Abstract
Early diagnosis and treatment of bladder cancer are crucial, and since inflammation plays a role in all stages of bladder cancer, this study aims to develop a model based on inflammation-related genes to accurately predict patient prognosis. The data were initially processed through differential analysis and prognostic correlation analysis, then a Least absolute shrinkage and selection operator (LASSO) regression model was constructed by M-cohort and a nomogram was designed to increase the model readability. The T-cohort was used for internal validation, with the GSE32894 and Imvigor210 cohorts used as external data to verify the model’s accuracy. The model’s predictive ability was verified for the prognosis of patients of different ages, gender, tumor stage, and tumour grade. The GSE3167, GSE13507 and GeneExpression Profiling Interactive Analysis (GEPIA) datasets and Human Protein Atlas (HPA) database were used to verify the expression of the inflammation-related genes, which were confirmed by real-time Polymerase Chain Reaction (PCR). A comprehensive analysis of the model’s inflammation-related genes, Gene Set Enrichment Analysis (GSEA), Gene Set Variation Analysis (GSVA) enrichment analysis, and immune-related analysis were also performed. Both internal and external data validations confirmed that the developed model can accurately predict the prognosis across different patient populations. Hierarchical validation results demonstrated that the model’s predictive power is reliable for various patient stratifications. The expression of inflammation-related genes was consistent across The Cancer Genome Atlas (TCGA) database, GSE3167 dataset, GSE13507 dataset, Gene Expression Profiling Interactive Analysis (GEPIA) database, and the Human Protein Atlas (HPA) database, and was further validated by real-time Polymerase Chain Reaction (PCR). Pathway enrichment analysis indicated that patients in the high-risk (H-risk) group exhibited a variety of tumors. Meanwhile, patients in the low-risk (L-risk) group may be candidates for immunotherapy, whereas those in the high-risk group are more likely to benefit from chemotherapy. The model of inflammation-related genes can accurately predict bladder cancer patient prognosis, with MEST, FASN, KRT6B, and RGS2 anticipated to become new prognostic bladder cancer markers.
Similar content being viewed by others
Introduction
Bladder cancer is a common tumour of the urinary system1,2 and is ranked among the top ten cancers globally3. It is a distinctive tumour with a higher incidence in men in developed regions4 and the number of new cases increasing worldwide annually5,6. Although most bladder cancers are non-muscle-invasive, the prevalence of bladder cancer has remained persistent due to its high recurrence rate7. Once muscular invasion occurs, bladder cancer easily metastasizes and has a poor prognosis8. At present, early bladder cancer can only be detected with some invasive tests but the development of bioinformatics and prognostic models are advantageous for predicting the prognosis of bladder cancer patients in the early stage of the disease9. Approximately 25% of bladder cancer patients require systemic chemotherapy and/or immunotherapy, curative therapy, or palliative care10, therefore, there is an urgent need to develop a model to predict patient prognosis for early intervention.
The tumour microenvironment comprises numerous stromal tissues, inflammatory cells, and inflammatory mediators11 and inflammation can induce tumorigenesis, with tumour sites initiating an inflammatory response that accelerates tumour progression11,12. For example, Helicobacter pylori infection is closely related to stomach cancer, schistosomiasis infection often leads to bladder cancer, and colitis may eventually lead to colon cancer13,14. Anti-inflammatory therapy can reduce tumour mortality15, so in recent years, the relationship between chronic inflammation and tumours has also received widespread attention16. Related studies found that JAK2/STAT3 pathway regulation can affect immune evasion in hepatocellular carcinoma17 and long noncoding RNAs associated with inflammation aid in the typing of bladder cancer18. Fibroblasts in anti-tumour immunity are also associated with inflammation. The inflammatory environment before surgery for bladder cancer is closely related to the prognosis after surgery19, with many studies on the inflammatory characteristics of bladder cancer showing that bladder cancer is inextricably linked to inflammation12,18.
There is no doubt that tumor development is closely linked to inflammation. This study aimed to develop a model based on inflammation-related genes to accurately predict the prognosis of bladder cancer patients. To enhance the model’s interpretability, a nomogram was constructed, and its reliability was validated using multiple datasets. Furthermore, the expression of inflammation-related genes was confirmed. A comprehensive analysis of these genes identified novel biomarkers for bladder cancer.
We present this article in accordance with the TRIPOD reporting checklist.
Methods
Data acquisition and preliminary analysis
The study process is illustrated in Fig. 1. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Inflammation-related genes used in this study were sourced from the GeneCards database (https://www.genecards.org/). Transcriptome data from 412 bladder cancer patients and 19 normal samples were obtained from The Cancer Genome Atlas (TCGA) (https://portal.gdc.cancer.gov/) database to identify a gene signature in bladder cancer patients. Additionally, copy number data and mutation data necessary for the analysis were acquired from the TCGA database.
Flowchart of this study. HPA, Human Protein Atlas; TCGA, The Cancer Genome Atlas; GEO, Gene Expression Omnibus; GSE, Gene Series; GEPIA, Gene Expression Profiling Interactive Analysis; M-cohort, Model cohort; T-cohort, Test cohort; LASSO, Least absolute shrinkage and selection operator; PCR, Polymerase Chain Reaction.
