Abstract
Guizhou Province is one of the most important karst regions of southwest China, with 22 Primulina species. These species are highly habitat-specialized and dependent on the soils of the karst region, and many inter-species classifications are unclear. Therefore, studying the chloroplast genomes and estimating the divergence times of there species can not only provide a better understanding of interspecific relationships but also help to know the species speciation and divergence in the karst environment. So, we sequenced and assembled the chloroplast genomes of nine Primulina species (including six endemic species of Guizhou) and conducted chloroplast genome comparison analysis and phylogenetic study. The chloroplast genome structures of the nine Primulina species were quadripartite with total lengths of 152,869–153,364 base pairs (bp) and GC content of 37.55–37.64%. There are 132 functional genes annotated, respectively. A total of 375 simple sequence repeats and 375 interspersed nuclear elements were identified. The 30 highly preferred codons identified were used at similar frequencies in different species, respectively. A phylogenetic tree constructed on the basis of the 38 chloroplast genomes showed that Primulina species form a monophyletic group. Eleven mutational hotspot regions that could serve as potential molecular markers were identified, of which two regions near the 3′ and 5′ ends of the ycf1 gene were of appropriate size and can serve as molecular markers for phylogenetic studies of Primulina. The results of molecular clock analyses indicate that the three major branches of Primulina begin to diverge in the Miocene, and the number of species proliferated in the Pliocene and Pleistocene. Most of the species of Primulina in Guizhou Province were formed in the Pleistocene and rapidly diverged within a short period of time. This research study enriches the genetic resource information of Primulina and deepens the understanding of the phylogenetic relationships of the genus.
Similar content being viewed by others
Introduction
Karst landforms are specialized landforms formed by the dissolution of soluble rocks on the surface and underground1. Due to the high surface relief, diverse topography, high soil calcium content, and lack of continuous soil coverage, a wide variety of microhabitats and "terrestrial island habitats" have been formed, with significant habitat segregation and heterogeneity, giving rise to many karst-exclusive species with a high degree of regional endemism2,3,4,5.
Primulina Hance is one of the most diverse taxa in karst regions, and most species of the genus are dependent on limestone soils, creating taxa of great diversity and endemism in segregated and heterogeneous habitats in karst regions6,7,8,9,10. To date, chloroplast gene fragments (atpB-rbcL, rpl32-trnL, trnL-trnF, rpl32-trnL, rps16 and matK, etc.) and nuclear gene fragments (ITS, 7FR, 13FR, 97FR, 117FR, 155FR, 165FR, 166FR, 248FR, 302FR and 383FR, etc.) have been used to construct phylogenetic trees to explore phylogenetic relationships in Primulina6,11,12,13,14,15,16,17. However, the support for some branch nodes in the phylogenetic tree of the above cited studies was not high, and the phylogenetic positions of some species remain uncertain, such as in the case of the phylogenetic positions of P. lunglinensis, P. lunglinensis var. amblyosepala, P. liguliformis, P. subulatisepala, P. swinglei and P. nandanensis were inconsistent in different studies. Therefore, screening high-resolution molecular markers is one of the main directions for phylogenetic studies in this genus.
Guizhou Province is one of the most important karst regions in China, and its special geological landscape makes it one of the main areas in which Primulina (Fig. 1), with 22 species (the 10 species of which are endemic to Guizhou)18,19,20. Species in the genus are highly habitat-specialized, and changes in the environment often lead to drastic reductions in the number of individuals in the population8,9,10, which is problematic for the phylogenetic studies of Primulina. For example, the caves of the P. flexusa type locality (Duyun city, Guizhou Province, China) collapsed due to quarrying, and only a few individuals remained.
The chloroplast genome, due to its simple structure, moderate evolutionary rate, conserved sequence, and primarily matrilineal inheritance, is not only effective in improving the support and resolution of phylogenetic trees in phylogenetic studies21,22 but also is effective for identifying natural hybridization, which is one of the very important data in phylogenetic analyses23,24. However, the chloroplast genomic information is under-reported of the Primulina species distributed in Guizhou Province. To date, Feng et al.25, Hsieh et al.26, Hou et al.27, Lu et al.28, Peng et al.29, and Wen et al.30 sequenced and analyzed the chloroplast genomes of several Primulina species. Only 31 chloroplast genomes of 28 Primulina species have been uploaded to the National Center for Biotechnology Information (NCBI) database (https://www.ncbi.nlm.nih.gov/), with most of these species being endemic to Guangxi (39.28%) or Guangdong (28.57%) provinces, China. There are fewer chloroplast genomes endemic to Guizhou Province that have been assembled.
To supplement and preserve molecular data on the endemic and understudied species, the chloroplast genomes of nine Primulina species (including six endemic species of Guizhou Province) (Fig. 2) were sequenced, assembled, and analyzed. Based on these data, (1) comparative analyses of the structure, characteristics, gene content, repetitive sequence content and distribution, and codon usage bias of nine chloroplast genomes were performed to increase knowledge of the Primulina chloroplast genomes; (2) comparative analysis of nucleotide differences in nine Primulina chloroplast genomes and screening of 11 nucleotide mutation hotspot regions that can be used as potential molecular markers were conducted; (3) a phylogenetic tree was constructed based on 36 Primulina chloroplast genomes and two Petrocodon chloroplast genomes, and the phylogenetic relationships of Primulina were briefly explored; and (4) estimated divergence times for these species are based on 36 Primulina chloroplast genomes and two Petrocodon chloroplast genomes.
Results
Chloroplast genome structure, features, and gene content
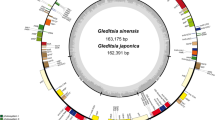
The chloroplast genomes of the nine Primulina species, ranging from 152,869 bp (P. obtusidentata) to 153,364 bp (P. silaniae), displayed a typical quadripartite structure: a large single-copy region (LSC) with a length of 84,333–84,709 bp, a small single-copy region (SSC) with a length of 17,648–17,885 bp, and two inverted repeat regions (IRs) with a length of 25,235–25,522 bp (Fig. 3; Table 1). The GC content of the nine chloroplast genomes were similar (37.55–37.64%), with the content of the LSC and SSC regions ranging from 35.50 to 35.60% and 31.18–31.39%, respectively, and those of the IRs ranging from 43.08 to 43.23%, which were significantly greater than those of the LSC and SSC regions (Table 1). The GC content of the coding sequences (CDSs) were 37.71–38.80%, with the first, second, and third positions of their codons (GC1, GC2, and GC3) being 45.27–45.39%, 38.10–38.19%, and 29.76–29.84%, respectively, showing the regular pattern of GC1 > GC2 > GC3. Notably, different base types were unevenly represented in the three positions of the codons.
