Abstract
Studies have shown that acute exercise alters the expression of circulating neutrophil and peripheral blood mononuclear cell (PBMC) genes, affecting a wide range of processes from cellular growth to the promotion of inflammatory and anti-inflammatory responses. In this study, we identified differential gene expression associated with acute exercise and PBMC in puberty children through the GEO database. We utilized bioinformatics technology to analyze the impact of acute exercise on PBMC and identified key genes involving the puberty children. Combining two datasets of gene expression, we identified 197 shared differentially expressed genes (DEGs) in PBMC before and after exercise. We conducted pathway analysis of these DEGs and built a gene co-expression network. The identification of hub genes was achieved through the utilization of protein-protein interaction (PPI) networks. 268 shared DEGs were identified in the GSE11761 dataset in pre- and post-exercise groups, while 352 differential genes were identified between the pre- and post-exercise groups of females in the GSE14642 dataset. A total of 197 shared genes were identified between the two datasets. Seven core genes were identified by MCODE and CytoHubba, namely IL2RB, KLRD1, GZMB, TBX21, PRF1, HAVCR2, and GZMA. Bioinformatics techniques are effective in screening and analyzing PBMC-associated DEGs in children undergoing acute pre- and post-exercise puberty. Microarray analysis of PBMCs in children undergoing puberty can identify molecules that may predict acute exercise responses. This study provides insights for further exploration of the mechanisms and targets of acute exercise on the organism.
Similar content being viewed by others
Introduction
Research has demonstrated that exercise-induced enhancements in immune function may play a pivotal role in the anti-senescence of the immune system and the development of chronic diseases. For several decades, research in the field of exercise immunology has demonstrated that the immune system exhibits robust responsiveness to both acute and chronic exercise training. Duggal et al. emphasize the benefits of exercise on improving immune function in adults of all ages1. It is well known that engaging in intense physical activity can significantly increase the number of peripheral blood mononuclear cells (PBMCs) in humans, including lymphocytes, monocytes, and natural killer cells, as well as neutrophils2. PBMCs are stimulated by exercise and contributed not only to host defense, but also to growth, repair, and disease pathogenesis3. Acute exercise can induce mitochondrial biogenesis through NF-kB activation and PGC1α gene expression4. In another study of healthy young people, it was found that intense physical activity boosts the number of active T-cells in the bloodstream, which may improve immune surveillance and reduce the need for a large number of specialized T-cells1.
Adolescence is a crucial period during human maturation, with puberty being associated with significant hormonal changes that impact not only an individual’s sexual and reproductive characteristics but also their musculoskeletal characteristics5. Approximately 5.3 million people worldwide die from physical inactivity, a major risk factor for many chronic diseases and cancers. Physical activity habits in childhood can influence adult lifestyles6. Studies have shown that adolescents have high rates of physical inactivity and that this sedentary lifestyle often continues into adulthood. Recent data show an increase in the prevalence of noncommunicable diseases and their risk factors, such as hypertension and obesity, among adolescents. The impact of physical inactivity on adolescent health is becoming increasingly clear7. Physical activity has been shown to be key to reducing the risk of chronic disease, helping to increase energy expenditure and balance the diet, and is an effective way to prevent chronic degenerative diseases8. The World Health Organization (WHO) recommends that children and adolescents perform at least 60 min of moderate to vigorous physical activity(MVPA )per day. purposeful repetitive exercise to maintain or increase fitness is recommended, with at least 30 min of MVPA-intensity exercise on five or more days per week, or at least 20 min of vigorous-intensity exercise on three consecutive days9. Physical activity among children and adolescents is associated with lower adiposity, improved cardio-metabolic health, and improved fitness. Worldwide, fewer than 30% of children and adolescents meet global physical activity recommendations of at least 60 min of moderate to vigorous physical activity per day5.