IMvigor210 is an open-label, multicenter, single-arm phase II clinical study, and the Imvigor210 cohort was used as external data to verify the model’s accuracy. Furthermore, the Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/) database was searched using ‘bladder cancer’, and datasets with complete survival information and a sample size of no less than 30 were selected for model construction. The GSE32894 dataset from the GEO database was used for external validation, including 308 bladder cancer samples, of which 224 samples contained complete follow-up information. Any data with incomplete survival information were excluded from all cohorts.
The GSE3167 and GSE13507 datasets were also obtained from the GEO database and used to validate differential gene expression patterns. The GSE3167 dataset consisted of 60 samples, including 46 tumor samples and 14 normal samples, while the GSE13507 dataset comprised 68 normal samples and 188 tumor samples. Bladder cancer-related data from the Gene Expression Profiling Interactive Analysis (GEPIA) (http://gepia.cancer-pku.cn/) database was used to further validate differential gene expression patterns. Additionally, protein expression of the inflammation-related genes was examined through the Human Protein Atlas (HPA) (https://www.proteinatlas.org/) database.
Preliminary data analysis using R software identified differentially expressed inflammation-related genes, which were then normalized using the SVA (Surrogate Variable Analysis) software package to remove batch effects (logFC = 1, P = 0.05). In total, 790 differentially expressed inflammation-related genes associated with bladder cancer prognosis were identified, which were subsequently screened to obtain signatures of inflammation-related genes in bladder cancer patients (P = 0.05).
The LASSO regression model and nomogram construction
The caret package was used to randomly divide the TCGA cohort into two groups: the M-cohort (Model cohort) and the T-cohort (Test cohort). The M-cohort was utilized to build the LASSO regression model, which offers advantages over traditional linear regression models due to its parameter selection and shrinkage capabilities. An optimized model was developed by methodically compressing less important parameters, and cross-validation was then performed to select the most appropriate lambda. This optimal lambda was used to create the final model, which assigned a score to each standardized sample.
All samples were divided into two groups based on the median M-cohort sample score: the H-risk (high risk) group, with scores above the median, and the L-risk (low risk) group, with scores below the median. A nomogram was constructed by combining model scores with common clinical indicators to enhance the interpretability of the model’s predictions. Principal Component Analysis (PCA) was performed to assess whether the model could effectively distinguish between patients in the H-risk and L-risk groups.
Finally, univariate and multivariate independent prognostic analyses were conducted to determine whether the risk scores and nomogram could serve as independent prognostic indicators, separate from other factors. The model’s predictive performance was validated using survival analysis, the area under the Receiver Operating Characteristic (ROC) curve, and survival status distribution plots of M-cohort patients.
Verification of the model accuracy with TCGA internal data
The T-cohort (Test cohort) samples were used for internal validation of the model. These samples were divided into two groups based on the median M-cohort (Model cohort) sample score for survival analysis and other operations to verify the model’s ability to predict patient prognosis. The accuracy of the nomogram was assessed by comparing the predicted results with the actual outcomes. The Area Under the Curve (AUC) was also used to evaluate the prediction accuracy of the nomogram.
Decision Curve Analysis (DCA) is a widely used method for assessing clinical predictive models and offers unique advantages over the Receiver Operating Characteristic (ROC) curve area. DCA can incorporate the preferences of decision-makers into the analysis, making it a more realistic evaluation tool.
Applicability of hierarchical validation models
It is crucial for the model to accurately predict the prognosis for all patients, as varying clinical features can significantly influence outcomes. To ensure the model’s robustness across different patient profiles, stratified validation was conducted. Initially, patients were divided into two age groups, with those over 60 years classified as the older age group, to assess the model’s predictive capability across different age ranges. Subsequently, patients were further stratified by gender, tumor stage, and grade, allowing for a comprehensive evaluation of the model’s performance across various clinical characteristics.
External data to verify the model accuracy
The model’s applicability is determined by its ability to predict prognosis across a broader population. To evaluate this capability, survival analysis was conducted using external datasets GSE32894 and Imvigor210, which allowed for an assessment of the model’s prognostic accuracy. The area under the ROC curve (AUC) quantified the model’s predictive performance on these external datasets. Furthermore, a comprehensive analysis of model-related genes was performed, with volcano plots illustrating the expression patterns of these genes in bladder cancer patients. Subsequently, we examined the correlation between model-related genes and their interaction with immune cells. A waterfall chart visualized gene mutations, while chromosomal circles depicted the locations and copy numbers of model-associated genes on the chromosomes.
Public data to verify the expression of differentially expressed inflammation-related genes
For external validation of the differential expression patterns of model-related genes, the GSE3167 and GSE13507 datasets from the GEO database were utilized, along with expression data from the Gene Expression Profiling Interactive Analysis (GEPIA) database (http://gepia.cancer-pku.cn/). Additionally, the Human Protein Atlas (HPA) database was consulted to investigate the protein expression of genes associated with the model.
