Abstract
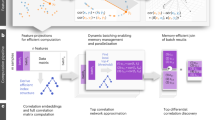
With the advent of multiomics, software capable of multidimensional enrichment analysis has become increasingly crucial for uncovering gene set variations in biological processes and disease pathways. This is essential for elucidating disease mechanisms and identifying potential therapeutic targets. clusterProfiler stands out for its comprehensive utilization of databases and advanced visualization features. Importantly, clusterProfiler supports various biological knowledge, including Gene Ontology and Kyoto Encyclopedia of Genes and Genomes, through performing over-representation and gene set enrichment analyses. A key feature is that clusterProfiler allows users to choose from various graphical outputs to visualize results, enhancing interpretability. This protocol describes innovative ways in which clusterProfiler has been used for integrating metabolomics and metagenomics analyses, identifying and characterizing transcription factors under stress conditions, and annotating cells in single-cell studies. In all cases, the computational steps can be completed within ~2 min. clusterProfiler is released through the Bioconductor project and can be accessed via https://bioconductor.org/packages/clusterProfiler/.
Key points
-
clusterProfiler is a software package for characterizing and interpreting omics data. Functional enrichment can be achieved using either over-representation or gene set enrichment analyses; it supports the use of a variety of databases, e.g., Gene Ontology and Kyoto Encyclopedia of Genes and Genomes.
-
Three procedures show specific R commands for example applications asking different research questions and having different graphical outputs. Advice is provided on how to modify the procedures for other applications.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
27,99 € / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
269,00 € per year
only 22,42 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
For the integrated analysis of metabolomics and metagenomics, the original metagenomic gene expression data and corresponding metadata, along with metabolomic metabolite expression profiles, were obtained from the supplementary materials of ref. 44. The data can be accessed through PubMed Central at https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6342642. For the transcriptomics analysis of PE, we obtained the raw FASTQ sequencing data from the CNGBdb database (https://db.cngb.org/search/project/CNP0002243/) and conducted alignment and quantification analyses to determine the expression levels of individual genes. In Procedure 3, we acquired Illumina NextSeq 500 sequencing data for 2,700 PBMCs from 10X Genomics (https://cf.10xgenomics.com/samples/cell/pbmc3k/pbmc3k_filtered_gene_bc_matrices.tar.gz). Source data are provided with this paper.
Code availability
The original data, processed data and source code, including those for processing the original data and demonstrated in the protocol, are all deposited in the GitHub repository, https://github.com/YuLab-SMU/clusterProfiler_protocol/.
References
Paczkowska, M. et al. Integrative pathway enrichment analysis of multivariate omics data. Nat. Commun. 11, 735 (2020).
Boyle, E. I. et al. GO:: TermFinder—open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 20, 3710–3715 (2004).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Xie, C., Jauhari, S. & Mora, A. Popularity and performance of bioinformatics software: the case of gene set analysis. BMC Bioinforma. 22, 191 (2021).
Liu, X., Xu, K., Tao, X., Bo, X. & Chang, C. EnrichMiner: a biologist-oriented web server for mining biological insights from functional enrichment analysis results. Preprint at bioRxiv https://doi.org/10.1101/2023.07.12.548786 (2023).
Zhao, K. & Rhee, S. Y. Interpreting omics data with pathway enrichment analysis. Trends Genet. 39, 308–319 (2023).
Yu, G., Wang, L.-G., Han, Y. & He, Q.-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012).
Wu, T. et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation 2, 100141 (2021).
Ding, J. & Zhang, Y. Analysis of key GO terms and KEGG pathways associated with carcinogenic chemicals. Comb. Chem. High. Throughput Screen. 20, 861–871 (2017).
Li, Z. et al. Prediction and analysis of retinoblastoma related genes through gene ontology and KEGG. Biomed. Res. Int. 2013, 304029 (2013).