Structural map of the nine Primulina chloroplast genomes. Genes outside the circle are transcribed clockwise, and genes inside the circle are transcribed counterclockwise. An photograph of P. flexusa was used as the background image and template. Other species are shown in Supplementary Fig. S1.
The nine chloroplast genomes were each annotated with 132 functional genes, including 87 protein-coding genes (PCGs), 37 transfer ribonucleic acids (tRNAs), and 8 ribosomal ribonucleic acids (rRNAs) (Table 1). Based on the different functions of the genes, these genes were classified into protein synthesis and DNA-replication genes (74), photosynthesis genes (45), other genes coding for proteins (6), and genes of unknown function (7) (Supplementary Table S1). Eighteen unique genes had introns, including clpP1 and ycf3, which had two introns. Due to the inverted repeat nature of the IR regions, the IRb/SSC boundaries had a pseudogene, ycf1, that was partially duplicated and has lost the ability to encode a complete protein. In addition, the rps12 gene in the chloroplast genome consists of two divided parts, 5′-rps12 and 3′-rps12. 5′-rps12 is generally located in the LSC region; because of the expansion or contraction of the IR regions, 3′-rps12 will be located in the LSC region or IR regions 31,32. In this study, 5′-rps12 was located in the LSC region, and 3′-rps12 was located in the IR regions with double copies (Fig. 3).
Identification of simple sequence repeats and interspersed nuclear elements
SSRs have strong polymorphism and good stability and are therefore ideal molecular markers that are widely used in species kinship identification, genetic diversity research, and molecular breeding 33,34. In this study, a total of 375 SSRs were identified (Supplementary Table S2), with 35 (Primulina silaniae) to 50 (P. obtusidentata) SSRs identified for each species, including 15–25 mononucleotide (Mono-) repeats, 6–13 dinucleotide (Di-) repeats, 2–5 trinucleotide (Tri-) repeats, 6–12 tetranucleotide (Tetra-) repeats, 0–2 pentanucleotide (Penta-) repeats, and 0–2 hexanucleotide (Hexa-) repeats (Fig. 4A). The repeat units were mainly composed of adenine (A) and thymine (T), with a predominance of mononucleotide repeats A/T, followed by dinucleotide repeats AT/AT (Fig. 4B). Among the nine species, all six types of SSRs were detected only in the chloroplast genome of P. silaniae (Fig. 4A). The SSRs were mainly distributed in the LSC region (Fig. 4C). The percentages of the length of SSRs in the LSC, SSC, and IR regions were calculated separately, and it was found that there was no regular pattern in the percentage of the length of SSRs in these regions (Fig. 4D).
Comparative analysis of SSRs and INEs in the nine Primulina chloroplast genomes. (A) Number of SSRs of six types; (B) Number of SSRs in different repeat units; (C) Number of SSRs in different regions; (D) Percentage of the length of SSRs in different regions; (E) Number of INEs of four types; (F) Length of four types of INEs, from left to right: forward repeats, palindromic repeats, reverse repeats, and complement repeats.
INEs were dispersed throughout the genome and were of four types: forward repeats, reverse repeats, palindromic repeats, and complement repeats 35. In this study, a total of 375 INEs were identified in nine chloroplast genomes (Supplementary Table S2), with the number of INEs in each species ranging from 29 (Primulina liboensis) to 53 (P. flexusa), including 12–28 forward repeats, 16–25 palindromic repeats, 0–2 reverse repeats, and 0–2 complement repeats (Fig. 4E). Forward repeats and palindromic repeats were the most numerous; reverse repeats and complement repeats were absent in some species (Fig. 4E). The INEs ranged from 30 to 99 bp in length, with only one forward repeat of more than 90 bp present in P. malingheensis (Fig. 4F).
Codon usage bias analysis
Codon usage bias refers to the presence of a particular codon or codons in a species that is used more frequently than its synonymous codon and can directly affect the efficiency and accuracy of protein synthesis 36,37. In this study, each species had 58 CDSs of no less than 300 bp in length, of which 64 codons were detected, with the total number ranging from 24,243 (Primulina malingheensis) to 24,714 (P. flexusa) (Supplementary Table S3). UGA (stop codon) and AUU (encoding isoleucine) were, respectively, the least and most used codons of the nine species; cysteine and leucine were, respectively, the least and most common amino acids of the nine species (Supplementary Table S3).
The nine chloroplast genomes were statistically analyzed and visualized based on relative synonymous codon usage (RSCU) (Fig. 5). Thirty of the 64 codons with RSCU > 1 were highly preferred codons, of which 29 ended in A/U, while 32 codons with RSCU < 1 were less preferred codons, of which 29 ended in G/C. Notably, methionine and tryptophan were encoded by the unique codons AUG and UGG only, with RSCU = 1, and without bias.
RSCU values for stop codons and codons encoding 20 amino acids in nine chloroplast genomes. Each column in the bar chart represents a species, from left to right: Primulina flexusa, P. fordii, P. liboensis, P. malingheensis, P. obtusidentata, P. secundiflora, P. silaniae, P. tenuituba, and P. vestita. "#" denotes codons with RSCU > 1.
Comparative analysis of SC/IR boundaries
The nine chloroplast genomes displayed a typical quadripartite structure with four boundaries between the LSC, SSC, and IR regions (SC/IR boundaries) (Fig. 6). Comparative analyses revealed that the SC/IR boundaries were relatively conserved among the nine species. The LSC/IRb boundary (JLB line) is located in the rps19 gene, the IRb/SSC boundary (JSB line) is located in the overlap between the ndhF gene and the pseudogene ycf1, the SSC/IRa boundary (JSA line) is located in the ycf1 gene, and the IRa/LSC boundary (JLA line) is located in the intergenic sequences of the rpl2 gene and the trnHGUG gene (Fig. 6).
At the LSC/IRb boundary, the rps19 gene spanned the JLB line, with very few base pairs extending into the IRb region. It can be divided into three categories: (1) 251 bp in the LSC region and 28 bp in the IRb region, with this category containing Primulina flexusa, P. malingheensis, P. secundiflora, P. tenuituba, and P. vestita; (2) 249 bp in the LSC region and 30 bp in the IRb region, with this category containing P. liboensis, P. fordii, and P. obtusidentata; and (3) 247 bp in the LSC region and 32 bp in the IRb region, with this category containing only P. silaniae.