Real-time PCR to validate the expression patterns of model-related genes
Real-time Polymerase Chain Reaction (PCR) was performed to verify the expression patterns of model-related genes using the 5637 cell line for bladder cancer cells and the SV-HUC-1 cell line for normal cells. The primer sequences were as follows: MEST F: AGATCGCCTCCGCAGGAT, R: CCACACCCACAGAGTCTGAA; FASN F: GCAAGCTGAAGGACCTGTCT, R: AATCTGGGTTGATGCCTCCG; KRT6B F: CTTCATCGACAAGGTGCGGT, R: CAGCTCCGAGTCCAGACGA; RGS2 F: GAGCGAGGAGAAGCGAGAAA, R: CTTCCTCAGGAGAAGGCTTGA.
GSVA and GSEA enrichment analyses
Gene Set Variation Analysis (GSVA) is a specialized gene set enrichment method that provides insights into the different prognoses of bladder cancer patients from a bioinformatics perspective. Data for this analysis were sourced from the C2 gene set of The Molecular Signatures Database (MSigDB). Additionally, Gene Set Enrichment Analysis (GSEA) is a widely used bioinformatics method to observe pathway enrichment in patients.
Analysis of the patient’s immune characteristics
Initially, seven common algorithms were employed to estimate the abundance of immune cell infiltration in bladder cancer patients. Subsequently, the correlation between immune cell infiltration abundance and the risk score was analyzed, with the results presented in a bubble chart. The tumor microenvironment of bladder cancer patients was also explored, and single sample gene set enrichment analysis (ssGSEA) was conducted to investigate differences in immune cells and immune-related functions across different patient groups. Finally, the differential expression of 37 immune checkpoint-related genes was evaluated among different patient risk groups.
Prediction of immunotherapy and chemotherapy outcomes
Immunotherapy and chemotherapy play crucial roles in the treatment of bladder cancer patients. Personalized and precise treatment approaches can enhance treatment outcomes and reduce side effects. First, we explored the sensitivity of different patient groups to various immune checkpoint inhibitors using data from The Cancer Immunome Atlas (TCIA) database and validated the findings with immunotherapy data from bladder cancer patients in the Imvigor210 cohort. The oncoPredict software package was used to predict patients’ sensitivity to common chemotherapy drugs. By predicting the effects of immunotherapy and chemotherapy, more precise treatment plans can be developed for patients.
Statistical analysis
All statistical analyses were performed using R software (version 4.2.2). A P-value < 0.05 was considered significant (∗ p = 0.05, ∗∗ p = 0.01, ∗∗∗ p = 0.001.)
Results
Construction of the LASSO regression model and nomogram
According to the coefficient distribution plot of LASSO regression and the cross-validation plot, the model of seven genes had the smallest error (Fig. 2A & B), so MEST, FASN, KRT6B, and RGS2 were selected to construct the optimal model as follows: EXP [(MEST* 0.120700130624973) + (FASN* 0.388886806141193) + (KRT6B* 0.0996952276302296) +(RGS2* 0.23199409391433)]. In order to make the model more accessible to clinicians, we integrated the risk scores with common clinical indicators to construct a nomogram. The use of this nomogram was demonstrated by applying it to the data of the sixth patient from the TCGA cohort. As shown in the figure, the patient’s 1-year, 3-year, and 5-year survival probabilities were 0.864, 0.603, and 0.484, respectively(Fig. 2C). In addition, the patient’s chance of survival gradually decreased as the length of their illness increased, in line with clinical observations.The principal component analysis revealed that the model-related genes enabled better differentiation of patients with different risk scores (Fig. 2D & E). According to the univariate independent prognostic analysis of risk scores, risk scores, age, and tumour stage were independent indicators of prognosis (Fig. 2F). In the univariate independent prognostic analysis of nomograms, the nomograms independently predicted patient outcomes without relying on other clinical indicators (Fig. 2G). Multivariate independent prognostic analyses also yielded consistent conclusions (Fig. 2H & I).
Construction of a prognostic model of bladder cancer with genes related to inflammatory. (A) Plot of coefficient distribution for LASSO regression; (B) Cross-test maps of penalty terms; (C) Nomogram predicts prognosis in patients with bladder cancer;
(D)PCA plot of inflammation-related genes; (E) PCA plot of model-related genes; (F) Univariate independent prognostic analysis of risk scores; (G) Univariate independent prognostic analysis of nomogram; (H) Multivariate independent prognostic analysis of risk scores; (I) Multivariate independent prognostic analysis of nomogram. PC1, Principal Component1; PC2, Principal Component2; ( “∗”, P < 0.05; “∗∗”, P < 0.01).
The model showed excellent predictive performance in internal data
M cohort patient survival analysis showed that the difference in prognosis between H-risk and L-risk patients was statistically significant, and the prognosis of patients in the L-risk group was better than that of patients in the H-risk group (Fig. 3A). The area under the ROC curve was higher than 0.65, indicating that the model had excellent prognostic predictive ability for bladder cancer patients (Fig. 3B). In the distribution of survival states of M cohort patients, the higher the risk score, the higher the mortality, suggesting that patients in the L-risk group had a better prognosis. In addition, the heat map showed that the expression of model-related genes in the H-risk group was significantly higher than in the L-risk group (Fig. 3C). Since both the T cohort and M cohort were obtained from the TCGA database, the T cohort was used to verify the accuracy of the model. Patients in the H-risk group had a similarly poorer prognosis in the T cohort (Fig. 3D). In addition, the area under the ROC curve in the T cohort was also greater than 0.65 (Fig. 3E). The distribution of survival states in the T cohort and the expression of model-associated genes were consistent with the M cohort (Fig. 3F). We also verified the accuracy of nomogram predictions, with the calibration curve showing that the nomogram has excellent consistency (Fig. 3G). The area under the ROC curve of the nomogram was 0.714, which was much higher than common clinical indicators such as age, sex, and tumour grade (Fig. 3H). The DCA curve showed that the nomogram had excellent decision-making power (Fig. 3I).