In recent years, microarray technology and bioinformatics analyses have been widely used to screen for genetic alterations at the genomic level. In this study, we first analyzed two raw microarray datasets downloaded from the Gene Expression Omnibus database (GEO, http://www.ncbi.nlm.nih.gov/geo) for DEGs between acute exercise and pbmc in adolescent children. Subsequently, the identified DEGs were analyzed by Gene Ontology (GO) analysis, Kyoto Encyclopedia of Genes and Genomes ( KEGG) pathway analysis and Protein-Protein Interaction (PPI) analysis on the identified DEGs, aiming to identify hub genes.
Materials and methods
Gene expression microarray data acquisition
Expression profiling data by array of exercise and PBMC gene expression profiles GSE11761 and GSE14642 were downloaded from the Gene Expression Omnibus (GEO) database(https://www.ncbi.nlm.nih.gov/geo/). The GSE11761 (Affymetrix Human Genome U133 Plus 2.0 Array) data collection consists of pre-exercise and post-exercise expression profiles of relevant PBMC genes in 20 males puberty (8–18 years old). GSE14642 data were collected from 20 healthy female puberty (8–17 years old) with pre- and post-exercise gene expression profiles using the Affymetrix Human Genome U133 Plus 2.0 Array. In the late-pubertal girls, we made no attempt to perform exercise selectively in either the luteal or follicular phase of the menstrual cycle. Each subject performed exercise consisting of ten 2-min bouts of constant-work-rate cycle ergometry, with a 1-min rest interval between each bout of exercise. An indwelling catheter was inserted into the anterior elbow vein, and baseline samples were collected 30 min before the commencement of exercise and before the initiation of the experimental protocol. A 30-minute waiting period was implemented to ensure that stress parameters, such as heart rate and blood pressure, reached a stable state. Subjects performed 10 two-minute sessions of constant work, and blood samples were collected immediately after exercise, resulting in a total of 40 samples. PBMCs were isolated using OptiPrep density gradient medium(Sigma). Standard and consistent practices were employed in an effort to minimize any potential changes in mRNA expression levels due to manipulation of PBMCs. The duration from blood draw to stabilization of RNA never exceeded 60 min.
Data processing
GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/), an online analysis tool, can be used to filter DEGs between the pre-exercise and post-exercise groups. The set conditions for DEGs are P< 0.05 and ∣log fold-change (FC)∣ > 0.58510. Utilizing Wei Sheng Xin (http://www.bioinformatics.com.cn/) platforms to create volcano and heat maps.
Venn analysis
Venn diagrams are widely used charts that show relationships between multiple sets and are probably the most intuitive form of data visualization (https://bioinformatics.psb.ugent.be/webtools/Venn/)11.
GO and KEGG enrichment analyses of DEGs
The function of genes in the two datasets was analyzed using the Metascape database (https://www.metascape.org/gp/index.html) and the DAVID database (https://david.ncifcrf.gov/). Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses, including biological processes (BP), cellular components (CC), and molecular functions (MF), were conducted.
PPI network construction and MCODE analysis
Constructing a protein-protein interaction (PPI) network using the STRING database to evaluate interactions between DEGs12, Identify relevant pathways and functions of shared DEGs. The network was clustered into a specified number of clusters using the k-means algorithm (the number of clusters: 3)10. The interaction score of 0.4 was established as the minimum requirement to construct a PPI network. Then, the PPI network data was loaded into Cytoscape (version 3.9.1) software for visual adjustment. The optimal module selection was performed using the APP molecular complex detection (MCODE) tool. The parameters employed in MCODE were as follows: degree cutoff of 2, cluster finding with a node score cutoff of 0.2, k-core of 2, and max depth of 100.
Identification of key gene
The highest interlocking hub genes in the network were identified using CytoHubba, a plug-in application in Cytoscape. The shared gene PPI network can be analyzed to identify the top 10 genes. The primary gene of interest is the common gene among the top 10 hub genes in the network. The KEGG pathway was further analyzed.
