Abstract
African lovebirds are popular pet parrots commonly hybridized by breeders leading to genetic admixture. Trade escapees and land use change have now led lovebirds to genetically admix in the wild. Sampling origin is therefore of utmost importance when deriving genetic data from lovebird species to reconstruct phylogeny or historical demographic events as the inclusion of taxa of hybrid origin is a source of spurious results. Here we present complete genomes of all nine currently recognized lovebird species. Each species is represented by an archival geo-referenced individual collected within their natural range.
Similar content being viewed by others
Background & Summary
Lovebirds are a group of small sub-Sahara African parrots that belong to the genus Agapornis. Currently nine species are recognized by avian checklists and the International Union of Conservation in Nature IUCN1. Multiple aspects of the phylogeny and historical biogeography of the genus remain poorly understood and genomics presents an opportunity to gain insights into a number of research questions regarding evolution and approaches to conservation2. Past phylogenetic studies based on genetics have either not included all known species3, relied on only one or two molecular markers3,4 or relied on samples from captive-bred individuals5,6,7,8,9,10. The most recent phylogenetics study8 included archival individuals (i.e. specimens held in museum collections) from the wild but for most species the ancestral heritage and precise geographical provenance of the specimens used, are unknown, except three A. canus. The use of specimens from captivity or with uncertain provenance is problematic given that lovebirds hybridize readily, and crossbreeding has been a common practice in some captive collections. Naturalised populations of lovebirds have also been established from introductions outside their natural ranges in multiple localities. As a result, cryptic hybrids may exist within captive and introduced populations11,12,13.
Here we present full genome data of all nine species of lovebird derived from archival geo-referenced individuals collected within the natural historical range of each species with the aim of presenting sequences of individuals with a high certainty of being free from artificial hybridisation. Each genome is linked to a voucher specimen housed in a recognized natural history collection (Table 1).
Methods
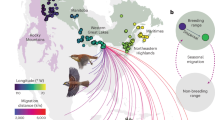
A toepad from representative wild-caught individuals (collected from within their natural range2, Fig. 1) of each species was sampled from the Academy of Natural Sciences of Drexel University (ANSP) and the Field Museum of Natural History (FMNH). To keep the possibility of hybrid material due to past human interference minimal, we took the following into account when selecting samples. (1) Locality (Fig. 1) - samples were collected from sites that were remote and geographically separate from known centres of human habitation where the risk of hybridisation as result of escaped captive birds is low. Samples were also collected from the core of species range and not from possible hybrid zones or areas where the distribution of species overlap. Agapornis swindernianus and A. pullarius distributions do overlap (Fig. 1), however, these two species are ecologically separated and are not known to hybridize12 (2) Age - most samples (apart from A. canus, collection in the 1996) were collected before the peak of exploitation and mass trade of lovebirds which occurred in 1980s. (3) Phenotypic characters – the specimens included in our study did not show phenotypic characteristics such as unusual plumage characters that might indicate hybridisation. DNA extraction was performed on the toe pad tissue using the Qiagen DNeasy genomic extraction kit (QIAGEN N.V.) using the standard protocol. Paired-end sequencing libraries were constructed using the Illumina TruSeq kit according to the manufacturer’s instructions. The library was sequenced in 50–100x coverage on an Illumina Hi-Seq platform in paired-end (haploid), 2 × 150 bp format by GENEWIZ (Azenta Life Sciences). The resulting fastq files were trimmed of adapter/primer sequence and low-quality regions using Trimmomatic v0.3314. The trimmed sequences were then assembled using SPAdes v2.515 followed by a finishing step performed in Zanfona16. Zanfona16 uses a series of related species that function as references for each other. The genome was assembled on scaffold-level. There was no single alignment where reads from one species were mapped to the chromosomes of another species.
Specimen sample locations and BirdLife distribution for each of the 9 species1.
Data Records
All raw reads and assembled genomes have been deposited and are available on GenBank (National Center for Biotechnology Information NCBI, Table 2).
Technical Validation
The specimens selected for sequencing were collected from the wild and accompanied by precise geo-referenced information (including precise Latitude and Longitude to the nearest minute). We used the default parameters for SPAdes15, Trimmomatic14, and Zanfona16 to assess the validity and quality of the data. Decontamination was performed using FSCR-gx17 and the NCBI’s in-house libraries. No gap closing was performed.
Code availability
Publicly available codes used are listed in Methods. No custom code was used for this study.
References
BirdLife International. IUCN Red List for birds. Downloaded from https://datazone.birdlife.org on 28/02/2025 (2025).
Dueker, S. et al. Conservation status and threats to lovebirds: Knowledge gaps and research priorities. Ostrich. 94(1), 1–27, https://doi.org/10.2989/00306525.2023.2206674 (2023).
Eberhard, J. R. Evolution of nest-building behavior in agapornis parrots. The Auk. 115(2), 455–464, https://doi.org/10.2307/4089204 (1998).
Manegold, A. & Podsiadlowski, L. On the systematic position of the black-collared lovebird agapornis swindernianus (agapornithinae, psittaciformes). Journal of Ornithology. 155(3), 581–589, https://doi.org/10.1007/s10336-013-1039-z (2014).
