Abstract
One of the biggest challenges encountered by the current generation is the evolution of antibiotic resistant bacteria as a result of excessive and inappropriate use of antibiotics. This problem has led to the development of alternative approaches to treat the diseases caused by these multidrug resistant bacteria (MDR). One of the most promising and novel approaches to combat these pathogens is utilization of nanomaterials as antimicrobial agents. In the current investigation, copper oxide nanoparticles (CuO NPs) were fabricated by green method using Dalbergia sissoo leaf extract. The fabricated nanoparticles were characterized through various techniques like UV–visible spectroscopy, scanning electron microscopy (SEM), Fourier transform infrared (FTIR) spectroscopy and X-ray diffraction (XRD). The UV–visible spectroscopy revealed an absorption peak at 290 nm. SEM micrograph revealed only few spherical nanoparticles (with average diameter of < 100 nm), whereas most of the CuO NPs were agglomerated and formed large clusters. FTIR indicated presence of different functional groups that were used as reducing and capping agents while XRD analysis showed crystalline phase structure for the nanoparticles. These nanoparticles exhibited significant growth inhibition in terms of maximum inhibitory zones of 24 mm with minimum inhibitory concentrations (MIC) ranging from 62.5 to 125 µg/ml against MDR bacteria such as Acinetobacter baumannii, Staphylococcus aureus, Escherichia coli and Klebsiella pneumoniae. The effect of different concentrations of nanoparticles on cell membrane disruption was also investigated and a significant increase (p < 0.05) in the leakage of cellular content such as DNA, proteins and reducing sugar was measured. These nanoparticles also showed antibiofilm potential and a significant increase (p < 0.05) in biofilm inhibition was observed by increasing the concentration of nanoparticles. It was noted that percentage of inhibition of biofilm was found to be 68.4–75.8% at the highest tested concentration. The combined effects of antibiotics and nanoparticles revealed a synergistic interaction between them against tested bacteria. In vitro antioxidant activity of fabricated nanoparticles revealed significant antioxidant potential (p < 0.05) by quenching free radicals such as DPPH (73.6%), ABTS (68%) and H2O2 (63%) in a dose-dependent manner.
Similar content being viewed by others
Introduction
In the current milieu, the global emergence of plethora of multidrug-resistant (MDR) organisms accounting for more than 16% of hospital-acquired infections has been reinforced by infectious disease exposure, self-medication and inappropriate use of drugs1. The situation is so alarming that World Health Organization (WHO) has marked MDR organisms as one of the three most challenging threats faced by humans2. Moreover, bacterial biofilms also pose a great challenge in the field of medicine as bacterial cells inside the biofilms often become more resistant to different antibiotics3. Thus, biofilm-forming MDR bacteria will become more challenging to treat, therefore, there is an increasing interest in creating novel bactericidal agents based on inorganic materials to replace existing organic agents, which have restricted applicability due to their low resistance to heat, short life, and high degradability4,5. In this context, nanotechnology offers the possibilities to modify surface properties of metallic nanoparticles having significant applications as antimicrobial agents6.
It is feasible to overcome antibiotic resistance with the use of nanoscale metals using green method that has a significant prospective in the field of medicine by combining nanotechnology and biotechnological approaches6,7. During the last few years nanoparticles (NPs) have emerged as novel antibacterial agents. Various classes of NPs are effective for the treatment of infections including those caused by resistant pathogens8. NPs provide better properties to traditional antibacterial agents for numerous reasons; one reason is that they exhibit high surface area, resulting in advent of new, magnetic, chemical, electro-optical, electrical, optical and mechanical properties of the NPs that are dissimilar from those of the bulk material9. These characteristics enable the NPs to interact intimately with bacterial cell membranes, preferably affected by the release of ions10.
The literature has widely reported the distinctive features and potential uses of iron, gold, palladium, silver, titanium, zinc, platinum metal oxide nanomaterials11,12,13,14,15, but copper oxide nanoparticles (CuO NPs) have received significant attention due to their promising scientific and industrial applications. Having unique structural and biological features, less toxicity and high biocompatibility, CuO NPs proved themselves as promising candidates to the scientific community16,17. In our body Cu is required for different metabolic and physicochemical functions such as serving as cofactors for numerous enzymes18. CuO NPs have wide range of biomedical applications such as their broad-spectrum antimicrobial and antioxidant potential, physicochemical stability, low cost, biocompatibility and being compatibility with other materials to form polymers19,20. Due to these properties, these NPs can be presented as an alternative to antibiotics for the treatment of infections caused by MDR pathogens. Like with other nanomaterials, CuO NPs can also access environment and human body through various routes. Excessive exposure to these NPs might have severe and adverse impact on biological systems. Studies have indicated that excess doses of CuO NPs can induce renal, hepatic, gastrointestinal, splenic and neuronal toxicity in rats via the generation of reactive oxygen species (ROS)21.
CuO NPs can be synthesized with conventional methods such as chemical and physical processes. However, these methods have lots of limitations that cannot be mitigated easily such as very high temperature and pressure conditions, critically longer reaction time, toxic chemicals utilization and adsorption of hazardous by-products on the surface of nanomaterials22,23. Green nano-preparation has become very popular in recent years as it provides benefits from its low cost, good yields, and rapid reaction times under standard reaction conditions24. Since plants are easily available and have a variety of metabolites that can be used during fabrication process, therefore, their utilization is safe. The existence of phytochemicals like carbohydrates, terpenoids, amino acids, flavonoids and saponins in extracts play a substantial part in the synthesis of NPs. The antioxidant attributes of the plant extracts enable them to function as electron acceptors, thereby acting as reductants for metal salts to form nanoparticles by quenching electrons25.