Result
DEGs screening result
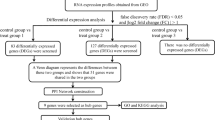
A visual representation in the form of a flowchart was developed to depict the essential and significant procedures of our research (refer to Fig. 1). To investigate the impact of exercise on the puberty children, we analyzed PBMC samples isolated from blood. In the GSE11761 (puberty males), a total of 268 DEGs were identified. This included 144 up-regulated genes and 124 down-regulated genes when comparing the expression of PBMC before and after exercise. In the GSE14642 (puberty females) data set, a total of 352 DEGs were identified, comprising 237 up-regulated genes and 115 down-regulated genes. Venn diagram revealed that 197 DEGs were shared in both the GSE11761 and GSE14642 datasets (Fig. 2).
DEGs expression map and venn diagram. (A) volcano plot showing DEGs expression of GSE11761 dataset. (B) volcano plot showing DEGs expression of GSE14642 dataset. Up-regulated and down-regulated DEGs are highlighted in red and blue, respectively. threshold: |logFC| >0.585 and P < 0.05. (C) venn diagram of shared 197 DEGs between GSE11761 and GSE14642 by comparing the PBMC before and after exercise. (D) Heatmap of top 10 DEGs of GSE11761. (E) Heatmap of top 10 DEGs of GSE14642.The color of red to blue indicates up-regulation to down-regulation.
GO and KEGG enrichment analyses of DEGs
GO enrichment analyses are frequently employed to demonstrate the roles of genes associated with particular terms, while KEGG enrichment analyses can elucidate pathways and functional patterns involved13. The 197 shared genes were subsequently analyzed for GO and KEGG enrichment. The top 10 enriched BP, CC, and MF terms were displayed in Fig. 3A-C, respectively, ranked according to their p-values. The GO enrichment analyses of BP terms revealed the top three enrichments (p-value < 0.001) in the following categories: immune response, killing of cells of other organisms, and cytolysis. As evident from the CC annotations, the top 3 components (p-value < 0.001) were cell part, integral component of the plasma membrane, and cytolytic granule. The top 3 MF (p-value < 0.001) included transmembrane signaling receptor activity, protein binding, MHC class I receptor activity. By analyzing the pathways, We find that the top three pathways are involved in natural killer cell-mediated cytotoxicity, graft-versus-host disease, and antigen processing and presentation(Fig. 3D).
GO and KEGG functional enrichment analysis of the share differentially expressed genes (DEGs). (A) Enriched biological process terms. (B) Enriched cellular component terms. (C) Enriched molecular function terms. (D) The top 10 enriched terms of the KEGG pathway. Cut off value is P value < 0.05. The magnitude of dots reflects the number of DEGs enriched within each term. The p-value, is indicated by the color of the dot.
PPI network and key genes selection of share DEGs
To unravel the critical mechanism of acute exercise in PBMCs of puberty children, it is crucial to explore key genes14. A comprehensive analysis of 197 share DEGs was conducted using STRING to investigate protein interactions. Figure 4A illustrates the interactions between the share DEGs in acute exercise and peripheral blood mononuclear cells of pubescent children. The obtained PPI network consisted of 169 nodes and 557 interactions. Significantly more interactions were observed in the network than expected and the PPI network was imported into the local version of Cytoscape, and the PPI network was constructed based on the degree size (Fig. 4D). Three sets of clusters were identified by MCODE. A total of 34 genes in the top two modules were recognized as key genes with a score of ≥ 5. The score of the first set of clusters is 19.04, consisting of 22 nodes and 200 edges (Fig. 4B). The score of the second set of clusters is 8.9, consisting of 12 nodes and 49 edges (Fig. 4C; Table 1).
PPI network and key gene analysis. (A) Protein-protein internecion (PPI) DEGs from the STRING database are represented by connecting lines of different thicknesses to indicate the strength of association between proteins. (B) Module 1 consisted of 22 nodes and 200 edges. (C) Module 2 consisted of 12 nodes and 49 edges. (D) The transcription factors in the top 20 are ranked based on their P values and their interactions with DEGS. Within the network diagram, the red circles denote the transcription factors exhibiting the 20 lowest P values, whereas the yellow circles signify the differentially expressed genes that exhibit correlation with these transcription factors. (E) Utilize five topological algorithms to predict and explore the central genes involved in these networks, presenting the results in a Venn diagram.