Morgan, M. Sequences, Genomes, and Genes in R/Bioconductor (2013); https://www.ebi.ac.uk/sites/ebi.ac.uk/files/content.ebi.ac.uk/materials/2013/131021_HTS/genesandgenomes.pdf
Abromeit, F., Fäth, C. & Glaser, L. Annohub–annotation metadata for linked data applications. In Proc. 7th Workshop on Linked Data in Linguistics (LDL-2020) 36–44 (2020).
Liberzon, A. et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740 (2011).
Martens, M. et al. WikiPathways: connecting communities. Nucleic Acids Res. 49, D613–D621 (2021).
Conesa, A. et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676 (2005).
Li, S. & Xu, J. KAAS: a keyword-aware attention abstractive summarization model for scientific articles. In International Conference on Database Systems for Advanced Applications 263–271 (Springer, 2022).
Yu, G. & He, Q.-Y. ReactomePA: an R/Bioconductor package for reactome pathway analysis and visualization. Mol. Biosyst. 12, 477–479 (2016).
Yu, G. Using meshes for MeSH term enrichment and semantic analyses. Bioinformatics 34, 3766–3767 (2018).
Yu, G., Wang, L.-G., Yan, G.-R. & He, Q.-Y. DOSE: an R/Bioconductor package for disease ontology semantic and enrichment analysis. Bioinformatics 31, 608–609 (2014).
Yu, G. et al. GOSemSim: an R package for measuring semantic similarity among GO terms and gene products. Bioinformatics 26, 976–978 (2010).
Yu, G., Wang, L.-G. & He, Q.-Y. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31, 2382–2383 (2015).
Wang, Q. et al. Exploring epigenomic datasets by ChIPseeker. Curr. Protoc. 2, e585 (2022).
Wickham, H. et al. Welcome to the Tidyverse. J. Open Source Softw. 4, 1686 (2019).
Yu, G. Enrichplot: visualization of functional enrichment result. R Package Version 1 (2021).
Wickham, H. in ggplot2 189–201 (Springer, 2016).
Merico, D., Isserlin, R., Stueker, O., Emili, A. & Bader, G. D. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PloS ONE 5, e13984 (2010).
Hoover, A. R. et al. Single‐cell RNA sequencing reveals localized tumour ablation and intratumoural immunostimulant delivery potentiate T cell mediated tumour killing. Clin. Transl. Med. 12, e937 (2022).
Tan, Z. et al. HSPB8 is a potential prognostic biomarker that correlates with immune cell infiltration in bladder cancer. Front. Genet. 13, 804858 (2022).
Liu, J. et al. Eleven genes associated with progression and prognosis of endometrial cancer (EC) identified by comprehensive bioinformatics analysis. Cancer Cell Int. 19, 136 (2019).
Kuleshov, M. V. et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 (2016).
Zhang, B., Kirov, S. & Snoddy, J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 33, W741–W748 (2005).
Gennady Korotkevich et al. Fast gene set enrichment analysis. Preprint at bioRxiv https://doi.org/10.1101/060012 (2021).
Falcon, S. & Gentleman, R. Using GOstats to test gene lists for GO term association. Bioinformatics 23, 257–258 (2007).
Kolberg, L., Raudvere, U., Kuzmin, I., Vilo, J. & Peterson, H. gprofiler2—an R package for gene list functional enrichment analysis and namespace conversion toolset g:Profiler. F1000Research 9, ELIXIR-709 (2020).
Dennis, G. et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4, R60 (2003).
Zhou, Y. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523 (2019).
Treveil, A. et al. ViralLink: an integrated workflow to investigate the effect of SARS-CoV-2 on intracellular signalling and regulatory pathways. PLOS Comput. Biol. 17, e1008685 (2021).
Jiang, A., Lehnert, K., You, L. & Snell, R. G. ICARUS, an interactive web server for single cell RNA-seq analysis. Nucleic Acids Res. 50, W427–W433 (2022).
Liu, J., Erenpreisa, J. & Sikora, E. Polyploid giant cancer cells: an emerging new field of cancer biology. Semin. Cancer Biol. 81, 1–4 (2022).
Cui, G. et al. A carbon–nitrogen negative feedback loop underlies the repeated evolution of cnidarian–Symbiodiniaceae symbioses. Nat. Commun. 14, 6949 (2023).