During green synthesis, NPs can be functionalized by the phytochemicals involved in the bio-reduction of the metal ions. These functionalized nanoparticles can then be easily complexed to polymers and drugs via interaction with the organic functional groups. Thus, by conjugating these functionalized NPs with antibiotics not only reduce the toxicity of antibiotics by lowering the dosage requirements, but also strengthen and improve their antimicrobial effects26.
Various plants have been documented to prepare and stabilize CuO NPs such as Euphorbia escula12, Lemongrass27 and Nerium oleander28. However, Dalbergia sissoo Roxb. ex DC. belonging to the family “Fabaceae” has not been utilized so far for the synthesis of these NPs. The plant is commonly recognized as ‘Shisham’ or ‘Indian Rose Wood’ and in medicine numerous parts of this plant have been used29,30. Dalbergia species possess a wide range of biomedical applications such as antimicrobial, antioxidant, anticancer, analgesic, antipyretic due to the existence of various components such as neoflavonoids, flavonoids, steroids, isoflavonoids and terpenoids31. The bioactive compounds present in the plant extract can be effectively utilized as reducing and capping agents for the synthesis of nanoparticles. Moreover, cytotoxicity and mutagenic profiling revealed that this plant and its extracts have non-toxic and non-mutagenic nature32. Thus D. sissoo extract was employed as a reductant and stabilizing medium for the fabrication of CuO NPs.
The current study employed a green, ecofriendly and cost-effective approach using Dalbergia sissoo leaf extract for the fabrication of CuO NPs. Moreover, organic solvents were avoided for metabolite extraction from plant and aqueous extract was utilized under appropriate conditions. The synthesized NPs were further tested against antibiotic resistant Gram positive and Gram negative bacteria and their antibiofilm effect was also explored. To determine the antibacterial activity of NPs, antibiofilm activity was also evaluated since most clinical isolates are antibiotic-resistant due to their biofilm formation. So, to validate the antibacterial activity, antibiofilm potential of nanoparticles was also determined. While for the further use of these nanoparticles in biomedical application, their antioxidant activity was also evaluated. In vitro antioxidant activity of nanoparticles was investigated using 2,2-diphenyl-1-picrylhydrazyl (DPPH), hydrogen peroxide (H2O2) and 2,2’-azino-bis (3-ethylbenzothiazoline-6-sulphonic acid) ABTS radical scavenging assays. Another significance of the current investigation was that the interaction of CuO NPs with commonly used antibiotics was also checked and synergism was observed for few combinations that could be beneficial if used in clinical practices.
Materials and methods
Bacterial strains and growth conditions
MDR clinical isolates such as Acinetobacter baumannii (Accession No. KY228372), Staphylococcus aureus (Accession No. KX685332), Escherichia coli (Accession No. KY305421) and Klebsiella pneumoniae (Accession No. MF953599) were obtained from the Institute of Microbiology, Government College University Faisalabad (GCUF), Pakistan. The bacterial cultures were then grown in Luria–Bertani (LB) broth and agar.
Plant extract preparation
Fresh leaves of Dalbergia sissoo (Shisham) were collected from the Botanic garden, GCUF, thoroughly washed and further authenticated from Department of Botany, GCUF. For extract preparation, 10 g of fresh leaves were added in deionized water (100 ml), which was then boiled for 30 min at 60–80 °C in a water bath. Later, the obtained extract was filtered and diluted to an optimal concentration (25%) using distilled water (the final volume was adjusted to 250 ml).
Copper oxide nanoparticles (CuO NPs) biofabrication and characterization
CuO NPs were fabricated using the procedure described by Acharyulu et al. with minor modifications33. A magnetic stirrer was used to dissolve 100 ml of analytical grade copper sulfate CuSO4 0.5H2O (5 mM) solution in 50 ml of aqueous extract of the plant. The mixture was kept under constant stirring at 130 °C for 7 h after complete dissolution, then cooled at room temperature, and supernatant was decanted. After thorough washing, the centrifugation of solid black product was done at 3500 rpm for 20 min and dried at 120 °C for 6 h. For further analysis, the dry precursor was converted into powder and placed in an airtight container.
For the characterization of fabricated nanoparticles, UV–visible absorption scanning was performed in 200–800 nm range. To observe shape and morphology of nanoparticles, SEM was performed, and for elemental composition, energy dispersive X-ray (EDX) spectroscopy was performed. The presence of different functional groups was confirmed through FTIR in the range of 4000–600 cm−1. The fabricated nanoparticles were further characterized by X-ray diffractometry to check their crystalline nature.
Antibacterial susceptibility testing of biofabricated copper oxide nanoparticles
The lowest concentration of an antibacterial agent that could hinder the bacterial apparent growth following overnight incubation is known as the Minimum Inhibitory Concentration (MIC). NPs antibacterial activity was measured using broth microdilution assay following the protocol of Hayat et al.34. CuO NPs (0–1000 µg/ml) twofold serial dilutions were made in Mueller–Hinton (MH) broth using microtiter plate. 100 µl of standardized bacterial inoculum (24 h old with turbidity adjusted to 0.5 McFarland) was dispensed into plate having 100 µl of MH broth containing nanoparticles following incubation (at 37 °C for 24 h). Following incubation, 30 µl of nitro-blue tetrazolium chloride (at concentration of 5 mg/ml), was dispensed into all wells to monitor cell’s viability. The yellow colour of NBT dye is reduced to blue colour to confirm cell’s viability. Thus, the lowest concentration of nanoparticles that prevented the colour change was MIC of NPs.