Hub gene selection
It is essential to investigate important proteins and their related subnetworks within the intricate PPI network. Five algorithms from the CytoHubba plugin in Cytoscape were utilized for this purpose. Screening of 7 genes at key positions by Venn diagram revealed IL2RB, KLRD1, GZMB, TBX21, PRF1, HAVCR2, and GZMA (Fig. 4E; Table 2 description from Gene Card) and analyzed the expression of these seven key genes (Fig. 5).
Discussion
The development of gene chip technology and high-throughput sequencing has enabled the simultaneous detection of thousands of genes at varying expression levels. The integration and reanalysis of microarray data has yielded valuable insights, including the identification of hub genes, biological functions, and signaling pathways. These findings have the potential to inform the development of novel exercise therapies for individuals15. In the present study, we conducted an analysis of two GEO datasets, GSE11761 and GSE14642, with the objective of identifying pivotal genes associated with acute exercise in adolescent children. By combining the two GEO datasets, a total of 197 hub genes were identified, with seven hub genes (IL2Rβ, KLRD1, GZMB, TBX21, PRF1, HAVCR2, and GZMA) exhibiting statistically significant differential expression between the pre- and post-samples. Previous studies did not present a comprehensive microarray analysis, and in this study, we reanalyzed the microarray data to elucidate the key genes involved in the molecular mechanisms of exercise in adolescents.
Interleukin-2 receptor β (IL2Rβ) is a receptor protein for interleukin-2. IL2RB plays a role in regulating T cell-mediated immune responses. This beta subunit is involved in receptor-mediated endocytosis and transduces the mitogenic signal of IL-216. The IL2RB chain regulates the slow dissociation rate of IL-2 with high-affinity receptors17. IL2RB potentially aids in the internalization of IL-2 and the transmission of signals via its long intracytoplasmic ___domain.Altered IL-2Rβ expression and signaling in T cells and NK cells may lead to functional defects in early-onset autoimmunity and immunodeficiency18. In patients with sepsis, differentially expressed genes were assessed to identify genes involved in T cell immunity. IL2RB was found to be inversely associated with sequential organ failure score and mortality19.
KLRD1 (Killer Cell Lectin Like Receptor D1) is a Protein Coding gene. This antigen is preferentiallyex pressedon NK cells and is categorized as a type II membrane protein due to its external C terminus.KLRD1 is associated with a number of diseases, including NK cell deficiency and chronic NK-cell lymphocytosis. KLRD1 is associated with NK cell immunisation19. The expression of KLRD1 on NK cells may serve as a potential biological marker for susceptibility to influenza. A negative correlation was observed between KLRD1 expression in the blood at baseline and the severity of influenza infection symptoms20. It was postulated that exercise may affect NK cell immunity by increasing the expression of KLRD1 in the blood.
TBX21 (T-Bet) is a member of a phylogenetically conserved gene family that share a DNA-binding ___domain. After a 12-week Tai Chi Chuan exercise program, there was a significant increase in the expression of TBX21 in patients with type 2 diabetes mellitus21. After 1 week of completing a strenuous exercise marathon, the expression of TBX21 was significantly lower22. We hypothesized that TBX21 may be related to exercise intensity.
The gene GZMB (Granzyme B) is a member of the granzyme subfamily of proteins, belonging to the serine protease peptidase S1 family. Diseases associated with GZMB include peripheral T-cell lymphoma and severe cutaneous adverse reactions. GZMB pronounced in the expression of cytotoxic activity23. Moderate exercise stimulates transcription activities of GZMB24. GZMB expression was different in different exercise intensities, with mice exhibiting higher proliferation and GZMB expression at moderate intensity in the presence of GZMB.
