel attention networks. In Proc. European Conference on Computer Vision (ECCV) 286–301 (IEEE, 2018).
Fuoli, D., Van Gool, L. & Timofte, R. Fourier space losses for efficient perceptual image super-resolution. In Proc. IEEE/CVF International Conference on Computer Vision 2360–2369 (IEEE, 2021).
Qiao, C. et al. Zero-shot learning enables instant denoising and super-resolution in optical fluorescence microscopy. Nat.Commun. 15, 4180 (2024).
Li, Y. et al. Incorporating the image formation process into deep learning improves network performance. Nat. Methods 19, 1427–1437 (2022).
Descloux, A., Grussmayer, K. S. & Radenovic, A. Parameter-free image resolution estimation based on decorrelation analysis. Nat. Methods 16, 918–924 (2019).
Otsu, N. A threshold selection method from gray-level histograms. Automatica 11, 23–27 (1975).
Wang, H. & Jiang, L. Transient expression and analysis of fluorescent reporter proteins in plant pollen tubes. Nat. Protoc. 6, 419–426 (2011).
Tse, Y. C. et al. Identification of multivesicular bodies as prevacuolar compartments in Nicotiana tabacum BY-2 cells[W]. Plant Cell 16, 672–693 (2004).
Brenner, S. The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974).
Köppen, M. et al. Cooperative regulation of AJM-1 controls junctional integrity in Caenorhabditis elegans epithelia. Nat. Cell Biol. 3, 983–991 (2001).
Li, Y. et al. The lysosomal membrane protein SCAV-3 maintains lysosome integrity and adult longevity. J. Cell Biol. 215, 167–185 (2016).
Chang, Q. & Yuhuan, L. BioSR for LLS-SIM. Zenodo https://doi.org/10.5281/zenodo.14322456 (2025).
Acknowledgements
The authors thank H. Wang for the donor pollen tubes and help in recording the growth process of them. This work was supported by grants from National Natural Science Foundation of China (32125024, 92254306, 32471517, 62088102, 62401156 and 62231018, to D.L., C.Q., Q.D. and Z.L.); the National Key R&D Program of China (2023YFC3402600 and 2023YFC3402602, to Q.D. and C.Q.); the Ministry of Science and Technology (2021YFA1300303 and 2020AA0105500, to D.L. and Q.D.); the Chinese Academy of Sciences (ZDBS-LY-SM004, to D.L.); Collaborative Research Fund of the Chinese Institute for Brain Research, Beijing (2021-NKX-XM-03, to Q.D.); China Postdoctoral Science Foundation (2022M721842 and 2023T160365, to C.Q.); the New Cornerstone Science Foundation (to D.L.); and the Shuimu Tsinghua Scholar Program (2022SM035, to C.Q.).
Author information
Authors and Affiliations
Contributions
D.L., C.Q. and Z.L. conceived the idea. D.L. and Q.D. supervised the research. D.L. and C.Q. designed the experiments. Yong Liu, S.Z., C.Q. and X.D. built and improved the microscope systems under the supervision of D.L. C.L., X.Y., J.G., W.F., X.W. and T.J. prepared samples and performed experiments. C.Q., Z.L., Z.W., Yuhuan Lin, Q.M. and Q.W. analyzed the data with conceptual advice from D.L. and C.Q. C.Q., Z.L., Z.W., Yuhuan Lin, Y.F. and W.X. composed the figures and videos. C.Q., Z.L. and D.L. wrote the paper with input from all authors. All authors discussed the results and commented on the paper.
Corresponding authors
Ethics declarations
Competing interests
D.L., C.Q., S.Z. and Z.L. have a pending patent on the presented frameworks. The other authors declare no competing interests.