For Minimum Bactericidal Concentration (MBC) determination, bacterial cultures from the wells having concentrations of nanoparticles equal to MIC and above were swabbed onto MH agar plates and further incubated at 37 °C for 24–48 h. Following incubation, the plates were examined for the presence of bacterial growth and the lowest concentration of the CuO NPs that prevented the bacteria to grow on the plates was considered MBC.
The antibacterial potential of CuO NPs was checked using agar well diffusion method35. Numerous concentrations (125–1000 µg/ml) of NPs in DMSO (Dimethyl sulfoxide; 0.1%) were prepared. 100 µl of standardized bacterial suspensions were spread on MH agar plates. The plates were dried for 5 min then wells were cut into the agar plate with a sterile well cutter. 100 µl of each concentration of CuO NPs was pipetted into the respective wells and incubated the plates at 37 °C (overnight). The antibacterial effect was demonstrated as inhibitory zones diameter (expressed in mm) against tested bacteria.
Growth kinetic assay
Bacterial growth curve analysis was performed at different time intervals using various concentrations of NPs (0.25 × MIC, 0.5 × MIC, 1 × MIC respective to each strain). Briefly, overnight bacterial cultures were taken and then standardized to 0.5 McFarland. The cultures were grown in presence of varying NPs concentrations in LB broth at incubated at 37 °C. After every 4 h, 100 µl of suspension was pipetted out and the optical density (OD) was monitored at 600 nm. Same procedure was used for the control cells (that were grown in the without NPs).
Cell membrane disruption assay
The cellular content leakage such as of DNA, proteins and reducing sugar was checked to determine the disintegration of the cellular envelope. For this purpose, bacterial inoculum standardized to 0.5 McFarland was added to LB broth having varying concentrations (0.5 × MIC and 1 × MIC and 2 × MIC related to each bacteria) of CuO NPs. Bacterial culture in LB media without NPs was used as control. The culture tubes were placed on shaking incubator for 24 h at 37 °C. After incubation, the culture was collected by centrifugation (10,000g, at 4° C) for 30 min and the supernatant was kept at -20 °C and was further analysed for the quantification of reducing sugars, proteins and DNA. Protein leakage was quantified using the protocol of Bradford36 whereas for the quantification of DNA, 10 µl of supernatant from each tube was added into 96 wells microtiter plate and then amount of DNA was determined by measuring absorbance at 260 nm37. Quantification of reducing sugar was done through phenol–sulphuric acid assay following the protocol of Mecozzi38.
Antibiofilm effect of CuO NPs
Effect of NPs on biofilm formation by antibiotic resistant bacteria was also checked. Briefly, overnight bacterial cultures were standardized to 0.5 McFarland. To 96 wells microtiter plate, 100 µl of LB having different concentrations of nanoparticles (0.25 × MIC, 0.5 × MIC and 1 × MIC) was added. Then 100 µl of standardized bacterial culture was added followed by incubation (37 °C; 24 h). After incubation, the planktonic cells were decanted followed by fixation with the addition of 200 µl of methanol in each well for 15 min. The adhered cells were then stained with 0.1% crystal violet (CV) solution for 10 min. Normal saline (0.85% sodium chloride) was used to wash the wells thrice and the adhered cells were then suspended in glacial acetic acid (33% v/v). The density of attached cells was measured by ELISA reader (575 nm). Control cells (without nanoparticles) were processed following the same protocol.
Percent inhibition was measured by comparing with untreated control that was considered as 100% biofilm formation39:
Synergistic studies
The interaction of CuO NPs and antibiotics (erythromycin and cefexime) was evaluated using synergistic assay. For this purpose, two-dimensional checkerboard titration was performed using the broth micro-dilution assay, where concentrations of antibiotic decreased in the vertical direction, and concentrations of CuO NPs decreased in the horizontal directions40. Serial two-fold dilutions of antibiotics and NPs were prepared in LB broth starting from the concentration of 1000 µg/ml. In column 12, only antibiotic was added with gradual decrease in concentration from A to F. In row H only NPs were added with gradual decrease in concentration from 1st to 11th well. In 96th well only LB broth and bacterial suspension was added which served as positive control. From column 1 to 11 and row A to G different combinations of antibiotics and NPs were added. Well 1 had the highest concentration of both antibiotic and NPs. In each well 20 µl of standardized bacterial inoculum (density equivalent to 0.5 McFarland standard) was added, and the plates were incubated for 24 h at 37 °C. The fractional inhibitory concentration index (FICI) was used to check the synergistic interaction and was calculated using the following formula:
where the FICI of antibiotic (A) and NPs (B) were calculated by using the following formulas:
The results were interpreted as a synergy if the FICI of the combination is ≤ 0.5, Additive when it was > 0.5 or ≤ 1, Indifferent if it was between > 1 and ≤ 4, or antagonism for FICI > 4.
Antioxidant activities of CuO NPs
CuO NPs were examined for their in vitro antioxidant activity using 2,2-diphenyl-1-picrylhydrazyl (DPPH), hydrogen peroxide (H2O2) and 2,2’-azino-bis (3-ethylbenzothiazoline-6-sulphonic acid) ABTS radical scavenging assays. Various concentrations (62.5–1000 µg/ml) of CuO NPs and the standards were made and the absorbance was measured spectrophotometrically against the corresponding blank solutions.