PRF1 (Perforin 1) is a Protein Coding gene. This gene encodes a protein that shares structural similarities with complement component C9, which plays a crucial role in immunity. This protein facilitates the formation of membrane pores, enabling the discharge of granzymes and subsequent cytolysis of target cells. Diseases associated with PRF1 include familial hemophagocytic lymphohistiocytosis (type 2) and aplastic anemia. PRF1 plays a crucial role in promoting cytolysis and apoptosis of target cells by facilitating the passage and uptake of cytotoxic granzymes. Its expression levels are closely linked to the inflammatory and toxic effects of NK cells, indicating its importance in regulating their activity and function. In an in vivo experiment involving mice, it was observed that after 30 min of swimming, NK cells showed increased infiltration rates along with heightened expression of PRF1. This suggests that the upregulation of PRF1 following exercise corresponds to the increased rate of NK cell infiltration, highlighting the dynamic relationship between PRF1 expression and NK cell activity25.
The gene HAVCR2 is classified as a Protein Coding gene. It is linked to various medical conditions such as T-cell lymphoma, subcutaneous Panniculitis-Like and hepatitis A. Linked pathways include immune cytokine signaling and interleukin-2 family signaling.In a study of elderly practitioners of physical activities increased expression of HAVCR2in CD4 + T lymphocytes26. A large amount of experimental data supports HAVCR2 as an immune checkpoint, and targeting HAVCR2 is a promising treatment method in current immunotherapy, especially the new combination of other immune checkpoint blockers27.
GZMA is a serine protease that exhibits specificity towards T cells and natural killer cells, and it is believed to be a crucial component in the lysis of target cells by cytotoxic T lymphocytes and natural killer cells. GZMA plays an important role in cell death, cytokine processing and inflammation28. In patients with peritoneal sepsis extracellular active form of GZMA functions as a pro-inflammatory agent in macrophages by inducing the TLR4-dependent production of IL-6 and TNFα29.
Using bioinformatics, we found seven core genes in which exercise affects PBMC in puberty children, among which KLRD1 is associated with NK cell immunity, HAVCR2 is a versatile immunomodulator associated with T cells, IL2RB and TBX21 are genes involved in T cells. PRF1, GZMB, and GZMA significantly express cytotoxic activity, linked to injuries caused by physical activity and the body’s inflammatory immune reaction.GZMB/PRF1 interaction is an important factor in the immunization process of circulating monocytes, which may be regulated by transient exercise, thus enhancing immune function.
We characterized the genes affected by exercise using gene pathways. Many of the pathways identified were involved in innate or early immune responses30.The majority of significant pathways are linked to inflammatory processes, stress responses, and programmed cell death, which aligns with prior observations on gene expression patterns following motility in human circulating leukocytes. Two alternative mechanisms might explain the robust effect of exercise on PBMC gene pathways: the first, a direct effect of exercise on gene expression within the population of circulating PBMCs; and the second, an indirect effect, the mobilization into the circulation of PBMCs that were expressing genes differently in their marginal pools or because of different maturational status of the mobilized PBMCs compared with those PBMCs already in the circulating blood3.
The objective of this study was to examine the pathways of peripheral blood mononuclear cell (PBMC) gene response to exercise in adolescent children.It has been well documented that both gender and puberty can influence immune function. Additionally, there is a growing body of literature that suggests that leukocyte gene expression may be affected by these factors as well31. The study found that exercise stimulates the gene expression of PBMCs in adolescent children and young adults. The genomic response will include pro-inflammatory and anti-inflammatory cytokines, stress factors, and alterations in growth mediators. Acute exercise is associated with increased numbers of immune cells and activation of key gene pathways during early and late adolescence. This suggests the existence of an immune cell pathway between acute exercise and health and disease in growing children. Nevertheless, there are some limitations to this study. First, it should be noted that only seven hub genes were involved in the current study. Secondly, the detailed molecular mechanisms for studying the hub genes and which changes in gene expression in PBMC lead to functional changes in immune cells have yet to be determined.