At the IRb/SSC boundary, the ndhF gene spanned the JSB line, with very few base pairs extending into the IRb region. The ndhF genes of Primulina flexusa, P. liboensis, P. secundiflora, P. tenuituba, and P. vestita all had 107 bp on the left side of the JSB line; on the right side of the JSB line, they had 2185 bp for P. liboensis and 2179 bp for the remaining four species. The ndhF genes of P. fordii, P. malingheensis, P. obtusidentata, and P. silaniae all had 2195 bp on the right side of the JSB line; on the left side of the JSB line, it was 70 bp for P. fordii and P. obtusidentata and 91 bp for P. malingheensis and P. silaniae.
At the SSC/IRa boundary, the ycf1 gene spanned the JSA line, with very few base pairs extending into the IRa region. The ycf1 genes of Primulina flexusa, P. liboensis, P. secundiflora, P. tenuituba, and P. vestita all had 802 bp on the left side of the JSA line; on the right side of the JSA line, the length ranges from 4454 to 4664 bp. The ycf1 genes of P. fordii, P. malingheensis, P. obtusidentata, and P. silaniae ranged in length from 765 to 786 bp on the right side of the JSA line and from 4644 to 4704 bp on the left side of the JSA line.
At the IRa/LSC boundary, the rpl2 gene was located to the left of the JLA line, at 83 bp (Primulina flexusa, P. malingheensis, P. secundiflora, P. tenuituba, and P. vestita), 85 bp (P. liboensis, P. fordii, and P. obtusidentata), or 87 bp (P. silaniae); the trnHGUG gene was located to the right of the JLA line, at 11 bp (P. liboensis), 8 bp (P. flexusa, P. malingheensis, P. secundiflora, P. tenuituba, and P. vestita), 4 bp (P. fordii and P. obtusidentata), or 2 bp (P. silaniae).
Comparative analysis of the chloroplast genomes
In this study, the chloroplast genomes of nine species were aligned and visualized using Primulina flexusa as the reference sequence (Fig. 7). The results showed that the nucleotide differences in the LSC and SSC regions were greater than those in the IR regions, and the noncoding regions were more differentiated than the coding regions. In the coding regions, the exons of all genes are relatively conserved except ycf1. Among the noncoding regions, the regions with large nucleotide differences were mainly the spacer region and introns of 5000–15,000 bp, 25,000–35,000 bp, 40,000–50,000 bp, 70,000–80,000 bp, and 110,000–120,000 bp.
To quantify the nucleotide polymorphisms in the nine chloroplast genomes in more detail and to identify the mutation hotspot regions among them, we calculated the nucleotide polymorphisms (Pi) in a window of 800 bp in length from the aligned matrices of the nine chloroplast genomes. The results showed that the Pi values ranged from 0 to 0.02267, with the lowest Pi value of 0.01583 in the window with the top 5% of the Pi values. A total of 11 mutational hotspot regions that could serve as potential molecular markers were identified (Pi > 0.01583): trnHGUG-psbA, trnKUUU-rps16, rps16-trnQUUG, petN-psbM, trnEUUC-trnTGGU-psbD, clpP1-psbB, ndhF-rpl32-trnLUAG, ndhD-psaC-ndhE, and 3 regions in ycf1 (0–338 bp, 1900–3892 bp, and 4292–4955 bp) (Fig. 8). All 11 mutation hotspot regions were located in the LSC and SSC regions, and the Pi values of the IR regions were lower and more conserved.
Nucleotide polymorphisms (Pi) of the nine Primulina chloroplast genomes (window length: 800 bp; step size: 200 bp). The red dashed line shows the lowest Pi value for the window with the top 5% of the Pi values, and regions with Pi values higher than the red dashed line are identified as mutation hotspot regions.
Phylogenetic analysis
The topology of the phylogenetic tree constructed using maximum likelihood (ML) and Bayesian inference (BI) based on the 38 chloroplast genomes in this study is consistent, and the two are shown together (Fig. 9). Thirty-six Primulina species were clustered in the phylogenetic tree as a fully supported clade. In the study of Kong et al.13, Primulina was classified into Clades A, B, C, and D. The phylogenetic tree constructed in the present study contained three major clades that received full support, roughly corresponding to Clades B, C, and D as delineated by Kong et al. The Kong et al. study differs from the present study in that P. secundiflora was located in Clade D and clustered with P. vestita in a branch that received little support (BS = 78; PP < 0.50) in the present study, whereas in Kong et al., P. secundiflora was located in Clade B and formed a fully supported branch with a branch comprising P. maguanensis and P. grandibracteata.
Phylogenetic tree constructed based on 38 complete chloroplast genomes. The BI tree and the ML tree are shown together, and posterior probabilities (PP) and bootstrap searches (BS) are displayed on the branch and are indicated by "-" for PP < 0.50 or BS < 50. The different background colors indicate the three main branches, which correspond to Clade B, Clade C, and Clade D in the study by Kong et al. 13. The chloroplast genomic data added in this study are shown in red font.
Four of the nine Primulina species analysed in this study are located in Clade B, one is in Clade C, and four are in Clade D, and the genetic distances among them further support this result (Fig. 10). Among the three new taxa of Primulina reported in Guizhou Province, China, in recent years, P. silaniae and P. malingheensis are located in Clade B, and P. flexusa is located in Clabe D. In the available chloroplast genome data, P. silaniae is located at the base of Clade B. Two points are worth noting: first, P. liboensis (PP954905) in Clade C did not cluster with P. liboensis (MF177039) but instead formed a fully supported clade with P. sclerophylla; second, P. tenuituba (PP954910) in Clade D did not cluster with P. tenuituba (MW245830) but instead formed a highly supported (BS = 99; PP = 0.94) clade with P. flexusa.
Divergence time estimation
Estimated divergence times for these species are based on 36 Primulina chloroplast genomes and two Petrocodon chloroplast genomes. Primulina and Petrocodon diverged about 14.57 million years ago (Mya), with a 95% highest posterior density (HPD) of 11.06–18.76 Mya (Fig. 11). The three major branches of Primulina begin to diverge in the Miocene, with most of the species of clade B and clade C formed in the Miocene and Pliocene, and most of the species of clade D formed in the Pleistocene. Species of Primulina in Guizhou Province diverged from 6.54 to 0.52 Mya, with most of them formed in the Pleistocene and diverging rapidly in a short period of time (divergence time: 2.48 to 0.52 Mya).