Nie, M. et al. Evolutionary metabolic landscape from preneoplasia to invasive lung adenocarcinoma. Nat. Commun. 12, 6479 (2021).
Xu, S. et al. MicrobiotaProcess: a comprehensive R package for deep mining microbiome. Innovation 4, 100388 (2023).
Chen, A. et al. Single-cell spatial transcriptome reveals cell-type organization in the macaque cortex. Cell 186, 3726–3743.e24 (2023).
Franzosa, E. A. et al. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat. Microbiol. 4, 293–305 (2019).
Nie, Y. et al. Innovations and stepwise evolution of CBFs/DREB1s and their regulatory networks in angiosperms. J. Integr. Plant Biol. 64, 2111–2125 (2022).
Vandereyken, K., Sifrim, A., Thienpont, B. & Voet, T. Methods and applications for single-cell and spatial multi-omics. Nat. Rev. Genet. 24, 494–515 (2023).
Castanza, A. S. et al. Extending support for mouse data in the Molecular Signatures Database (MSigDB). Nat. Methods 20, 1619–1620 (2023).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2009).
Fernandes, A. D., Macklaim, J. M., Linn, T. G., Reid, G. & Gloor, G. B. ANOVA-like differential expression (ALDEx) analysis for mixed population RNA-seq. PLoS ONE 8, e67019 (2013).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000).
Wishart, D. S. et al. HMDB: the human metabolome database. Nucleic Acids Res. 35, D521–D526 (2007).
Jewison, T. et al. SMPDB 2.0: big improvements to the small molecule pathway database. Nucleic Acids Res. 42, D478–D484 (2013).
Jin, J. et al. PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 45, D1040–D1045 (2016).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Zheng, G. X. Y. et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017).
McCarthy, D. J., Campbell, K. R., Lun, A. T. L. & Wills, Q. F. Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics 33, 1179–1186 (2017).
Lun, A., McCarthy, D. & Marioni, J. A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Research 5, 2122 (2016).
Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018).
Cortal, A., Martignetti, L., Six, E. & Rausell, A. Gene signature extraction and cell identity recognition at the single-cell level with Cell-ID. Nat. Biotechnol. 39, 1095–1102 (2021).
Wadi, L., Meyer, M., Weiser, J., Stein, L. D. & Reimand, J. Impact of outdated gene annotations on pathway enrichment analysis. Nat. Methods 13, 705–706 (2016).
Pritykin, Y., Ghersi, D. & Singh, M. Genome-wide detection and analysis of multifunctional genes. PLoS Comput. Biol. 11, e1004467 (2015).
Tian, F., Yang, D.-C., Meng, Y.-Q., Jin, J. & Gao, G. PlantRegMap: charting functional regulatory maps in plants. Nucleic Acids Res. 48, D1104–D1113 (2019).
Anders, S. et al. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat. Protoc. 8, 1765–1786 (2013).
Satija, R., Farrell, J. A., Gennert, D., Schier, A. F. & Regev, A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502 (2015).
Buie, M. J. et al. Global hospitalization trends for Crohn’s Disease and ulcerative colitis in the 21st century: a systematic review with temporal analyses. Clin. Gastroenterol. Hepatol. 21, 2211–2221 (2023).
Scoville, E. A. et al. Alterations in lipid, amino acid, and energy metabolism distinguish Crohn’s disease from ulcerative colitis and control subjects by serum metabolomic profiling. Metabolomics 14, 17 (2018).
Duboc, H. et al. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut 62, 531 (2013).
Moura, J. C. M. S., Bonine, C. A. V., De Oliveira Fernandes Viana, J., Dornelas, M. C. & Mazzafera, P. Abiotic and biotic stresses and changes in the lignin content and composition in plants. J. Integr. Plant Biol. 52, 360–376 (2010).
Lv, K. et al. Overexpression of an AP2/ERF family gene, BpERF13, in birch enhances cold tolerance through upregulating CBF genes and mitigating reactive oxygen species. Plant Sci. 292, 110375 (2020).