Internal validation of model accuracy. (A) Survival analysis curve of M-cohort; (B) ROC curve of M-cohort; (C) Distribution of survival states of M-cohort; (D) Survival analysis curve of T-cohort; (E) ROC curve of T-cohort; (F) Distribution of survival states of T-cohort; (G) Nomogram’s calibration curve; (H) Nomogram’s ROC curve; (I) Nomogram’s DCA curve. M-cohort, Model cohort; T-cohort, Test cohort; ROC, Receiver Operating Characteristic; DCA, Decision Curve Analysis.
The predictive ability of the model was also excellent for different at-risk patients
Equally important was the ability to predict the prognosis of patients with different stratifications including survival status, age, gender, tumour grade and tumour stage. The analysis revealed that patients in the H-risk group had higher mortality, were older and had a tumour grade but there was little difference in gender between patients in the H-risk group and the L-risk group (Fig. 4A). The survival analysis showed that the prognosis of patients in the H-risk group was poorer in the older group but the model could not predict the prognosis of patients in the younger group (Fig. 4B). The predictive power of the model was excellent for both male and female patients (Fig. 4C) and had a better prognosis prediction for patients with a higher tumour grade (Fig. 4D). The model also had excellent predictive power regardless of the tumour stage (Fig. 4E).
The results of hierarchical validation of the model. (A)TCGA dataset Bladder cancer patient clinical information pie chart; (B) The ability of the model to predict the prognosis of patients of different ages; (C) The ability of the model to predict prognosis for patients of different sexes; (D) The ability of the model to predict the prognosis of patients with different grades; (E) The ability of the model to predict the prognosis of patients with different stages. TCGA, The Cancer Genome Atlas.
The predictive power of the model was confirmed by external data
The model had a good ability to predict the prognosis of bladder cancer patients in the GSE32894 and Imvigor210 cohorts (Fig. 5A-F). The stability of the model was verified by external data. KRT6B was strongly correlated with immune cells, with 5 of the 21 immune cells negatively correlated with KRT6B and 6 positively correlated with KRT6B (Fig. 5G). FASN had the most mutations, mainly missense mutations (Fig. 5H). FASN, KRT6B, and RGS2 had increased copy numbers whereas MEST had decreased copy numbers in bladder cancer. RGS2 is located on chromosome 1, MEST on chromosome 7, KRT6B on chromosome 12, and FASN on chromosome 17 (Fig. 5I).
External validation of model accuracy and comprehensive analysis of model-related genes.(A) Survival analysis curve of GSE32894-cohort; (B) ROC curve of GSE32894-cohort; (C) Distribution of survival states of GSE32894-cohort; (D) Survival analysis curve of Imvigor210-cohort; (E)ROC curve of Imvigor210-cohort; (F) Distribution of survival states of Imvigor210-cohort; (G) Heat map of the correlation of model-associated genes with immune cells; (H) Chart waterfall chart of model-related genes; (I) Location of model-related genes on chromosomes. ROC, Receiver Operating Characteristic.
The expression of model-related genes in multiple datasets was consistent with the experimental results
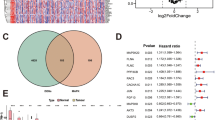
The results of the differential analysis of the TCGA cohort showed that only RGS2 was highly expressed in normal tissues, with the remaining model-associated genes highly expressed in tumour tissues (Fig. 6A). The differential expression patterns of model-associated genes were validated in the GSE3167 and GSE13507 datasets (Fig. 6B) and consistent in the GEPIA database (Fig. 6C). There was only immunohistochemical data for FASN and KRT6B in the HPA database but the data was consistent with gene expression (Fig. 6D). Finally, the experimental results were consistent with the results of the public data analysis (Fig. 6E).
Differential expression validation of model-related genes. (A)Expression patterns of model-related genes in the TCGA database; (B) Expression patterns of model-associated genes in the GSE3167 + GSE13507 dataset; (C) Expression patterns of model-related genes in the GEPIA database; (D) Immunohistochemistry images of model-related genes; (E) Experimental validation of model-related gene expression patterns. TCGA, The Cancer Genome Atlas; GEPIA, Gene Expression Profiling Interactive Analysis; The staining method used is DAB staining. All images in Fig. 5D are magnified by a factor of 40. ( “∗”, P < 0.05)
FASN.
Tumor cell: https://www.proteinatlas.org/ENSG00000169710-FASN/pathology/urothelial+cancer#ihc.
Urothelial cell: https://www.proteinatlas.org/ENSG00000169710-FASN/tissue/urinary+bladder.