Discussion
In this study, the chloroplast genomes of the nine Primulina species were relatively conserved in size, structure, gene type, and content, and all nine chloroplast genomes, ranging from 152,869 to 153,364 bp, displayed a quadripartite structure, which aligns with the general range of chloroplast genome sizes of 115–165 kp for photosynthesizing plants 38. Each chloroplast genome was annotated with 87 PCGs, 37 tRNAs, and 8 rRNAs, of which 23 genes possessed introns, in agreement with the findings of Hsieh et al. 26. Notably, Feng et al. annotated the chloroplast genomes of three Primulina species [P. eburnea (MF177038), P. huaijiensis (MF472012), and P. linearifolia (MF472013)] 25. In contrast to the findings of the present study, Feng et al. found that the rps19 and trnGUCC genes were absent, and only one exon was present in the rpl16, petB, and petD genes according to their annotation results for the three chloroplast genomes. Annotating the chloroplast gene sequences of these three species using the reference genomes of the present studyrevealed that they included the rps19 and trnGUCC genes, and that the rpl16, petB and petD genes had two exons and one intron. Differences in the reference genome annotation information are likely responsible for the differences in the annotation results.
SSRs are the most commonly used and effective molecular markers34. The SSRs identified in the nine chloroplast genomes in this study were dominated by mononucleotide, dinucleotide, trinucleotide, and tetranucleotide repeats. The repetitive units were mainly composed of either A or T, with A/T repeats and AT/AT repeats predominating, consistent with the major SSR repetitive unit types identified in the chloroplast genomes of other species in Gesneriaceae26,39. In contrast to the chloroplast genomic SSRs identified by previous research on Primulina26,39, SSRs of all six length types were detected in only P. silaniae. This may be related to the particular systematic position of P. silaniae, which is at the base of Clade B in the available Primulina chloroplast genomes (Fig. 9)13, and which may be a transitional taxon between Clades A and B. At present, there is a lack of data on the chloroplast genomes of the taxa in Clade A, as delineated by Kong et al. In the future, the chloroplast genomes of taxa in this clade could be added to study the similarities and differences of SSRs among different clades. The SSR loci identified in this study were mainly distributed in the LSC region, but there was no pattern in the percentage of the length in the LSC, SSC, and IR regions, and there was no preference for the LSC region. The distribution of SSRs in the LSC, SSC, and IR regions is likely to be random.
Codon usage bias analysis revealed that the frequencies of the 64 codons in the nine chloroplast genomes were essentially similar, with 30 highly preferred codons, 2 without biased codons, and 32 fewer preferred codons detected. Highly preferred codons ended in A/U, except for UUG (encoding leucine), while less preferred codons mostly ended in C/G, corroborating the lower GC3 observation (Table 1). These results are similar to those of previous studies36,39,40, supporting a high degree of consistency in chloroplast genome codon usage bias in Gesneriaceae, as suggested by Hsieh et al.26, and further supporting the hypothesis that codons in higher plants tend to have A/T(U) endings41,42,43.
Expansion, contraction, and loss of the IR regions cause changes in the size of the chloroplast genome, leading to gene duplication, gene deletion, and the generation of pseudogenes44,45,46,47. The SC/IR boundaries of the nine chloroplast genomes in this study were relatively conserved, and no obvious expansion or contraction of the IR regions was detected, which is generally consistent with the results of SC/IR boundary analyses in other species of Gesneriaceae39. Although the SC/IR boundaries of the nine chloroplast genomes were relatively conserved, they remain distinct in some aspects and are broadly consistent with the phylogenetic relationships. The SC/IR boundaries of Primulina flexusa, P. secundiflora, P. tenuituba, and P. vestita were nearly identical and distinguishable from those of the other five species, perhaps because of the close affinity of these four species.
Due to the simple structure, moderate evolutionary rate, and conserved sequence of the chloroplast genome, many genes, introns, and spacer regions have been exploited to construct phylogenetic relationships at different taxonomic levels48,49,50. In studies of Primulina, trunk branches of phylogenetic trees constructed using common molecular markers did not receive high support13, and the development of additional molecular markers is therefore imperative. In this study, the alignment of the nine chloroplast genomes showed that the IR regions were relatively conserved, and the regions with large nucleotide differences were the ycf1 gene and the noncoding regions of the LSC and SSC regions. After further quantification of nucleotide polymorphisms, eleven mutational hotspot regions were identified: trnHGUG-psbA, trnKUUU-rps16, rps16-trnQUUG, petN-psbM, trnEUUC-trnTGGU-psbD, clpP1-psbB, ndhF-rpl32-trnLUAG, ndhD-psaC-ndhE, and three regions of ycf1. The extent of the three highly mutated regions of the ycf1 gene was localized, of which the 0–338 bp and 4292–4955 bp regions were in general agreement with the findings of Hsieh et al.26. Both regions were < 800 bp in size, which is convenient for amplification and sequencing and is conducive to the development of molecular markers for Primulina.
The phylogenetic relationships of the 36 Primulina species in this study are largely consistent with those of Kong et al.13, and the three main clades correspond roughly to Clades B, C, and D, as delineated by Kong et al.; however, there are still some differences. Primulina secundiflora was located in Clade D in this study, with the P. secundiflora material collected from its type locality, Kweichow, Ching-Che, Hwa-Chiao (now Huaqiao village, Baihuahu town, Guanshanhu District, Guizhou Province, China)51. Kong et al. reported that P. secundiflora was located in clade B and formed a fully supported branch with branches comprising P. maguanensis and P. grandibracteata. However, the small corolla and small linear bracts of P. secundiflora differ markedly from the larger corolla and large, suborbicular, or broadly ovate to narrowly ovate bracts of P. maguanensis and P. grandibracteata52,53. During the compilation of Plants of Gesneriaceae in China and Gesneriaceae of South China, researchers identified a plant collected in Xingyi city, Guizhou Province, China, as P. secundiflora54,55, which was published by our research team as a new taxon and named P. malingheensis20 because of its larger corolla and larger and ovate bracts, which are clearly different from those of P. secundiflora. Primulina malingheensis is morphologically more similar to P. maguanensis and P. grandibracteata, and in this study, it is located in Clade B13. Kong et al. likely misidentified P. malingheensis as P. secundiflora because of the records of Plants of Gesneriaceae in China and Gesneriaceae of South China. There are many species in Primulina, and the classification is confusing. Li14 divided the species in the genus that are phylogenetically close and have similar morphological characteristics into several complexes, but the definition of species within the complexes is still highly problematic. According to Li14, P. secundiflora, P. tenuituba, P. vestita, and P. flexusa in this study should belong to the P. secundiflora complex, while P. liboensis and P. sclerophylla should belong to the P. sclerophylla complex. Within the P. secundiflora complex, P. tenuituba is widely distributed in Guizhou Province, Hunan Province, and Hubei Province, China, and there are some differences in the morphological characteristics of P. tenuituba from different localities, with those from Shennongjia Forestry District, Hubei Province being more similar to P. vestita and slightly different from P. tenuituba from Huayuan County, Hunan province. In this study, P. tenuituba was represented by only two populations from Guizhou Province (Guiyang city and Tongren city) and did not form a monophyletic lineage. The interspecific relationships of the P. secundiflora complex need to be investigated by adding additional samples of P. tenuituba from Hunan province and Hubei province. The P. sclerophylla complex is widely distributed in the Guangxi Zhuang Autonomous Region and southwestern Guizhou Province, China, and contains 20 species, the vast majority of which are distinguished from each other using continuously variable traits14. In this study, the P. sclerophylla complex involved only two P. liboensis populations from Pingtang County and southeastern Guizhou Province and one P. sclerophylla population from Hechi city, Guangxi Zhuang Autonomous Region, and the two P. liboensis populations did not form a monophyletic lineage. The interspecific relationships within the P. sclerophylla complex need to be investigated by adding additional samples from the type locality of the species within the complex.