Peer review
Peer review information
Nature Methods thanks Jonathan Ventura, Niall Geoghegan, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Rita Strack, in collaboration with the Nature Methods team. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Comparison of VSI-SR models with inputs/outputs (IO) of single slices or multiple slices.
a–c, Representative images (MIP) acquired and generated by LLSM, conventional LLS-SIM, and VSI-SR models with single-slice or multi-slices IO, which were trained by datasets of peroxisomes (a), lysosomes (b) and ER (c), respectively. Scale bar, 5 μm (full FOV images), 1 μm (horizontal bars in magnified regions), and 3 μm (vertical bars in magnified regions). d,e, Box-and-whisker plots of the PSNR (d) and SSIM (e) for images reconstructed by VSI-SR models using the dataset of peroxisome, ER and lysosomes (n = 20 for each sample). Central line, medians; limits, 75% and 25%; whiskers, maximum and minimum.
Extended Data Fig. 2 Comparison of VSI-SR models constituted with 2D and 3D convolutional architecture.
a–c, Representative images (MIP) acquired and generated by LLSM, conventional LLS-SIM, and VSI-SR models constructed with 2D and 3D convolutional architectures, which were trained by datasets of peroxisomes (a), lysosomes (b), and F-actin (c), respectively. Scale bar, 5 μm (full FOV images), 1 μm (horizontal bars in magnified regions), and 3 μm (vertical bars in magnified regions). d,e, Box-and-whisker plots of the PSNR (d) and SSIM (e) for images reconstructed by VSI-SR models (n = 20 for each specimen). Central line, medians; limits, 75% and 25%; whiskers, maximum and minimum.
Extended Data Fig. 3 Comparison of coherent LLS-SIM-based VSI-SR scheme with photo-reassignment-based laterally isotropic SR reconstruction.
a, In the training phase, raw SIM data were first collected with 6-phase illumination and 3-phase lattice illumination by each approach, and then reconstructed by applying the digital photo-reassignment algorithm and SR-SIM reconstruction algorithm, respectively, to generate 1D SR training data. The detection NA was set to 1.0 for both approaches, while the effective excitation NAs of structure illumination generation were set to 0.4 for LLS-SIM and 1.0 for incoherent photo-reassignment. Although using lower excitation NA, LLS-SIM achieved larger spatial frequency extension and finer structural details compared with those of photo-reassignment. b, In the inference phase, different neural network models, that is, RCAN and VSI-SR, were trained, which were both used to predict 1D SR outputs along multiple orientations from the diffraction-limited inputs. The 1D SR outputs were further assembled to produce the lateral isotropic reconstruction using Fourier projection or a generalized Wiener filter. The MIP of spatial profiles and corresponding Fourier spectrum are presented for each step, and a magnified inset and the line profile are displayed to demonstrate the resolution comparison of the two methods. Scale bar, 1 μm (excitation patterns), 5 μm (full FOV images) and 2 μm (magnified regions).
Extended Data Fig. 4
Meta-training workflow of the Meta-VSI-SR model.
Extended Data Fig. 5 Comparison of RL-DFN with existing axial resolution enhancement methods.
a-d, Representative x-y slices and x-z slices of simulated spherical shells (a), simulated tubular structures (b), experimentally acquired MTs (c), and experimentally acquired ER (d), reconstructed by multiple axial resolution enhancement methods, including ID-Net19, DL-ARE20, Self-Net21, multi-view RL deconvolution27, and the proposed RL-DFN. Anisotropic inputs, 3D rendering of the inputs, and the GT images are shown for reference. f,g, Statistical comparisons of ID-Net, DL-ARE, Self-Net, multi-view RL deconvolution, and RL-DFN in terms of PSNR and SSIM (n = 100) using simulated data of spherical shells (f) and tubular structures (g). Central line, medians; limits, 75% and 25%; whiskers, maximum and minimum. Scale bar, 2 μm (3D rendering images in a-d), and 1 μm (slice-images in a-d).