KRT6B.
Tumor cell: https://www.proteinatlas.org/ENSG00000185479-KRT6B/pathology/urothelial+cancer#ihc.
Urothelial cell: https://www.proteinatlas.org/ENSG00000185479-KRT6B/tissue/urinary+bladder.
Enrichment analysis
The GSVA enrichment analysis revealed that 90% of the functional enrichment was in the H-risk group, with only taurine and hypotaurine metabolism functions enriched in the L-risk group. The analysis showed that many cell division-related functions were associated with patients in the H-risk group. The WNT signalling pathway may be closely related to the poor prognosis of bladder cancer. In addition, acute myeloid leukaemia, renal cell carcinoma, colorectal cancer, uterine cancer, and glioma were strongly associated with patients in the H-risk group (Fig. 7A). The GSEA enrichment analysis of the C5 gene set revealed that patients in the H-risk group were mainly enriched in gene sets associated with cell components including the synthesis of keratin filaments, intermediate filament cytoskeleton, intermediate filaments, and keratinised envelope. The bulk of GO was enriched in patients in the L-risk group (Fig. 7B). The GSEA enrichment analysis results of the C2 gene set showed that the gene sets enriched in patients in the H-risk group were adhesive plaque, melanoma, actin cytoskeleton regulation, and arrhythmogenic right ventricular cardiomyopathy. The gene sets enriched into patients in the L-risk group are mostly immune-related (Fig. 7C).
Pathway and functional enrichment analysis results of bladder cancer patients in different risk groups. (A)GSVA enrichment analysis results. (B)Results of GO’s GSEA enrichment analysis; (C)Results of GSEA enrichment analysis of pathways. GO, Gene Ontology; GSEA, Gene Set Enrichment Analysis; GSVA, Gene Set Variation Analysis.
Immune characteristics of different groups of patients
The correlation between immune cell infiltration abundance and risk score obtained by seven mainstream algorithms are displayed through bubble charts with the different colours representing the different algorithms. The immune cells on the left are negatively correlated with the risk score and the immune cells on the right are positively correlated with the risk score (Fig. 8A). The invasion of immune cells and stromal cells in the tumour microenvironment was higher in the H-risk group than in the L-risk group (Fig. 8B), with aDCs, macrophages, Tfh, Th1_cells, and Treg being more invasive in patients in the H-risk group, whereas Th2_cells were more invasive in patients in the L-risk group (Fig. 8C). Almost all immune-related functions were more active in patients in the H-risk group (Fig. 8D). The expression of 37 immune checkpoint-associated genes is shown in the histogram in Fig. 8E.
Results of immune-related analyses. (A)Bubble plot of the correlation between risk score and immune cell infiltration abundance; (B) Results of tumor microenvironment analysis; (C) ssGSEA analysis results of immune cells; (D) ssGSEA analysis results for immune-related functions; (E) Expression of immune checkpoint-associated genes in patients with different risk groups. ssGSEA, single sample gene set enrichment analysis; ( “∗”, P < 0.05; “∗∗”, P < 0.01; “∗∗∗”, P < 0.001)
Immunotherapy and chemotherapy result prediction in patients of different groups
The analysis of the TCIA database showed that patients in the L-risk group were more sensitive to immunotherapy (Fig. 9A). Similarly, L-risk patients in the Imvigor210 cohort also responded better to immunotherapy (Fig. 9B). The chemotherapy drug susceptibility analysis showed that patients in the H-risk group were sensitive to common chemotherapy drugs (Fig. 9C). Treatment predictive analysis showed that patients in the L-risk group may be better candidates for immunotherapy and patients in the H-risk group may be better candidates for chemotherapy.
Discussion
In this study, we developed a model for predicting the prognosis of patients with bladder cancer using inflammation-related genes. The internal data and external data verification confirmed that the model can accurately predict the prognosis of different patient populations, and the hierarchical validation showed that the predictive power of the model was also reliable for different patients. Thus, this model formula can predict the prognosis of bladder cancer patients. Previous studies have proposed some useful scoring formulas for the prognosis of bladder cancer patients. For example, Francesco et al. found that the preoperative albumin-to-fibrinogen ratio (AFR) has a high prognostic value in patients undergoing radical cystectomy for bladder cancer20. In addition, the Controlled Nutritional Status (CONUT) score they proposed could serve as a comprehensive, individualised risk stratification tool to identify patients who might benefit from an intensive supportive care regimen21. Makito et al. found that inflammation and nutrition are significant markers for risk assessment through the comprehensive evaluation of inflammation, nutrition and other indicators of muscle-invasive bladder cancer patients who underwent radical cystectomy22. Victor et al. reported that the preoperative serum albumin globulin ratio had a great impact on the prognosis of bladder cancer after radical cystectomy23. Other studies identified some meaningful biomarkers of bladder cancer24,25,26,27.
The expression patterns of model-associated genes in the TCGA database, GSE3167 dataset, GSE13507 dataset, GEPIA database and HPA database were consistent and confirmed by real-time PCR. The pathway enrichment analysis showed that patients in the H-risk group were associated with a variety of tumours, L-risk patients may be candidates for immunotherapy, and patients in the H-risk group were more likely to benefit from chemotherapy.