DPPH radical scavenging assay
The free radical scavenging assay of CuO NPs was examined by the method of Sowndhararajan and Kang, with minor alterations41. 10 µl of each concentration of CuO NPs was pipetted into microtiter plate and then 200 µl of 0.1 mM solution of DPPH was added. The suspension was incubated at 37 °C for 30 min in dark. After incubation, the optical density (OD) was measured at 517 nm. Ascorbic acid was used as a standard and was run through the same procedure. A change in colour of DPPH solution (from deep violet to yellow) in the presence of NPs indicated the radical reduction of. The antioxidant activity was calculated using the following equation:
ABTS radical scavenging activity
The ABTS scavenging activity was checked using the protocol of Re et al.42 The working solution of ABTS was prepared by mixing equal amounts of ABTS (7 mM) and potassium persulfate (245 mM) and the reaction mixture was kept for 12 h at room temperature. The prepared ABTS+ solution was diluted using methanol to reach an absorbance of 0.7 at 743 nm. The solution was then mixed with 0.1 ml of different concentrations of nanoparticles and incubated for 6 min (at room temperature) followed by measuring absorbance at 734 nm. Ascorbic acid as a standard was run using the same protocol. Percentage radical scavenging activity was calculated using the same formula as mentioned above.
Hydrogen peroxide radical scavenging assay
The hydrogen peroxide assay was performed according to the protocol of Keshari et al.43 0.1 ml of nanoparticles in phosphate buffer (pH 7.4) was mixed with 0.6 ml of hydrogen peroxide (2 mM H2O2) solution in phosphate buffer. After 10 min absorbance was measured at 230 nm. Ascorbic acid was used as a standard during the assay. Percentage radical scavenging activity was calculated using the same formula as mentioned for DPPH scavenging activity.
Statistical analysis
All the experiments were performed in triplicates and the results were expressed as means ± SE. In order to check the significance of each experiment, Student’s t-test and ANOVA (analysis of variance) were performed using Microsoft Excel. A value of p < 0.05 was considered to be statistically significant.
Results
Characterization of biofabricated CuO NPs
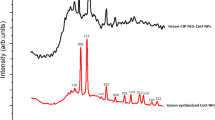
CuO NPs were synthesized by green method using aqueous leaf extract of Dalbergia sissoo. UV–vis spectroscopy revealed the greatest absorbance at 290 nm indicating the production of CuO NPs (Fig. 1A). In XRD studies, diffraction peaks were noticed for biofabricated nanoparticles at 2θ values of Brags angle for 31.36°, 43.51°, 45.31°, 51.47°and 60.35° (Fig. 1B). In Fig. 1C, FTIR spectrum of biofabricated nanoparticles has been shown in which various peaks can be clearly seen at 3296, 2920, 1728, 1642, 1392, 1128, 828 and 610 cm−1. The peak at 3296 cm−1 revealed the presence of O–H group of phenol that might be present in plant extract. The peaks at 1642 cm−1 and 1392 cm−1 correspond to alkene group and amines (C–N stretching vibration) respectively. The peak at 610 cm−1 indicated the Cu–O stretching vibrations confirming the synthesis of CuO NPs. Synthesis of CuO NPs was also checked by EDX where absorption peaks at 0.3, 1 and 8 keV confirmed the generation of these nanoparticles (Fig. 1D). SEM micrograph revealed only few spherical nanoparticles (with average diameter of < 100 nm), whereas most of the CuO NPs were agglomerated due to the oxidation of metallic nanoparticles (Fig. 1E).
Antibacterial susceptibility testing of biofabricated CuO NPs
The antibacterial activity of biofabricated CuO NPs was investigated by measuring MIC and MBC using broth microdilution method. As mentioned in Table 1, the maximum antibacterial activity was observed for K. pneumoniae with MIC value of 62.5 µg/ml, whereas for remaining MDR strains the value was found to be 125 µg/ml. Similarly, MBC value was 500 µg/ml against A. baumannii and E. coli, whereas 250 µg/ml represented MBC for K. pneumoniae and S. aureus.
Antibacterial potential of CuO NPs was also evaluated by agar well diffusion method against MDR bacteria by measuring inhibitory zones diameter. The results as presented in Table 2, revealed a dose dependent inhibitory potential of CuO NPs and inhibitory zones diameter exhibited an increasing trend by increasing the nanoparticles concentration. The diameter of inhibitory zone was in the range of 8–24 mm. Maximum antibacterial effect (24 mm) was observed against K. pneumoniae at a concentration of 1000 µg/ml followed by E. coli (22 mm), S. aureus (20 mm) and A. baumannii (19 mm) as shown in Fig. 2.
Growth kinetic assay
After treatment with various concentrations of nanoparticles, growth pattern of bacteria was analyzed by measuring absorbance (600 nm) at regular intervals. As depicted in Fig. 3, maximum reduction in growth was observed when bacterial cells were treated with 1 × MIC of CuO NPs. As shown in Fig. 3B, in control (untreated) cells of A. baumannii, the highest cell density was observed in comparison with other target bacteria. The results also revealed that a comparatively increased absorbance was measured for the cells treated with 0.25 × MIC as compared to other concentrations, and by increasing the concentration of nanoparticles a reduction in growth was observed.
Cell membrane disruption assay
Cellular contents release such as DNA, reducing sugars and proteins was checked to determine the disintegration of the cellular envelope. As depicted in Fig. 4A, the amount of DNA liberated into the supernatant following the treatment of cells with 2 × MIC of NPs was significantly higher (p < 0.05) in comparison to control and those treated with 0.5 × MIC concentrations of NPs. The range of DNA content released after disruption of cell membrane was 30–72 µg/ml. As indicated by the results, a significant increase (p < 0.05) in protein leakage of cells treated with CuO NPs (at 2 × MIC) was observed compared to the control and the cells treated with 0.5 × MIC of NPs. However, no discernible variations in the amount of protein leakage was detected between control and cells treated with NPs (at 0.5 × MIC) (Fig. 4B). Leakage of protein content following treatment of bacterial cells with CuO NPs was in the range of 5.6–12.2 µg/ml. Moreover, the quantity of reducing sugar liberated in supernatant was in the range of 11.4–69.1 µg/ml (Fig. 4C).