Conclusion
In conclusion, our current study bioinformatically analyzed DEGs of PBMC in puberty children before and after exercise to obtain pivotal genes, and the functions of these genes need to be further explored to determine the biological mechanisms underlying the effects of exercise therapy on PBMC in puberty children.
Data availability
The datasets presented in this study can be found in online repositories. All the data were acquired from the Gene Expression Omnibus (GEO) database.GSE11761_series_matrix.File(s): The GSE11761 series based on the GPL570 [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array.(https://www.ncbi.nlm.nih.gov/geo/).GSE14642_series_matrix.File(s): The GSE14642 series based on the GPL570 [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array.(https://www.ncbi.nlm.nih.gov/geo/).
References
Theall, B. et al. Impact of acute exercise on peripheral blood mononuclear cells nutrient sensing and mitochondrial oxidative capacity in healthy young adults[J]. Physiol. Rep. 9 (23), e15147 (2021).
Connolly, P. H. et al. Effects of exercise on gene expression in human peripheral blood mononuclear cells[J]. J Appl Physiol (1985). 97(4), 1461–1469 (2004).
Radom-Aizik, S. et al. A brief bout of exercise alters gene expression and distinct gene pathways in peripheral blood mononuclear cells of early- and late-pubertal females[J]. J. Appl. Physiol. (1985). 107 (1), 168–175 (2009).
Busquets-Cortés, C. et al. Training and acute exercise modulates mitochondrial dynamics in football players’ blood mononuclear cells[J]. Eur. J. Appl. Physiol. 117 (10), 1977–1987 (2017).
Neil-Sztramko, S. E., Caldwell, H. & Dobbins, M. School-based physical activity programs for promoting physical activity and fitness in children and adolescents aged 6 to 18[J]. Cochrane Database Syst. Rev. 9 (9), CD7651 (2021).
Avloniti, A. et al. Exercise-induced oxidative stress responses in the pediatric population[J]. Antioxid. (Basel) 6(1), 112–114, (2017).
van Sluijs, E. et al. Physical activity behaviours in adolescence: current evidence and opportunities for intervention[J]. Lancet. 398 (10298), 429–442 (2021).
Lippo, B. R. et al. Determinants of physical inactivity among urban adolescents[J]. J. Pediatr. (Rio J). 86 (6), 520–524 (2010).
Varghese, T. et al. Physical activity in the prevention of coronary heart disease: implications for the clinician[J]. Heart. 102 (12), 904–909 (2016).
Zheng, Q. et al. SARS-CoV-2 induces cytokine storm hyperinflammatory responses in RA patients through pyroptosis[J]. Front. Immunol. 13, 1058884 (2022).
Kong, C. et al. Six potential biomarkers in septic shock: a deep bioinformatics and prospective observational study[J]. Front. Immunol. 14, 1184700 (2023).
Yu, C. et al. Gender Differences in Rheumatoid Arthritis: Interleukin-4 Plays an Important Role[J]. J Immunol Res. 2020, 4121524 (2020).
Kanehisa, M. & Goto, S. KEGG: kyoto encyclopedia of genes and genomes[J]. Nucleic Acids Res. 28 (1), 27–30 (2000).
Zhang, Y. et al. Identifying key genes related to the peritubular capillary rarefaction in renal interstitial fibrosis by bioinformatics[J]. Sci. Rep. 13 (1), 19611 (2023).
Lv, J. & Li, L. Hub Genes and Key Pathway Identification in Colorectal Cancer Based on Bioinformatic Analysis[J]. Biomed Res Int. 2019, 1545680 (2019).
Ratthe, C. & Girard, D. Interleukin-15 enhances human neutrophil phagocytosis by a syk-dependent mechanism: importance of the IL-15Ralpha chain[J]. J. Leukoc. Biol. 76 (1), 162–168 (2004).