Previous studies have shown that model-associated genes play an important role in bladder cancer. RGS2, a member of the G protein signalling regulator family, is widely involved in various signalling pathways in the human body28,29. RGS2 is not only closely related to the activities of nerve cells and lymphocytes but is also involved in tumorigenesis30. The polymorphic properties of RGS2 were found to regulate the prognosis of bladder cancer28. ZHX3 and UHRF1 can influence the progression of bladder cancer through the regulation of RGS231,32. MicroRNA-494-3p can also accelerate the progression of bladder cancer by affecting the KLF9/RGS2 axis33.
MEST functions are complex, and its role in life activities is not yet clear34. Recently, studies have shown that MEST expression is associated with tumour progression35,36,37. Similarly, MEST promotes the proliferation and migration of bladder cancer38. In a recent prognostic model, MEST is involved in predicting overall survival in bladder patients39. KRT6B also plays an important role in the metastasis and invasion of bladder cancer40,41,42.
Reducing FSAN inhibits tumour metabolism and proliferation43. In addition, multiple studies have demonstrated that FSAN plays a key role in bladder cancer44,45,46. Metformin through the SREBP-1c/FASN axis can inhibit bladder cancer progression47. FASN plays an important role in tumour initiation and progression, with 95% of the fatty acids in tumour cells synthesised by FASN48,49. FASN can be detected in various tumour cells50,51,52 and is abundant in bladder cancer tissues. The prognosis of bladder cancer patients tends to be worse when FASN is overexpressed, while tumour metastasis and recurrence are limited when FASN is suppressed, and the sensitivity of chemotherapy drugs can be improved53,54,55,56,57. This indicates that FASN has further research value and is expected to provide new therapeutic targets for bladder cancer.
The bioinformatics analysis used in this study relies on published genomic, transcriptomic, or proteomic data. This method allows for more comprehensive utilization of previously conducted research, saving significant research funding and manpower. However, this research method also has certain limitations. Firstly, these data may contain biases or be incomplete. Additionally, the complexity of biological systems makes it difficult for purely computational analyses to capture all biological phenomena. To address the shortcomings of bioinformatics analysis, we used a large amount of public data to validate our analysis results. We also conducted PCR experiments to verify the bioinformatics analysis results. The public data and experimental results were consistent with our analysis, thus confirming the reliability of our conclusions. Furthermore, in future research, we will design corresponding experiments to validate and supplement these findings. In addition, integrating the findings of previous researchers into this model would make it more valuable for clinical application7,58,59. Further exploration of personalised treatment is our next research direction60,61.
Conclusion
In conclusion, the model developed to predict the prognosis of bladder cancer patients using inflammation-related genes (MEST, FASN, KRT6B, and RGS2) demonstrated good predictive accuracy across different cohorts and stratified patient groups. The analysis of the immune profile of bladder cancer patients and treatment prediction suggests that patients with higher risk scores may be more suitable for chemotherapy, while those with lower risk scores are expected to benefit from immunotherapy.
Data availability
The data used in this study are available in public databases. All data are attributed to the source.
References
Jubber, I. et al. Epidemiology of bladder Cancer in 2023: A systematic review of risk factors. Eur. Urol. 84 (2), 176–190 (2023).
Lv, J. et al. HNRNPL induced circFAM13B increased bladder cancer immunotherapy sensitivity via inhibiting glycolysis through IGF2BP1/PKM2 pathway. J. Experimental Clin. Cancer Res. ;42(1). (2023).
Bray, F. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68 (6), 394–424 (2018).
Ferlay, J. et al. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer. 136 (5), E359–E86 (2015).
Chen, H. et al. Neoadjuvant immunotherapy and chemoimmunotherapy for stage II-III muscle invasive bladder cancer. Front. Immunol. 13, 986359 (2022).
Lenis, A. T. et al. Bladder cancer: A review. JAMA ;324(19):1980–1991. (2020).
Catto, J. W. F. et al. Radical cystectomy against intravesical BCG for high-risk high-grade nonmuscle invasive bladder cancer: results from the randomized controlled BRAVO-feasibility study. J. Clin. Oncol. 39 (3), 202–214 (2021).
Malmström, P. U. et al. Five-year followup of a prospective trial of radical cystectomy and neoadjuvant chemotherapy: Nordic Cystectomy Trial I. The Nordic Cooperative bladder Cancer Study Group. J. Urol. 155 (6), 1903–1906 (1996).
Chen, H. et al. Leveraging a disulfidptosis-based signature to improve the survival and drug sensitivity of bladder cancer patients. Front. Immunol. 14, 1198878 (2023).
Ramirez, D. et al. Microscopic haematuria at time of diagnosis is associated with lower disease stage in patients with newly diagnosed bladder cancer. BJU Int. 117 (5), 783–786 (2016).
Mantovani, A. et al. Cancer-related inflammation. Nature. 454 (7203), 436–444 (2008).
Deng, Z. et al. Inflammation-related research within the field of bladder cancer: A bibliometric analysis. Front. Oncol. 13, 1126897 (2023).