Antibiofilm effect of CuO NPs
The effect of CuO NPs on biofilm formation was also evaluated by growing biofilms in the presence of different concentrations of nanoparticles followed by staining of adhered cells with CV. As depicted in Fig. 5, a dose dependent decrease in biofilm forming capacity of target bacteria was observed and an increase in NPs concentration significantly (p < 0.05) reduced biofilm formation. The order of percentage inhibition of biofilm was as follows K. pneumoniae > A. baumannii > S. aureus > E. coli. It was noted that even subinhibitory concentrations (0.5 × MIC) reduced the biofilm formation in the range of 38.9–49.3%, whereas at 1 × MIC, percentage biofilm inhibition was 68.4–75.8%.
Synergistic studies
In this study, the degree of interaction between CuO NPs and selected antibiotics (cefexime and erythromycin) was also investigated using checkerboard assay and by calculating the FIC index. The combination of erythromycin and nanoparticles exhibited synergism with FICI values of 0.49 and 0.375 against S. aureus and E. coli respectively. Whereas, against A. baumannii additive and for K. pneumoniae, indifferent effects were observed (Table 3).
As mentioned in Table 3, CuO NPs and cefixime combination executed synergism against A. baumannii, S. aureus and E. coli with FICI values of 0.5, 0.375 and 0.5 respectively. Whereas, for K. pneumoniae additive interaction was observed.
Antioxidant activities of CuO NPs
To determine the antioxidant capacity of CuO NPs, DPPH, ABTS and H2O2 scavenging assays were performed. It was observed that CuO NPs showed good antioxidant activity and a noteworthy increase in scavenging of free radicals by NPs was observed in a dose dependant manner (Table 4). Maximum DPPH scavanging activity (73.6%) of CuO NPs was observed at the highest concentration tested (1000 μg/ml), whereas, the minimum activity was observed at 62.5 μg/ml. In ABTS assay, the maximum antioxidant activity of (68%) was found at a concentration of 1000 µg/ml of CuO NPs while the minimum activity (18%) was produced at (62.5 µg/ml). H2O2 scavenging activity of CuO NPs was in the range of 20.3–63%.
Discussion
Over the past few decades, global health has been threatened by the emergence of novel pathogens and multidrug resistant strains44. Although promising efforts have been made in designing and developing novel antimicrobials yet the rapid surge in the number of infections caused by MDR pathogens has been observed45,46. In this regard, nanotechnology offers alternatives against MDR pathogens in terms of nanoparticles that substantiated their biological potential because of smaller size and high surface area to volume ratio. Metallic oxide nanoparticles, synthesized by various means, have shown their promising antibacterial activity in overcoming the emerging multidrug resistance problems47,48.
In the current investigation, CuO NPs were fabricated using green approach and characterized by using different techniques. Green route for the synthesis of NPs has now been appreciated since harmless biological compounds are being utilized during the process and this approach can also improve the activity and biocompatibility of nanoparticles. Improved biomedical applications of CuO NPs synthesized from leaf extract of Salacia reticulate were observed in comparison with nanoparticles obtained by chemical means49. Normally, plant extracts have variety of metabolites like sugars, terpenoids, tannins, phenols and flavonoids that serve as reducing and capping agents for the nanoparticle’s synthesis. In the current study, aqueous leaf extract of D. sissoo was used for the biofabrication of CuO NPs and a solid black product was obtained. The mechanism of biofabrication involves production of Cu (II) cations from CuSO4.5H2O dissolved in deionized water. These dissolved Cu (II) cations reacted with nucleophilic moieties i.e., hydroxyl and carboxylate groups of polyphenolic compounds from D. sissoo as evident from FTIR spectrum. For example, we noted a peak at 3296 cm−1 revealing the presence of O–H group of phenol that might be present in plant extract. Numerous other studies have also described the involvement of bioactive compounds of plant extracts and their involvement in reduction of metallic ions and stabilization of CuO NPs21,50.
UV–Vis absorption spectrometry can be employed to characterize metallic nanoparticles on the basis of their surface plasmon resonance51. In our study, CuO NPs exhibited characteristic absorption peak at 290 nm that correlates with Tshireletso et al. who also found maximum absorption peak at 290 nm for CuO NPs fabricated from citrus peel extracts.52 Numerous studies have reported the absorption peaks in the range of 250–300 nm for CuO NPs. Shanker and coworkers found UV–Visible spectra of CuO NPs mediated from papaya leaf extract spanning between 250 and 300 nm53. These discrepancies might be attributed to different forms of CuO nanomaterials and method employed for nanoparticles fabrication. EDX spectrum confirmed the synthesis of CuO NPs whereas, SEM micrograph showed the presence of few spherical nanoparticles, whereas, most of the particles were present in agglomerated form due to the oxidation of metallic nanoparticles. Ramazan et al. also biofabricated CuO NPs and found the agglomerated nanoparticles in SEM images54. Agglomeration of nanoparticles is a key factor influencing their biological activities. The morphology of green synthesized nanoparticles is also influenced by various factors such as temperature, pH, reaction time and the concentration of reactants. Temperature is one of the most important factors that can challenge the form and dimension of nanoparticles during fabrication process55. It is possible that agglomerates formation of CuO NPs was also increased as we have used very high temperature and reaction time during the synthesis process.