Extended Data Fig. 6 Comparison of RL-DFN with its variant architectures.
a–c, Schematic of three compared network architectures, that is, RL-DFN (a), DFN (b), and RLN-DFN (c). Details of the RL deconvolution module and rotation sectioning module are schematically illustrated. d-g, Reconstruction results of F-actin images using multi-view RL deconvolution (d), RLN-DFN (e), DFN (f), and RL-DFN (g). x-y MIPs and two different x-z slices along the dashed arrows are shown. Scale bars, 3 μm (full FOV images in d-g) and 1 μm (magnified regions in d-g).
Extended Data Fig. 7 Pipeline of Meta-rLLS-VSIM reconstruction.
a, Illustration of the imaging process of reflective lattice light-sheet microscopy (rLLSM). b, Step-by-step processing procedures for laterally isotropic SR reconstruction from wide-field input by VSI-SR models. c, Step-by-step processing procedures for isotropic 3D SR reconstruction by RL-DFN from the anisotropic SR views.
Extended Data Fig. 8 Meta-rLLS-VSIM reconstruction of simulated neuronal structures.
a-e, Representative 3D neuron images (max intensity projections) imaged by conventional lattice light-sheet microscopy (a), Meta-LLS-VSIM with laterally isotropic super-resolution (b), Meta-LLS-VSIM after rotation and view splitting (c), Meta-rLLS-VSIM with near-isotropic 3D super-resolution (d) and corresponding ground-truth (e). The coordinates are labeled in the detection view in a, b and in the sample view in c-e. Scale bar, 5 μm (a-e), 1 μm (zoom-in regions of c-e).
Extended Data Fig. 9 Resolution characterization for LLSM, Meta-LLS-VSIM, and Meta-rLLS-VSIM.
a–c, LLSM (a), Meta-LLS-VSIM (b), and Meta-rLLS-VSIM (c) images (MIP) of beads shown in the x-y and x-z plane and corresponding line profiles indicated by yellow arrows in both lateral (n = 12) and axial (n = 9). The FWHM measured from the averaged profiles indicates that the resolution of Meta-rLLS-VSIM reaches 119 nm in lateral and 157 nm in axial. Central points, mean; whiskers, minimum and maximum. Scale bar, 1 μm.
Extended Data Fig. 10 Rare biological behaviors of Lyso-Mito interactions revealed by Meta-rLLS-VSIM imaging.
a,b, Time-lapse max intensity projections (upper row) and surface rendering (lower row) of mitochondria and lysosomes showcasing that a moving Lyso-generated mechanical force to induce mitochondrial fission by pulling out (a) or pushing against (b) Mito membrane. c, Time-lapse max intensity projections (upper row) and surface rendering (lower row) of mitochondria and lysosomes showcasing a mitochondrion reformed itself from tubular into doughnut-like structure that corralled the Lyso to regulate its movement. Scale bar, 2 μm.
Supplementary information
Supplementary Video 1
Demonstration of online finetuning of the Meta-VSI-SR model to achieve quick adaptation to a new biological structure. After working on the developed graphical user interface to select three ROIs of the target biological sample, an automatic streamline including data acquisition, LLS-SIM SR image reconstruction and finetuning of the Meta-VSI-SR was produced. As the finetuning goes on, the reconstruction performance of the meta-model was substantially improved within few iterations. All of above operations typically take less than 2.5 min as shown in this video.
Supplementary Video 2
Entire process of contractile ring formation, contraction and disassembly during mitosis imaged by Meta-LLS-VSIM. Three-color Meta-LLS-VSIM imaging for 357 time points at 8-s volume intervals of a HeLa cell stably expressing Lifeact–mEmerald (F-actin in green), KDEL–mCherry (ER in magenta) and Lamp1–Halo (lysosomes in yellow).
Supplementary Video 3
The dynamics of mitochondria in a mouse embryo imaged by Meta-LLS-VSIM. Meta-LLS-VSIM imaging in the rolling-shutter confocal slit-scan mode of a developing mouse embryo with Mito labeled by TOMM20–mEmerald across a thick area of 95 × 95 × 96 μm3 for 100 time points at 30-s intervals. The trajectory of Mito is delineated.