Elinav, E. et al. Inflammation-induced cancer: Crosstalk between tumours, immune cells and microorganisms. Nat. Rev. Cancer. 13 (11), 759–771 (2013).
Chan, A. T., Ogino, S. & Fuchs, C. S. Aspirin and the risk of colorectal cancer in relation to the expression of COX-2. N Engl. J. Med. 356 (21), 2131–2142 (2007).
Koehne, C-H. & Dubois, R. N. COX-2 inhibition and colorectal cancer. Semin Oncol. 31 (2 Suppl 7), 12–21 (2004).
Nigam, M. et al. Evaluation of the association of chronic inflammation and cancer: Insights and implications. Biomed. Pharmacother. 164, 115015 (2023).
Liu, Y. et al. HERC2 promotes inflammation-driven cancer stemness and immune evasion in hepatocellular carcinoma by activating STAT3 pathway. J. Exp. Clin. Cancer Res. 42 (1), 38 (2023).
Xiong, X. et al. Identification of a novel defined inflammation-related long noncoding RNA signature contributes to predicting prognosis and distinction between the cold and hot tumors in bladder cancer. Front. Oncol. 13, 972558 (2023).
Ding, L. et al. Preoperative systemic immune-inflammation index as a significant prognostic factor after TURBT in patients with non-muscle-invasive bladder cancer: a retrospective study based on propensity score matching analysis. Cancer Med. 12 (6), 7019–7028 (2023).
Claps, F. et al. Prognostic value of preoperative albumin-to-fibrinogen ratio (AFR) in patients with bladder cancer treated with radical cystectomy. Urol. Oncol. ;39(12). (2021).
Claps, F. et al. Impact of the controlling nutritional status (CONUT) score on perioperative morbidity and oncological outcomes in patients with bladder cancer treated with radical cystectomy. Urol Oncol. ;41(1):49.e13-49.e22. (2023).
Miyake, M. et al. Integrative assessment of pretreatment inflammation-, nutrition-, and muscle-based prognostic markers in patients with muscle-invasive bladder cancer undergoing radical cystectomy. Oncology. 93 (4), 259–269 (2017).
Schuettfort, V. M., Andrea, D. & Quhal, D. F, et al. Impact of preoperative serum albumin-globulin ratio on disease outcome after radical cystectomy for urothelial carcinoma of the bladder. Urol. Oncol. ;39(4). (2021).
Laukhtina, E. et al. Prognostic blood-based biomarkers in patients treated with neoadjuvant chemotherapy for urothelial carcinoma of the bladder: A systematic review. Urol. Oncol. 39 (8), 471–479 (2021).
Mertens, L. S. et al. Prognostic markers in invasive bladder cancer: FGFR3 mutation status versus P53 and KI-67 expression: A multi-center, multi-laboratory analysis in 1058 radical cystectomy patients. Urol. Oncol. 40 (3), 110.e1–e9 (2022).
Mir, M. C. et al. Adjuvant systemic therapy for high-risk muscle-invasive bladder cancer after radical cystectomy: Current options and future opportunities. Eur. Urol. Oncol. 5 (6), 726–731 (2022).
Psutka, S. P. et al. Staging the host: Personalizing risk assessment for radical cystectomy patients. Eur. Urol. Oncol. 1 (4), 292–304 (2018).
Berman, D. M. et al. A functional polymorphism in RGS6 modulates the risk of bladder cancer. Cancer Res. 64 (18), 6820–6826 (2004).
Ross, E. M. & Wilkie, T. M. GTPase-activating proteins for heterotrimeric G proteins: Regulators of G protein signaling (RGS) and RGS-like proteins. Annu. Rev. Biochem. 69, 795–827 (2000).
Hollinger, S. & Hepler, J. R. Cellular regulation of RGS proteins: Modulators and integrators of G protein signaling. Pharmacol. Rev. 54 (3), 527–559 (2002).
Ying, L. et al. Epigenetic repression of regulator of G-protein signaling 2 by ubiquitin-like with PHD and ring-finger ___domain 1 promotes bladder cancer progression. FEBS J. 282 (1), 174–182 (2015).
Deng, M. et al. ZHX3 promotes the progression of urothelial carcinoma of the bladder via repressing of RGS2 and is a novel substrate of TRIM21. Cancer Sci. 112 (5), 1758–1771 (2021).
Xu, X. H. et al. MicroRNA-494-3p facilitates the progression of bladder cancer by mediating the KLF9/RGS2 axis. Kaohsiung J. Med. Sci. 38 (11), 1070–1079 (2022).
Lefebvre, L. et al. Genomic structure and parent-of-origin-specific methylation of Peg1. Hum. Mol. Genet. 6 (11), 1907–1915 (1997).
Boot, A. et al. Imprinted survival genes preclude loss of heterozygosity of chromosome 7 in cancer cells. J. Pathol. 240 (1), 72–83 (2016).
Nishihara, S. et al. Multipoint imprinting analysis in sporadic colorectal cancers with and without microsatellite instability. Int. J. Oncol. 17 (2), 317–322 (2000).
Kohda, M. et al. Frequent loss of imprinting of IGF2 and MEST in lung adenocarcinoma. Mol. Carcinog. 31 (4), 184–191 (2001).