In this study, the divergence time of Primulina was 14.57 Mya (with a 95% HPD of 11.06–18.76 Mya) (Fig. 11), which is much smaller than that of previous studies 56,57. We speculate that the reason for this discrepancy may be that the studies of Woo et al.56 and Perret et al.57 did not include close relatives of Primulina in their analyses. In their study, the sister taxa of Primulina are species of Streptocarpus, which is more distantly related to Primulina, and much data on other genera is missing between the two genera8, leading to the speculation of an older divergence time of Primulina. In addition, we hypothesize that because the species of Clade A, which diverged earlier, was missing from the present study, a younger crown group age of Primulina (13.06 Mya, with a 95% HPD of 9.90–16.37 Mya) was obtained than in the previous study (14.14 Mya, with a 95% HPD of 10.79–19.09 Mya) 13. In subsequent studies, the chloroplast genome of the species of Clade A could be added to estimate the divergence time to verify whether the speculation is correct.
Conclusion
In this study, the complete chloroplast genomes of nine Primulina species were assembled, of which the chloroplast genomes of seven species were reported for the first time. A total of 375 SSRs and 375 INEs were identified in nine chloroplast genomes, providing additional molecular data for Primulina that can be used to analyze genetic diversity. The usage frequency of codons was broadly similar across the nine chloroplast genomes, with highly preferred codons of the same 30 types, which is similar to the results of a previous study. The SC/IR boundaries were relatively conserved, and no obvious expansion or contraction of the IR regions was detected. In addition, a total of 11 mutational hotspot regions that could serve as potential molecular markers were identified: trnHGUG-psbA, trnKUUU-rps16, rps16-trnQUUG, petN-psbM, trnEUUC-trnTGGU-psbD, clpP1-psbB, ndhF-rpl32-trnLUAG, ndhD-psaC-ndhE, and three regions of ycf1 (0 bp–338 bp, 1,900 bp–3,892 bp, and 4,292 bp–4,955 bp). The two mutational hotspot regions near the 3′ and 5′ ends of the ycf1 gene were of appropriate size and are recommended as molecular markers for phylogenetic studies of Primulina. The phylogenetic tree based on complete chloroplast genomes strongly supports Primulina as a monophyletic taxon, but the interspecific relationships of the P. secundiflora complex and the P. sclerophylla complex still need to be investigated by adding more samples from more provenances. The results of molecular clock analyses indicate that the three major branches of Primulina begin to diverge in the Miocene, and the number of species proliferated in the Pliocene and Pleistocene. Most of the species of Primulina in Guizhou Province were formed in the Pleistocene and rapidly diverged within a short period of time.
Materials and methods
Plant materials and DNA extraction
The materials of the nine species were collected from Guizhou Province, China (Table 2). Mature leaves free of pests and diseases were collected in the field, washed, cleaned, sealed, and stored in a refrigerator at − 40 °C. The collection of plant materials was collected following international, national, and local legislation. Dr. Xinxiang Bai identified these plants, and the voucher specimens were stored in the Tree Herbarium of Guizhou University College of Forestry (GZAC). Total DNA was extracted from each of the nine samples using the GTAB method. DNA quality and purity were tested using 1% agarose gel electrophoresis and a fluorescence quantification instrument (Thermo Qubit 4.0).
Chloroplast genome sequencing, assembly and annotation
Sangon Biotech Co., Ltd. (Shanghai, China) used the Hieff NGS® MaxUp II DNA Library Prep Kit for Illumina® (Yeasen Biotechnology Co., Ltd., Shanghai, China) to prepare DNA libraries based on DNA samples of acceptable purity. The concentrations of the DNA libraries were determined using a fluorescence quantification instrument (Thermo Qubit 4.0). DNA libraries were sequenced using the Illumina MiSeq System (Illumina) to obtain the raw data. The raw data were cropped and filtered using FASTQ 0.3658 software to obtain clean data. Comparison of clean data with published sequences of Primulina in the NCBI database. De novo genome assembly was performed from the clean data using SOAPdenovo 259 and NOVOPlasty60. Primulina liboensis (GenBank accession number: NC_036101) was used as the reference genome, and the assembled chloroplast genome was annotated using the GeSeq online tool (https://chlorobox.mpimp-golm.mpg.de/geseq.html)61. CDS start and stop codons in the annotation file were manually checked and adjusted using Geneious 9.0.262. The type, length, and anticodon name of the tRNA genes in the annotation file were manually checked and adjusted using tRNAscan SE (http://lowelab.ucsc.edu/tRNAscan-SE/)63. Annotation information was visualized using OGDRAW v.1.3.16364.
Repeat sequence analysis
SSRs were identified using Krait65 with the following parameter settings: mononucleotide ≥ 10, dinucleotide ≥ 5, trinucleotide ≥ 4, tetranucleotide ≥ 3, pentanucleotide ≥ 3, and hexanucleotide ≥ 3. INEs were identified using the REPuter online tool (https://bibiserv.cebitec.uni-bielefeld.de/reputer)66, with the minimum repeat size set to 30 bp and a sequence identity of 90% (Hamming distance of three).
Codon usage bias analysis
The relative synonymous codon usage (RSCU) is the ratio between the number of real and theoretical uses of a codon67. An RSCU < 1 indicates that the codon is used infrequently and is a less preferred codon; an RSCU > 1 indicates that the codon is used frequently and is a highly preferred codon; and an RSCU = 1 indicates that the codon is used without bias. Since short sequences cannot correctly calculate the effective codon number68, to reduce errors, CDSs with sequence lengths of no less than 300 bp were selected for this study and analyzed for RSCU using CodonW 1.4.269.