Supplementary Video 4
The spatiotemporal dynamics of cytoskeleton during growing of pollen tubes. LLSM and Meta-LLS-VSIM recording a growing pollen tube labeled with Lifeact–GFP at a high speed of 4.125 Hz for 1,000 time points.
Supplementary Video 5
Plasma membrane fusion process in C. elegans embryo development by two-color Meta-LLS-VSIM. Two-color volumetric recording of the epithelial cell fusion process in a C. elegans embryo with apical junction (cyan) and lysosomes (orange) labeled by Meta-LLS-VSIM with a time duration of 66 min at 1-min intervals.
Supplementary Video 6
Long-term 4D SR imaging of the whole cell by Meta-rLLS-VSIM. Three-color, long-term, near-isotropic SR 4D imaging of a Cos-7 cell transferred with GFP–SKL (peroxisomes in cyan), ER–mCherry (ER in magenta) and Lyso–Halo (lysosomes in yellow) for 400 time points at 12-s intervals by Meta-rLLS-VSIM.
Supplementary Video 7
Illustration of near-isotropic SR imaging by dual-view fusion in Meta-rLLS-VSIM. Near-isotropic 3D SR reconstruction from dual-view volumes by Meta-rLLS-VSIM of a Cos-7 cell transferred with GFP–SKL (peroxisomes in cyan), ER–mCherry (ER in magenta) and Lyso–Halo (lysosomes in yellow) for 721 time points at a speed of 8 s per three-color whole-cell volume.
Supplementary Video 8
Photobleaching and phototoxicity comparison between conventional LLS-SIM and Meta-rLLS-VSIM. Left half, Normalized intensity curves of the three channels (top row), averaged raw LLS-SIM images (middle row), and conventional LLS-SIM reconstruction (bottom row) for a video of a Cos-7 cell transferred with GFP–SKL (peroxisomes in cyan), ER–mCherry (ER in magenta) and Lyso–Halo (lysosomes in yellow). Right half, Normalized intensity curves (top row), view-A of raw rLLSM images (middle row) and Meta-rLLS-VSIM reconstruction (bottom row) for a video of another Cos-7 cell with the same labeling.
Supplementary Video 9
Long-term 4D subcellular imaging of multi-organelles and cytoskeleton. Meta-rLLS-VSIM records the spatiotemporal coordination and interactions between multiple organelles and cytoskeleton in a Cos-7 cell labeled with 3×mEmerald–Ensconsin (microtubules in cyan), mCherry–SKL (peroxisomes in magenta) and Lamp1–Halo (lysosomes in yellow) for 812 time points at a speed of 8 s per three-color cell volume.
Supplementary Video 10
Rare biological behaviors of Lyso-Mito interactions revealed by Meta-rLLS-VSIM imaging. Meta-rLLS-VSIM records several rare biological events showcasing a mitochondrion reformed itself from tubular into doughnut-like structure that corralled the Lyso to regulate its movement, and a moving Lyso-generated mechanical force to induce Mito fission by pulling out or pushing against Mito membrane, which were observed in Cos-7 cells with Mito (green) and lysosomes (magenta) labeled by TOMM20–mEmerald and Lamp1–Halo.
Source data
Source Data Fig. 2
Statistical Source Data.
Source Data Fig. 3
Statistical Source Data.
Source Data Fig. 4
Statistical Source Data.
Source Data Fig. 5
Statistical Source Data.
Source Data Extended Data Fig. 1
Statistical Source Data.
Source Data Extended Data Fig. 2
Statistical Source Data.
Source Data Extended Data Fig. 5
Statistical Source Data.
Source Data Extended Data Fig. 9
Statistical Source Data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Qiao, C., Li, Z., Wang, Z. et al. Fast-adaptive super-resolution lattice light-sheet microscopy for rapid, long-term, near-isotropic subcellular imaging. Nat Methods 22, 1059–1069 (2025). https://doi.org/10.1038/s41592-025-02678-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-025-02678-3