Zhao, C. et al. MEST promotes bladder cancer cell proliferation, migration and invasion via STAT3/Twist-1-mediated EMT. Transl Cancer Res. 9 (10), 6178–6188 (2020).
Wang, Y. et al. A novel 17 apoptosis-related genes signature could predict overall survival for bladder cancer and its associations with immune infiltration. Heliyon. 8 (11), e11343 (2022).
Song, Q. et al. Bladder cancer-derived exosomal KRT6B promotes invasion and metastasis by inducing EMT and regulating the immune microenvironment. J. Transl Med. 20 (1), 308 (2022).
Liu, Y-R. et al. Enhanced metastatic potential in the MB49 urothelial carcinoma model. Sci. Rep. 9 (1), 7425 (2019).
Hoggarth, Z. E. et al. Enrichment of genes associated with squamous differentiation in cancer initiating cells isolated from urothelial cells transformed by the environmental toxicant arsenite. Toxicol. Appl. Pharmacol. 374, 41–52 (2019).
Massari, F. et al. Metabolic phenotype of bladder cancer. Cancer Treat. Rev. 45, 46–57 (2016).
Dong, Y. et al. Clinical value of anoikis-related genes and molecular subtypes identification in bladder urothelial carcinoma and in vitro validation. Front. Immunol. 14, 1122570 (2023).
Xiong, Q. et al. Fatty acid synthase is the key regulator of fatty acid metabolism and is related to immunotherapy in bladder Cancer. Front. Immunol. 13, 836939 (2022).
Li, X. et al. Identification of a novel metabolism-related gene signature associated with the survival of bladder cancer. BMC Cancer. 21 (1), 1267 (2021).
Deng, J. et al. Metformin targets clusterin to control lipogenesis and inhibit the growth of bladder cancer cells through SREBP-1c/FASN axis. Signal. Transduct. Target. Ther. 6 (1), 98 (2021).
Jones, S. F. & Infante, J. R. Molecular pathways: Fatty acid synthase. Clin. Cancer Res. 21 (24), 5434–5438 (2015).
Ookhtens, M. et al. Liver and adipose tissue contributions to newly formed fatty acids in an ascites tumor. Am. J. Physiol. 247 (1 Pt 2), R146–R53 (1984).
Cai, Y. et al. Expressions of fatty acid synthase and HER2 are correlated with poor prognosis of ovarian cancer. Med. Oncol. 32 (1), 391 (2015).
Walter, K. et al. Serum fatty acid synthase as a marker of pancreatic neoplasia. Cancer Epidemiol. Biomarkers Prev. 18 (9), 2380–2385 (2009).
Uddin, S. et al. Fatty acid synthase and AKT pathway signaling in a subset of papillary thyroid cancers. J. Clin. Endocrinol. Metab. 93 (10), 4088–4097 (2008).
Humbert, M. et al. Reducing FASN expression sensitizes acute myeloid leukemia cells to differentiation therapy. Cell. Death Differ. 28 (8), 2465–2481 (2021).
Gruslova, A. et al. FASN inhibition as a potential treatment for endocrine-resistant breast cancer. Breast Cancer Res. Treat. 187 (2), 375–386 (2021).
Shahid, M. et al. S-palmitoylation as a functional regulator of proteins associated with cisplatin resistance in bladder cancer. Int. J. Biol. Sci. 16 (14), 2490–2505 (2020).
Tao, T. et al. Down-regulation of PKM2 decreases FASN expression in bladder cancer cells through AKT/mTOR/SREBP-1c axis. J. Cell. Physiol. 234 (3), 3088–3104 (2019).
Abdelrahman, A. E. et al. Fatty acid synthase, Her2/neu, and E2F1 as prognostic markers of progression in non-muscle invasive bladder cancer. Ann. Diagn. Pathol. 39, 42–52 (2019).
Barone, B. et al. Preoperative fibrinogen-to-albumin ratio as potential predictor of bladder cancer: A monocentric retrospective study. Med. (Kaunas) ;58(10). (2022).
Crocetto, F. et al. A comparative study of the Triglycerides/HDL ratio and pseudocholinesterase levels in patients with bladder Cancer. Diagnostics (Basel) ;12(2). (2022).
Claps, F. et al. Prognostic impact of variant histologies in urothelial bladder cancer treated with radical cystectomy. BJU Int. 132 (2), 170–180 (2023).
Necchi, A. et al. Updated results of PURE-01 with preliminary activity of Neoadjuvant Pembrolizumab in patients with muscle-invasive bladder carcinoma with variant histologies. Eur. Urol. 77 (4), 439–446 (2020).
Author information
Authors and Affiliations
Contributions
Author Contributions (I) Conception and design: YX (II) Administrative support: XL and LL (III) Provision of study materials or patients: DX (IV) Collection and assembly of data: EB (V) Data analysis and interpretation: ZL, XZ, XL (VI) Manuscript writing: All authors. (VII) Final approval of manuscript: All authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Xiao, Y., Xu, D., Bao, E. et al. Identification of inflammation related gene signatures for bladder cancer prognosis prediction. Sci Rep 14, 28867 (2024). https://doi.org/10.1038/s41598-024-79942-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-79942-7