SC/IR boundary analysis, chloroplast genome comparison analysis, and mutational hotspot region identification
Comparison and visualization of the SC/IR boundaries of nine chloroplast genomes were performed using CPJSdraw70. The nine chloroplast genomes were aligned using the mVISTA online tool (https://genome.lbl.gov/vista/mvista/submit.shtml)71, Primulina flexusa as a reference, and the alignment program LAGAN. Nucleotide polymorphisms (Pi) of the aligned sequence matrix were analyzed in DnaSP v.6.12.033272 using sliding windows with a window length of 800 bp and a step size of 200 bp. Windows in the top 5% of Pi values were considered mutational hotspot regions.
Phylogenetic analysis
Two species of the closely related genus Petrocodon Hance, P. multiflorus and P. coriaceifolius, were used as outgroups to analyze the phylogenetic relationships of the chloroplast genomes of 36 species of Primulina. Except for the nine chloroplast genomes included in this study, the remaining chloroplast genomes used samples downloaded from GenBank (Supplementary Table S4). Sequence alignment was performed using MAFFT v.7.5.1.173 and manually adjusted using MEGA_11.0.1374 to obtain the chloroplast genome matrix. The index of substitution saturation (Iss) of the chloroplast genome matrix was assessed using DAMBE v5.3.1 75, which showed that the matrix was not saturated and could be used to construct a phylogenetic tree with Iss (0.0800) < Iss.c (0.8610), P = 0.0000 < 0.0500. The optimal models for ML and BI (TVM + F + I + I + R6 and GTR + F + I + G4, respectively) were assembled using Phylosuite v.1.2.376 according to the AICc criterion. ML and BI were also performed using Phylosuite v.1.2.3. ML performed 1,000 standardized BS to assess branch support. BI was performed for at least 10 million generations, samples were collected every 1,000 generations, and PP were calculated to assess branch reliability. Genetic distances were assembled by MEGA_11.0.1374, and BS was performed 1,000 times.
Divergence time estimation
Since no records of any fossil collections of the family Bitternidae were searched in the Paleobiology Database (https://paleobiodb.org/#/, accessed 17 October 2024), secondary calibrations were used to calibrate the clock in this study. Calibration of the clock using the crown group age of Clade B (5.49–10.56 Mya), Clade C (6.26–11.92 Mya), and Clade D (7.11–13.01 Mya) in the study by Kong et al.13. We estimated the divergence times based on the 36 Primulina chloroplast genomes and two Petrocodon chloroplast genomes using BEAST 1.8.377 under Bayesian relaxed clock. The outgroups were two species of Petrocodon, with nucleotide substitution modeled as GTR + G + I and the tree priori selecting the Yule process. Two independent analyses were performed in BEAST 1.8.3 on 100 million generations of Markov chain Monte Carlo (MCMC), sampled once per 10,000 generations. Convergence was assessed using TRACER 1.678 and checked that the effective sample size (ESS) for each parameter was greater than 200. Discard the first 25% of trees as burn-in.
Data availability
The data generated in the study were uploaded to the NCBI database (https://www.ncbi.nlm.nih.gov/) under the accession number PP954903-PP954911.
References
Lu, Y. R. Karst in China—A world of improbable peaks and wonderful caves 25–44 (Higher Education Press, 2012).
Crowther, J. Ecological observations in a tropical karst terrain, west Malaysia. I. Variations in topography, soils and vegetation. J. Biogeogr. 9(1), 65–78 (1982).
Cao, J. H., Yuan, D. X. & Pan, G. X. Some soil features in karst ecosystem. Adv. Earth Sci. 18(1), 37–44 (2003).
Clements, R., Sodhi, N. S., Schilthuizen, M. & Ng, P. K. Limestone karsts of southeast Asia: Imperiled arks of biodiversity. Bioscience 56(9), 733–742 (2006).
Zhu, H. The karst ecosystem of southern China and its biodiversity. Trop. For. 35(S1), 44–47 (2007).
Xu, M. Z., Yang, L. H., Kong, H. H., Wen, F. & Kang, M. Congruent spatial patterns of species richness and phylogenetic diversity in karst flora: Case study of Primulina (Gesneriaceae). J. Syst. Evol. 59(2), 251–261 (2021).
Wen, F. et al. The checklist of Gesneriaceae in China. Facilitated by the Gesneriad Conservation Center of China. Published on the Internet; http://gccc.gxib.cn/cn/about-68.aspx. Retrieved 20 July 2024 (2024).
Weber, A. et al. Molecular systematics and remodeling of Chirita and associated genera (Gesneriaceae). Taxon 60(3), 767–790 (2011).
Qi, Q. W., Hao, Z., Tao, J. J. & Kang, M. Diversity of calcium speciation in leaves of Primulina species (Gesneriaceae). Biodivers. Sci. 21(6), 715–722 (2013).
Xu, W. B., Guo, J., Pan, B., Liu, Q. & Liu, Y. Diversity and distribution of Gesneriaceae in China. Guihaia 37(10), 1219–1226 (2017).
Yang, G. W. Systematics of Primulina sensu lato (Gesneriaceae) (Henan Agricultural University, 2016).
Guo, J., Pan, B., Liu, J., Xu, W. B. & Chung, K. F. Three new species of Primulina (Gesneriaceae) from limestone karsts of China based on morphological and molecular evidence. Bot. Stud. 56(1), 34–46 (2015).
Kong, H. H. et al. Both temperature fluctuations and east Asian monsoons have driven plant diversification in the karst ecosystems from southern China. Mol. Ecol. 26(22), 6414–6429 (2017).
Li, P. W. A systematic study of the genus Primulina (Gesneriaceae) (University of Chinese Academy of Sciences, 2019).
Xu, W. B., Chang, H., Huang, J. & Chung, K. F. Molecular systematics of Chiritopsis–like Primulina (Gesneriaceae): one new species, one new name, two new combinations, and new synonyms. Bot. Stud. 60(1), 18–38 (2019).
Yang, L. H., Kong, H. H., Huang, J. P. & Kang, M. Different species or genetically divergent populations? Integrative species delimitation of the Primulina hochiensis complex from isolated karst habitats. Mol. Phylogenet. Evolut. 132, 219–231 (2019).
Ning, Z. N. Systematic taxonomyand resource conservation on the Primulina Hance (Gesneriaceae) (South China Agricultural University, 2017).
Xiong, K. N. et al. A typical remote sensing–GIS Study of karst desertification–A case study of Guizhou Province (Geology Press, 2002).