Kuziel, W. A. & Greene, W. C. Interleukin-2 and the IL-2 receptor: new insights into structure and function[J]. J. Invest. Dermatol. 94 (6 Suppl), 27S–32S (1990).
Fernandez, I. Z. et al. A novel human IL2RB mutation results in T and NK cell-driven immune dysregulation[J]. J. Exp. Med. 216 (6), 1255–1267 (2019).
Almansa, R. et al. Transcriptomic correlates of organ failure extent in sepsis[J]. J. Infect. 70 (5), 445–456 (2015).
Bongen, E. et al. KLRD1-expressing natural killer cells predict influenza susceptibility[J]. Genome Med. 10 (1), 45 (2018).
Yeh, S. H. et al. Regular Tai Chi Chuan exercise improves T cell helper function of patients with type 2 diabetes mellitus with an increase in T-bet transcription factor and IL-12 production[J]. Br. J. Sports Med. 43 (11), 845–850 (2009).
Xiang, L., Rehm, K. E. & Marshall, G. J. Effects of strenuous exercise on Th1/Th2 gene expression from human peripheral blood mononuclear cells of marathon participants[J]. Mol. Immunol. 60 (2), 129–134 (2014).
Bassez, A. et al. A single-cell map of intratumoral changes during anti-PD1 treatment of patients with breast cancer[J]. Nat. Med. 27 (5), 820–832 (2021).
Shleptsova, V. A. et al. Expression of KIR2DL3 and KIR2DS2 natural killer receptors in exercise[J]. Bull. Exp. Biol. Med. 149 (6), 755–758 (2010).
Jiang, F. et al. Effects of a natural nutritional supplement on immune cell infiltration and immune gene expression in exercise-induced injury[J]. Front. Nutr. 9, 987545 (2022).
de Carvalho-Costa, T. M. et al. Immunophenotypic Analysis of T Lymphocytes and Cytokine Production in Elderly Practicing Physical Activities and Its Relationship with Quality of Life and Depression[J]. Oxid Med Cell Longev. 2022, 7985596 (2022).
Zhao, L. et al. TIM-3: an update on immunotherapy[J]. Int. Immunopharmacol. 99, 107933 (2021).
Cheng, Q. et al. Three hematologic/immune system-specific expressed genes are considered as the potential biomarkers for the diagnosis of early rheumatoid arthritis through bioinformatics analysis[J]. J. Transl Med. 19 (1), 18 (2021).
Garzon-Tituana, M. et al. Granzyme a inhibition reduces inflammation and increases survival during abdominal sepsis[J]. Theranostics. 11 (8), 3781–3795 (2021).
Radom-Aizik, S. et al. Brief bout of exercise alters gene expression in peripheral blood mononuclear cells of early- and late-pubertal males[J]. Pediatr. Res. 65 (4), 447–452 (2009).
Béghin, L. et al. Gender influences physical activity changes during adolescence: the HELENA study[J]. Clin. Nutr. 38 (6), 2900–2905 (2019).
Author information
Authors and Affiliations
Contributions
Conceptualization and design of the study: L-X.T; Data analysis: L-J.D and Y-H.S; Result interpretation: L-J.D and Y-H.S; Methodology, Supervision, and Text Correction: J-Y.L; Writing of the manuscript: L-J.D. All the authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Du, LJ., Song, YH. & Tang, LX. Effect of acute exercise on gene expression in peripheral blood mononuclear cells of puberty children. Sci Rep 14, 27977 (2024). https://doi.org/10.1038/s41598-024-77242-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-77242-8
Keywords
This article is cited by
-
Bioinformatics analysis and experimental validation of potential targets and pathways in chronic kidney disease associated with renal fibrosis
Journal of Translational Medicine (2025)
-
CAT and CXCL8 are crucial cofactors for the progression of nonalcoholic steatohepatitis to hepatocellular carcinoma, the immune infiltration and prognosis of hepatocellular carcinoma
Discover Oncology (2025)