Various peaks (3296, 2920, 1728, 1642, 1392, 1128, 828 and 610 cm−1) were observed in FTIR spectrum of CuO NPs confirming the contribution of phytochemicals of plant extract in the process of fabrication of nanoparticles. Our results are in accordance with previous studies in which similar peaks in FTIR spectrum of CuO NPs have been mentioned56,57. The XRD pattern disclosed the crystalline nature of biofabricated CuO NPs. The observed peaks are in accordance with Ali et al. (2021), who also demonstrated crystalline nature of biogenic CuO NPs58. It has been reported that sharp and clear peaks in XRD spectrum revealed the highly crystalline nature of nanomaterials as observed in current study21.
CuO is black and dark brown crystalline solid while CuO NPs are shiny brown/greenish with high surface area and reactivity as compared to the bulk CuO. Normally, CuO are insoluble in water but CuO NPs are more reactive with altered solubility. This makes them more suitable for catalytic, antimicrobial, and environmental applications59. Thus, in the current study, we investigated different biological activities of CuO NPs synthesized by green route. Antibacterial activity of biofabricated CuO NPs was checked against antibiotic resistant bacteria and the results revealed significant antibacterial potential of these NPs. Previous studies also demonstrated antimicrobial potential of green synthesized CuO NPs20,60,61. The antimicrobial effects of Cu based nanoparticles usually involves generation of reactive oxygen species, destruction of cell membranes and cell wall and further interaction with proteins and DNA62.
Well diffusion method and broth microdilution assay were performed for in vitro susceptibility testing of green synthesized CuO NPs. The results demonstrated that MIC value was in the range of 62.5–125 µg/ml against test organisms. Biofabricated CuO NPs could execute different antibacterial activities against some bacteria depending upon the synergy of bioactive metabolites involved in the fabrication process. Padil and Cernik, worked on the antibacterial potential of CuO NPs and reported the MIC value of 120 and 10 µg/ml for S. aureus and E. coli respectively63 whereas, Ahamed and coworkers found MIC values of 62.5, 31.25 and 250 µg/ml against S. aureus, E. coli and K. pneumoniae respectively4. The diameter of inhibitory zone in agar well diffusion method was found to be 9–24 mm. Effectiveness of CuO NPs against both Gram-negative and Gram-positive bacteria indicated their wide spectrum antibacterial potential. Biogenic CuO NPs from the extract of Pterocarpus marsupium demonstrated inhibitory zones of 25 mm against K. pneumoniae, 24 mm for E. coli and 20 mm for S. aureus after incubation for 24 h64.
Nanoparticles can interact with cell surfaces and cause damage to the cell membrane that can result in release of cellular contents such as nucleic acid and proteins. In this study, membrane disruption assay was performed to quantify the cellular content leakage such as proteins, reducing sugar and nucleic acids after exposure to different concentration of CuO NPs. It was elucidated that amount of reducing sugars and proteins released was in the range of 11.4–69.1 µg/ml and 5.6–12.2 µg/ml respectively. The leakage in cellular content was the highest against S. aureus that showed that amount of reducing sugar and proteins present in supernatant was 69.1 and 12.2 µg/ml respectively after treatment with 250 µg/ml concentration of NPs. The results are in accordance to the findings of Azam et al., who quantified the reducing sugar and proteins from S. aureus after treatment with CdO NPs65. The results showed that the leakage of reducing sugar and protein was 60 µg/mg and 6.2 µg/mg respectively at a concentration (150 µg/ml) of CdO NPs. Additionally, Asadzadegan et al. also reported the disruption of cell membrane in Gram positive (S. aureus, Streptococcus pyogenes) and Gram negative bacteria (E. coli, Proteus mirabilis) after treatment with silver nanoparticles that resulted in increased leakage of cellular proteins66.
One of the main reasons for the emergence of antibiotic resistance among bacterial pathogens is the formation of biofilms. In order to treat infections caused by these biofilm formers, disruption/ eradication of biofilm is necessary since these pathogens do not respond to conventional antibiotics making it essential to go for novel biofilm targets67. Therefore, in the current study in addition to antibacterial potential of CuO NPs, their antibiofilm potential was also investigated against MDR target bacteria. The results revealed that CuO NPs significantly inhibited biofilm formation by target bacteria in a dose dependent way. Various studies have revealed the potential of CuO NPs as biofilm inhibitors11,68. In current study, percentage biofilm inhibition was found to be 68.4–75.8% at the highest tested concentration. The obtained results agreed with Shehabeldine et al. who also reported antibiofilm effect of mycogenic CuO NPs and found 59% and 49% inhibition of biofilms against K. oxytoca and E. coli at subinhibitory concentrations of NPs68. Chari et al. also reported > 60 inhibiton of biofilm formation in all tested aquaculture pathogens by CuO NPs synthesized by using one-pot strategy69.
Synergistic approaches can be used to combine two or more substances in order to get comparatively improved efficacy as compared to individual substances. The combination of metallic nanoparticles with other antimicrobials may contribute towards their enhanced activity. The drug combination is a commonly used strategy in health sector, and therapeutic success has been achieved for various ailments such as acquired immunodeficiency syndrome (AIDS), cardiovascular disease, cancer and microbial infections70. In this study combined effects of CuO NPs with some conventional antibiotics was checked and the obtained results indicated the synergistic interaction between NPs and antibiotics in most of the cases. Similar to our observations, Alazavi reported the synergistic interaction between CuO NPs and chloramphenicol to reduce the expression of mexA gene in MDR P. aeruginosa71. El-Sherbiny also reported the synergistic interaction between biogenic CuO NPs and selected antibiotics against methicillin resistant Staphylococcus aureus (MRSA) and found good synergy between NPs and antibiotics such as cefoxitin and polymyxin B72. Kaur also noted good synergism between CuO NPs with erythromycin, azithromycin and norfloxacin especially against Staphylococcus, Klebsiella and Pseudomonas73. Effective combination strategies must be designed in order to treat infection, overcome antibiotic resistance and for the protection of natural microbiota74.