Guo, Z. et al. Genome-wide analysis of the rhododendron AP2/ERF gene family: identification and expression profiles in response to cold, salt and drought stress. Plants 12, 994 (2023).
Ding, Y. & Yang, S. Surviving and thriving: how plants perceive and respond to temperature stress. Dev. Cell 57, 947–958 (2022).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Truong, D. T. et al. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat. Methods 12, 902–903 (2015).
Franzosa, E. A. et al. Species-level functional profiling of metagenomes and metatranscriptomes. Nat. Methods 15, 962–968 (2018).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Trapnell, C., Pachter, L. & Salzberg, S. L. TopHat: discovering splice junctions with RNA-seq. Bioinformatics 25, 1105–1111 (2009).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Kim, D., Paggi, J. M., Park, C., Bennett, C. & Salzberg, S. L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915 (2019).
Ghosh, S. & Chan, C.-K. K. in Plant Bioinformatics (ed. Edwards, D.) vol. 1374, 339–361 (Springer, 2016).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinforma. 12, 323 (2011).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Anders, S., Pyl, P. T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Consortium, G. O. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 32, D258–D261 (2004).
Schriml, L. M. et al. Disease Ontology: a backbone for disease semantic integration. Nucleic Acids Res. 40, D940–D946 (2011).
Piñero, J. et al. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 45, D833–D839 (2016).
Repana, D. et al. The Network of Cancer Genes (NCG): a comprehensive catalogue of known and candidate cancer genes from cancer sequencing screens. Genome Biol. 20, 1 (2019).
Fabregat, A. et al. The reactome pathway knowledgebase. Nucleic Acids Res. 46, D649–D655 (2017).
Acknowledgements
This study was supported by the National Natural Science Foundation of China (32270677). We appreciate the feedback and support from clusterProfiler users.
Author information
Authors and Affiliations
Contributions
S.X., E.H. and Y.C. wrote the main manuscript and discussed the cases. S.X. and E.H. improved the code. Z.X. and X.L. conducted the pipeline and analyzed the results. L.Z., W.T., Q.W. and B.L. edited the paper for improvement. R.W., W.X., T.W. and L.X. reviewed the paper. G.Y. supervised the project, conducted the analysis and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks Juri Reimand, Jianguo Xia and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Yu, G. et al. OMICS 16, 284–287 (2012): https://doi.org/10.1089/omi.2011.0118
Wu, T. et al. Innovation 2, 100141 (2021): https://doi.org/10.1016/j.xinn.2021.100141
Ne, M. et al. Nat. Commun. 12, 6479 (2021): https://doi.org/10.1038/s41467-021-26685-y
Alexandre, P. A. et al. Genome Biol. 22, 273 (2021): https://doi.org/10.1186/s13059-021-02489-7
Sankowski, R. et al. Nat. Med. 30, 186–198 (2024): https://doi.org/10.1038/s41591-023-02673-1
Supplementary information
Supplementary Information
Supplementary Notes 1 and 2.
Supplementary Table 1
Comparisons of clusterProfiler with other tools for enrichment analysis
Source data
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xu, S., Hu, E., Cai, Y. et al. Using clusterProfiler to characterize multiomics data. Nat Protoc 19, 3292–3320 (2024). https://doi.org/10.1038/s41596-024-01020-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-024-01020-z
This article is cited by
-
Multi-tool copy number detection highlights common body size-associated variants in miniature pig breeds from different geographical regions
BMC Genomics (2025)
-
Biological characterization and clinical significance of cuproptosis-related genes in lung adenocarcinoma
BMC Pulmonary Medicine (2025)
-
Chemical reprogramming culture for the expansion of salivary gland epithelial basal progenitor cells
Stem Cell Research & Therapy (2025)
-
Enhancing TREM2 expression activates microglia and modestly mitigates tau pathology and neurodegeneration
Journal of Neuroinflammation (2025)
-
Dysregulation of transposable elements and PIWI-interacting RNAs in myelodysplastic neoplasms
Biomarker Research (2025)