Huang, M., Li, M. J., Huang, H., Zhang, J. Q. & Bai, X. X. Species diversity and geographic distribution of wild Gesneriaceae in Guizhou. Guihaia 42(2), 210–219 (2022).
Zhou, Y. L., Wen, F., Li, Q. Y., Wang, J. Q. & Bai, X. X. Primulina malingheensis (Gesneriaceae), a new species from a karst area in Guizhou province, China. Annales Botanici Fennici 60(1), 61–65 (2023).
Kress, W. J. & Erickson, D. L. A two–locus global DNA barcode for land plants: The coding rbcL gene complements the non–coding trnH–psbA spacer region. Plos One 2(6), e508. https://doi.org/10.1371/journal.pone.0000508 (2007).
Möller, M., Forrest, A., Wei, Y. G. & Weber, A. A molecular phylogenetic assessment of the advanced Asiatic and malesian didymocarpoid gesneriaceae with focus on non–monophyletic and monotypic genera. Plant Syst. Evolut. 292(3), 223–248 (2011).
Ke, S. J. et al. Cymbidium × shangrilaense (Orchidaceae; Epidendroideae), a new natural hybrid from China: Evidence from morphology and molecular analyses. Phytotaxa 561(2), 183–190 (2022).
Tang, W. Natural hybridization between Sorbus pohuashanensis Hedl. and S. discolor Maxim (Chinese Academy of Forestry, 2016).
Feng, C., Xu, M., Feng, C., Von Wettberg, E. J. & Kang, M. The complete chloroplast genome of Primulina and two novel strategies for development of high polymorphic loci for population genetic and phylogenetic studies. BMC Evolut. Biol. 17(1), 224. https://doi.org/10.1186/s12862-017-1067-z (2017).
Hsieh, C. L., Xu, W. B. & Chung, K. F. Plastomes of limestone karst gesneriad genera Petrocodon and Primulina, and the comparative plastid phylogenomics of Gesneriaceae. Sci. Rep. 12(1), 15800. https://doi.org/10.1038/s41598-022-19812-2 (2022).
Hou, N., Wang, G., Li, C. R. & Luo, Y. Characterization of the complete chloroplast genomes of three Chirita species (C. brachytricha, C. eburnea & C. liboensis) endemic to China. Conserv. Genet. Resour. 10(4), 597–600 (2018).
Lu, Y. B., Qin, X. M., Huang, Z. P., Zhang, Q. & Shen, L. N. The complete chloroplast genome of the extremely drought–resistant plant Primulina ophiopogoides. Mitochondrial DNA Part B 5(3), 2271–2272 (2020).
Peng, T. et al. The complete chloroplast genome of a karst–dwelling plant Primulina tenuituba (Gesneriaceae). Mitochondrial DNA Part B 7(3), 507–509 (2022).
Wen, H. C., Su, Y. J., Xie, A., Lin, C. & Wei, W. Characteristics and phylogenetic analysis of the complete chloroplast genome of Primulina hedyotidea. Mitochondrial DNA Part B 8(9), 1007–1011 (2023).
Zaita, N., Torazawa, K., Shinozaki, K. & Sugiura, M. Trans splicing in vivo: Joining of transcripts from the ‘divided’ gene for ribosomal protein S12 in the chloroplasts of tobacco. Febs Lett. 210(2), 153–156 (1987).
Ping, J. Y., Zhu, M., Su, Y. J. & Wang, T. Molecular evolution of chloroplast gene rps12 in ferns. Plant Sci. J. 38(1), 1–9 (2020).
Powell, W., Machray, G. C. & Provan, J. Polymorphism revealed by simple sequence repeats. Trends Plant Sci. 1(7), 215–222 (1996).
Rajeev, K. V., Andreas, G. & Mark, E. S. Genic microsatellite markers in plants: Features and applications. Trends Biotechnol. 23(1), 48–55 (2005).
Jelinek, W. R. et al. Ubiquitous, interspersed repeated sequences in mammalian genomes. Proc. Natl. Acad. Sci. 77(3), 1398–1402 (1980).
Sharp, P. M., Stenico, M., Peden, J. F. & Lloyd, A. T. Codon usage: mutational bias, translational selection, or both?. Biochem. Soc. Trans. 21(4), 835–841 (1993).
Zhou, Z. P. et al. Codon usage is an important determinant of gene expression levels largely through its effects on transcription. Proc. Natl. Acad. Sci. 113(41), E6117–E6125 (2016).
Jansen, R. K. et al. Methods for obtaining and analyzing whole chloroplast genome sequences. Methods Enzymol. 395, 348–384 (2005).
Cui, Y. F. et al. Plastome evolution and phylogenomics of Trichosporeae (Gesneriaceae) with its morphological characters appraisal. Front. Plant Sci. 14, 1160535. https://doi.org/10.3389/fpls.2023.1160535 (2023).
Gu, L., Su, T., An, M. T. & Hu, G. X. The complete chloroplast genome of the vulnerable Oreocharis esquirolii (Gesneriaceae): Structural features, comparative and phylogenetic analysis. Plants 9(12), 1692. https://doi.org/10.3390/plants9121692 (2020).
Campbell, W. H. & Gowri, G. Codon usage in higher plants, green algae, and cyanobacteria. Plant Physiol. 92(1), 1–11 (1990).
Yan, R. R. et al. Comparative analyses of Linderniaceae plastomes, with implications for its phylogeny and evolution. Front. Plant Sci. 14, 1265641. https://doi.org/10.3389/fpls.2023.1265641 (2023).
Yan, R. R., Gu, L., Qu, L., Wang, X. Y. & Hu, G. X. New insights into phylogenetic relationship of Hydrocotyle (Araliaceae) based on plastid genomes. Int. J. Mol. Sci. 24(23), 16629. https://doi.org/10.3390/ijms242316629 (2023).
Tsudzuki, J. et al. Chloroplast DNA of black pine retains a residual inverted repeat lacking rRNA genes: Nucleotide sequences of trnQ, trnK, psbA, trnI and trnH and the absence of rps16. Mol. Gener. Genet. 232(2), 206–214 (1992).
Saski, C. et al. Complete chloroplast genome sequence of Gycine max and comparative analyses with other legume genomes. Plant Mol. Biol. 59(2), 309–322 (2005).
Chumley, T. W. et al. The complete chloroplast genome sequence of Pelargonium × hortorum: Organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol. Biol. Evolut. 23(11), 2175–2190 (2006).
Zhu, Q. L. et al. Advances on mitochondria and chloroplast genomes of cucurbit crop. China Cucurbits Veg. 29(8), 1–8 (2016).