Various metabolic process taking place in our body is responsible for generating free radicals that are unstable and cause cellular damage by releasing ROS. Thus, generation of free radicals led to various degenerative disorders and reduced immunity. Antioxidants play a significant role in the prevention of these disorders by scavenging free radicals via accepting or donating electrons from reactive oxygen species75. The antioxidant potential of biofabricated CuO NPs was also checked by using DPPH, ABTS and H2O2 scavenging assays and the results demonstrated a dose dependent scavenging potential of NPs. Numerous studies have also reported the antioxidant potential of biogenic CuO NPs particularly fabricated using plant sources76,77,78. Since plant extracts contain a variety of secondary metabolites such as phenolics, alkaloids, flavonoids etc. they might contribute to the enhanced antioxidant potential of NPs. Similar to our study, Muthuvel and coworkers reported the antioxidant activity of CuO NPs by using the DPPH scavenging and hydroxal radical assays and observed the antioxidant activity in the range of 19–90% and 17–93% measured by DPPH and hydroxyl radical assays respectively79.
Conclusions
The study clearly demonstrated the cost-effective way for synthesis of CuO NPs by using aqueous leaf extract of Dalbergia sissoo. The findings of this study highlight the marvellous potential of CuO NPs not only as antioxidant and antibacterial agents but also as promising antibiofilm agent in order to combat emerging threat of antimicrobial resistance. Further research work is needed to extend this antibacterial and antibiofilm potential of these NPs against other human pathogens. It is recommended that in future, research to understand their molecular mechanisms, signal pathway and in vivo studies should be carried out in order to validate their potential applications in practical settings. However, the performance of nanoparticles is intricately linked to environmental conditions used during the fabrication process such as temperature, pH, solvent concentration etc. So, these conditions must be optimized during fabrication process as they can influence the various parameters of nanoparticles such as stability, size, morphology and chemical reactivity.
Data availability
The data used in this study can be provided from the corresponding author upon request.
References
Mulani, M. S., Kamble, E. E., Kumkar, S. N., Tawre, M. S. & Pardesi, K. R. Emerging strategies to combat ESKAPE pathogens in the era of antimicrobial resistance: A review. Front. Microbiol. 10, 539 (2019).
Who, W. H. O. Priority pathogens list for R&D of new antibiotics (WHO, 2017).
Singh, A. K. et al. Prevalence of antibiotic resistance in commensal Escherichia coli among the children in rural hill communities of Northeast India. PLOS ONE 13, e0199179 (2018).
Ahamed, M., Alhadlaq, H. A., Khan, M. A. M., Karuppiah, P. & Al-Dhabi, N. A. Synthesis, characterization, and antimicrobial activity of copper oxide nanoparticles. J. Nanomater. 2014, e637858 (2014).
Naika, H. R. et al. Green synthesis of CuO nanoparticles using Gloriosa superba L. extract and their antibacterial activity. J. Taibah Univ. Sci. 9, 7–12 (2015).
Hsueh, P.-R. et al. Consensus statement on the adherence to Clinical and Laboratory Standards Institute (CLSI) Antimicrobial Susceptibility Testing Guidelines (CLSI-2010 and CLSI-2010-update) for Enterobacteriaceae in clinical microbiology laboratories in Taiwan. J. Microbiol. Immunol. Infect. 43, 452–455 (2010).
Poole, K. Mechanisms of bacterial biocide and antibiotic resistance. J. Appl. Microbiol. 92(Suppl), 55S-64S (2002).
Huh, A. J. & Kwon, Y. J. ‘Nanoantibiotics’: A new paradigm for treating infectious diseases using nanomaterials in the antibiotics resistant era. J. Control Rel. 156, 128–145 (2011).
Whiteside, A., Fisher, C. A. J., Parker, S. C. & Islam, M. S. Particle shapes and surface structures of olivine NaFePO4 in comparison to LiFePO4. Phys. Chem. Chem. Phys. 16, 21788–21794 (2014).
Morones, J. R. et al. The bactericidal effect of silver nanoparticles. Nanotechnology 16, 2346–2353 (2005).
Mansoor, A. et al. Anti-bacterial effect of titanium-oxide nanoparticles and their application as alternative to antibiotics. Pak. Vet. J. 43(2), 269–275 (2023).
Nasrollahzadeh, M., Sajjadi, M., Iravani, S. & Varma, R. S. Green-synthesized nanocatalysts and nanomaterials for water treatment: Current challenges and future perspectives. J. Hazard Mater. 401, 123401 (2021).
Huq, M. A. & Akter, S. Biosynthesis, characterization and antibacterial application of novel silver nanoparticles against drug resistant pathogenic Klebsiella pneumoniae and Salmonella Enteritidis. Molecules 26, 5996 (2021).
Ostovar, N., Mohammadi, N. & Khodadadeh, F. Photocatalytic, antioxidant and antibacterial potential of bio-synthesized ZnO nanoparticles derived from espresso spent coffee grounds: optimization by central composite design. Inorganic Nano-Metal Chem. https://doi.org/10.1080/24701556.2023.2187419 (2023).
Montiel Schneider, M. G. et al. Biomedical applications of iron oxide nanoparticles: Current insights progress and perspectives. Pharmaceutics 14, 204 (2022).
Yugandhar, P., Vasavi, T., Uma Maheswari Devi, P. & Savithramma, N. Bioinspired green synthesis of copper oxide nanoparticles from Syzygium alternifolium (Wt.) Walp: characterization and evaluation of its synergistic antimicrobial and anticancer activity. Appl. Nanosci. 7, 417–427 (2017).