Chen, Y. X., Hou, S. L., Zhou, Y. W. & Huang, Y. L. Characterization of the complete mitochondrial genome of agapornis personatus and its phylogenetic analysis. Mitochondrial DNA Part B. 4(2), 3772–3773, https://doi.org/10.1080/23802359.2019.1681325 (2019a).
Chen, Y. X., Huang, Y. L., Liu, J. Q., Zhou, Y. W. & Hou, S. L. Characterization and phylogenetic relationship of the complete mitochondrial genome of black-cheeked lovebird, agapornis nigrigenis. Mitochondrial DNA Part B. 4(2), 3589–3590, https://doi.org/10.1080/23802359.2019.1677195 (2019b).
Chen, Y. X., Zhou, Y. W., Hou, S. L. & Huang, Y. L. Complete mitochondrial genome of agapornis lilianae (psittaciformes: Psittacidae), with its phylogenetic analysis. Mitochondrial DNA Part B. 4(2), 3536–3537, https://doi.org/10.1080/23802359.2019.1675552 (2019c).
Huynh, S., Cloutier, A. & Sin, S. Y. W. Museomics and phylogenomics of lovebirds (psittaciformes, psittaculidae, agapornis) using low-coverage whole-genome sequencing. Molecular Phylogenetics and Evolution. 185, 107822, https://doi.org/10.1016/j.ympev.2023.107822 (2023).
Liu, H., Jin, K. & Li, L. The complete mitochondrial genome of the fischer’s lovebird agapornis fischeri (psittaciformes: Psittacidae). Mitochondrial DNA Part B. 4(1), 1217–1218, https://doi.org/10.1080/23802359.2019.1591186 (2019).
Van der Zwan, H., Van der Westhuizen, F., Visser, C. & Van der Sluis, R. Draft de novo genome sequence of agapornis roseicollis for application in avian breeding. Animal biotechnology. 29(4), 241–246, https://doi.org/10.1080/10495398.2017.1367692 (2018).
Lantermann, W. Verbreitung und status der ostafrikanischen papageien agapornis personatus reichenow, 1887 und agapornis fischeri reichenow, 1887. Bonner zoologische Beiträge. 52(1/2), 95–100, https://doi.org/10.5040/9781472927002.0020 (2004).
McCarthy, E. M. Handbook of avian hybrids of the world. (Oxford university press, 2006).
Mori, E. et al. Lovebirds in the air: Trade patterns, establishment success and niche shifts of agapornis parrots within their non-native range. Biological Invasions. 1–15, https://doi.org/10.1007/s10530-019-02100-y (2019).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 30(15), 2114–20, https://doi.org/10.1093/bioinformatics/btu170 (2014).
Bankevich, A. et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19(5), 455–77, https://doi.org/10.1089/cmb.2012.0021 (2012).
Kieras, M. Zanfona, a genome finishing process for use with paired-end short reads. https://github.com/zanfona734/zanfona (2021).
Astashyn, A. et al. Rapid and sensitive detection of genome contamination at scale with FCS-GX. Genome Biol. 25, 60, https://doi.org/10.5281/zenodo.10651084 (2024).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRP540681 (2024).
NCBI GenBank https://identifiers.org/ncbi/insdc.gca:GCA_036873685.1 (2024).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRP455935 (2023).
NCBI GenBank https://identifiers.org/ncbi/insdc.gca:GCA_034782895.1 (2023).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRP462750 (2023).
NCBI GenBank https://identifiers.org/ncbi/insdc.gca:GCA_035222285.1 (2023).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRP460808 (2023).
NCBI GenBank https://identifiers.org/ncbi/insdc.gca:GCA_035041115.1 (2023).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRP455936 (2023).
NCBI GenBank https://identifiers.org/ncbi/insdc.gca:GCA_034783155.1 (2023).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRP455938 (2023).
NCBI GenBank https://identifiers.org/ncbi/insdc.gca:GCA_034783915.1 (2023).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRP292226 (2021).
NCBI GenBank https://identifiers.org/ncbi/insdc.gca:GCA_024337125.1 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRP455914 (2023).
NCBI GenBank https://identifiers.org/ncbi/insdc.gca:GCA_034783435.1 (2023).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRP455937 (2023).
NCBI GenBank https://identifiers.org/ncbi/insdc.gca:GCA_034783195.1 (2023).
Acknowledgements
Funding was provided by Iridian Genomes, grant# IRGEN_RG_2021–1345. Sascha Dueker and Rowan Martin are supported by the World Parrot Trust.
Author information
Authors and Affiliations
Contributions
All authors discussed the results and contributed to the final manuscript. Taylor Hains and Sascha Dueker selected specimens for inclusion in the study. Stacy Pirro carried DNA extraction and sequencing. Sascha Dueker wrote the manuscript with support from Sandi Willows-Munro and Rowan Martin.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Dueker, S., Martin, R., Hains, T. et al. Full genomes of all nine currently recognized lovebird species (genus Agapornis) sampled from wild populations. Sci Data 12, 1057 (2025). https://doi.org/10.1038/s41597-025-05316-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-025-05316-x