Nock, C. J. et al. Chloroplast genome sequences from total DNA for plant identification. Plant Biotechnol. J. 9(3), 328–333 (2011).
Su, T. et al. Chloroplast genome of Salvia sect. Drymosphace: Comparative and phylogenetic analysis. Diversity 14(5), 324. https://doi.org/10.3390/d14050324 (2022).
Hu, G. X. et al. Integrating DNA sequences with morphological analysis clarifies phylogenetic position of Salvia grandifolia (Lamiaceae): An enigmatic species endemic to southwestern China. Int. J. Plant Sci. 181(8), 787–799 (2020).
Chun, W. Gesneriacearum plantae novae sinicarum. Sunyatsenia 6(3–4), 293–294 (1946).
Xu, H., Li, Z. Y. & Jiang, H. A new species of Chirita (Gesneriaceae) from Yunnan, China. Bot. J. Linn. Soc. 158(2), 269–273 (2008).
Li, J. M. & Möller, M. Chirita grandibracteata (Gesneriaceae), a new species from Yunnan, China. Annales Botanici Fennici 46(2), 125–129 (2009).
Li, Z. Y. & Wang, Y. Z. Plants of Gesneriaceae in China (Henan Sciences and Technology Publishing Press, 2005).
Wei, Y. G. et al. Gesneriaceae of South China (Guangxi Sciences and Technology Publishing House, 2010).
Woo, V. L., Funke, M. M., Smith, J. F., Lockhart, P. J. & Garnock-Jones, P. J. New World origins of southwest Pacific Gesneriaceae: Multiple movements across and within the South Pacific. Int. J. Plant Sci. 172(3), 434–457 (2011).
Perret, M., Chautems, A., De Araujo, A. O. & Salamin, N. Temporal and spatial origin of Gesneriaceae in the new world inferred from plastid DNA sequences. Bot. J. Linn. Soc. 171(1), 61–79 (2013).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. Fastp: An ultra–fast all–in–one FASTQ preprocessor. Bioinformatics 34(17), 884–890 (2018).
Luo, R. B. et al. SOAPdenovo2: An empirically improved memory–efficient short–read de novo assembler. Gigascience 1(1), 18. https://doi.org/10.1186/2047-217X-1-18 (2012).
Dierckxsens, N., Mardulyn, P. & Smits, G. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4), e18. https://doi.org/10.1093/nar/gkw955 (2017).
Tillich, M. et al. GeSeq–versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1), W6–W11 (2017).
Kearse, M. et al. Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28(12), 1647–1649 (2012).
Chan, P. P. & Lowe, T. M. tRNAscan–SE: Searching for tRNA genes in genomic sequences. Methods Mol. Biol. 1962, 1–14 (2019).
Greiner, S., Lehwark, P. & Bock, R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1), W59–W64 (2019).
Du, L., Zhang, C., Liu, Q., Zhang, X. & Yue, B. Krait: An ultrafast tool for genome–wide survey of microsatellites and primer design. Bioinformatics 34(4), 681–683 (2018).
Kurtz, S. et al. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 29(22), 4633–4642 (2001).
Sharp, P. M. & Li, W. H. The codon adaptation index–a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 15(3), 1281–1295 (1987).
Liu, X. E. A more accurate relationship between ‘effective number of codons’ and GC3s under assumptions of no selection. Comput. Biol. Chem. 42, 35–39 (2013).
Shield, D. C. & Sharp, P. M. Synonymous codon usage in Bacillus subtilis reflects both translational selection and mutational biases. Nucleic Acids Res. 15(19), 8023–8040 (1987).
Li, H. et al. CPJSdraw: Analysis and visualization of junction sites of chloroplast genomes. Peerj 11, e15326. https://doi.org/10.7717/peerj.15326 (2023).
Frazer, K. A., Pachter, L., Poliakov, A., Rubin, E. M. & Dubchak, I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 32(W1), W273–W279 (2004).
Rozas, J. et al. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evolut. 34(12), 3299–3302 (2017).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evolut. 30(4), 772–780 (2013).
Tamura, K., Stecher, G. & Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evolut. 38(7), 3022–3027 (2021).
Xia, X. H. DAMBE5: A comprehensive software package for data analysis in molecular biology and evolution. Mol. Biol. Evolut. 30(7), 1720–1728 (2013).
Zhang, D. et al. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Biol. Evolut. 20(1), 348–355 (2020).
Drummond, A. J., Suchard, M. A., Xie, D. & Rambaut, A. Bayesian phylogenetics with BEAUTI and the BEAST 1.7. Mol. Biol. Evolut. 29(8), 1969–1973 (2012).
Jamin, M. & Lautenbacher, M. E. TRACER version 1.1: A mathematica package for γ-algebra in arbitrary dimensions. Comput. Phys. Commun. 74(2), 265–288 (1993).
Acknowledgements
This study was supported by the National Natural Science Foundation of China (32260782), the Key Science & Technology Research and Development Project of Guangxi (Guike ZY21195050), the Natural Science Foundation of Guizhou Province (Qiankehezhongyindi [2023] 029), the Capacity-building project of SBR and CAS (KFJ-BRP-017-68), and the Special fund for innovation capacity construction of Guizhou research institution (Qiankehefuqi [2024]013).
Funding
National Natural Science Foundation of China, 32260782, Key Science & Technology Research and Development Project of Guangxi, Guike ZY21195050, Natural Science Foundation of Guizhou Province, Qiankehezhongyindi [2023] 029, Capacity-building project of SBR and CAS, KFJ-BRP-017-68, Special fund for innovation capacity construction of Guizhou research institution, Qiankehefuqi [2024]013.
Author information
Authors and Affiliations
Contributions
X.X.B. and Z.L. conceived the study. X.X.B., J.M.G. and M.J.L. collected the plant samples. J.M.G. and X.X.B. conducted the experiments, performed the bioinformatics analyses, and wrote and revised the manuscript. F.W., K.T., S.T.H., Z.L., M.J.L. and G.X.H. participated in the writing and revision of the manuscript. All authors have read and approved the submitted manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The plants collected in the study are not listed on the List of National Key Protected Wild Plants in China or the IUCN red list of threatened soecies, and were collected following international, national and local legislation, with no special permission required for sample collection.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Gu, J., Li, M., He, S. et al. Comparative chloroplast genomes analysis of nine Primulina (Gesneriaceae) rare species, from karst region of southwest China. Sci Rep 14, 30256 (2024). https://doi.org/10.1038/s41598-024-81524-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-81524-6