Javed, R., Ahmed, M., Haq, I. U., Nisa, S. & Zia, M. PVP and PEG doped CuO nanoparticles are more biologically active: Antibacterial, antioxidant, antidiabetic and cytotoxic perspective. Mater. Sci. Eng. C Mater. Biol. Appl. 79, 108–115 (2017).
Crisan, M. C., Teodora, M. & Lucian, M. Copper nanoparticles: Synthesis and characterization, physiology. Toxic. Antimicrob. Appl. Appl. Sci. 12, 141 (2022).
Potbhare, A. K. et al. Phytosynthesis of nearly monodisperse CuO nanospheres using Phyllanthus reticulatus/Conyza bonariensis and its antioxidant/antibacterial assays. Mater. Sci. Eng. C Mater. Biol. Appl. 99, 783–793 (2019).
Amin, F. et al. Green synthesis of copper oxide nanoparticles using Aerva javanica leaf extract and their characterization and investigation of in vitro antimicrobial potential and cytotoxic activities. Evid. Based Complement Altern. Med. 2021, 5589703 (2021).
Bugata, L. S. P. et al. Acute and subacute oral toxicity of copper oxide nanoparticles in female albino Wistar rats. J. Appl. Toxicol. 39(5), 702–716 (2019).
Buazar, F., Sweidi, S., Badri, M. & Kroushawi, F. Biofabrication of highly pure copper oxide nanoparticles using wheat seed extract and their catalytic activity: A mechanistic approach. Green Process. Synth. 8, 691–702 (2019).
Sukumar, S., Rudrasenan, A. & Padmanabhan Nambiar, D. Green-Synthesized rice-shaped copper oxide nanoparticles using Caesalpinia bonducella seed extract and their applications. ACS Omega 5, 1040–1051 (2020).
Mahboub, H. H. et al. Protective effects of Allium hirtifolium extract against foodborne toxicity of Zinc oxide nanoparticles in Common carp (Cyprinus carpio). Comp. Biochem. Physiol. C Toxicol. Pharmacol. 257, 109345 (2022).
Fafal, T., Tastan, P., Tuzun, B. S., Ozyazici, M. & Kivcak, B. Synthesis, characterization and studies on antioxidant activity of silver nanoparticles using Asphodelus aestivus Brot. aerial part extract. https://doi.org/10.1016/j.sajb.2017.06.019 (2017).
Ssekatawa, K. et al. Phyto-mediated copper oxide nanoparticles for antibacterial, antioxidant and photocatalytic performances. Front. Bioeng. Biotechnol. 10, (2022).
Tu, H. L. Biosynthesis, characterization and photocatalytic activity of copper/copper oxide nanoparticles produced using aqueous extract of Lemongrass leaf. Compos. Mater. 3, 30 (2019).
Sebeia, N., Jabli, M., Ghith, A., El Ghoul, Y. & Alminderej, F. M. Populus tremula, Nerium oleander and Pergularia tomentosa seed fibers as sources of cellulose and lignin for the bio-sorption of methylene blue. Int. J. Biol. Macromol. 121, 655–665 (2019).
Dixit, P. et al. Constituents of Dalbergia sissoo Roxb. leaves with osteogenic activity. Bioorg. Med. Chem. Lett. 22, 890–897 (2012).
Pund, K. V., Vyawahare, N., Gadakh, R. T. & Murkute, V. K. Antidiabetic Evaluation of Dalbergia Sissoo against alloxan induceddiabetes mellitus in wistar albino rats. J. Nat. Prod. Plant Resour. (2012).
Awais, M., Gulfraz, M., Asad, M. J., Kabir, F. & Khan, K. S. Mesophilic anaerobic co-digestion of cattle manure with malus domestica and Dalbergia sissoo during biomethane potential assays (2018).
Majeed, F., Munir, H., Rashid, R. & Zubair, M. Antimicrobial, cytotoxicity, mutagenicity and anti-epileptic potential of ethanol extracts of a multipurpose medicinal plant Dalbergia sissoo. Biocatal. Agric. Biotechnol. 19, 101155 (2019).
Acharyulu, N. et al. Green synthesis of CuO Nanoparticles using Phyllanthus amarus leaf extract and their antibacterial activity against multidrug resistance bacteria green synthesis of CuO nanoparticles. Int. J. Eng. Res. Technol. 3, 639 (2014).
Hayat, S. et al. Biofabrication of ZnO nanoparticles using Acacia arabica leaf extract and their antibiofilm and antioxidant potential against foodborne pathogens. PLOS ONE 17, e0259190 (2022).
Buszewski, B. et al. Antimicrobial activity of biosilver nanoparticles produced by a novel Streptacidiphilus durhamensis strain. J. Microbiol. Immunol. Infect. 51, 45–54 (2018).
Bradford, M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 (1976).
Green, M. R., & Sambrook, J. Isolation and quantification of DNA. Cold Spring Harbor Protocols 6, pdb-top093336 (2018).
Mecozzi, M. Estimation of total carbohydrate amount in environmental samples by the phenolsulphuric acid method assisted by multivariate calibration. Chemometr. Intell. Lab. Syst. 1–2, 84–90 (2005).
Verduzco-Chavira, K. et al. Antibacterial and antibiofilm activity of chemically and biologically synthesized silver nanoparticles. Antibiotics 12, 1084 (2023).
Pourmand, M. R., Yousefi, M., Salami, S. A. & Amini, M. Evaluation of expression of NorA efflux pump in ciprofloxacin resistant Staphylococcus aureus against hexahydroquinoline derivative by real-time PCR. Acta Med. Iran 52, 424–429 (2014).
Sowndhararajan, K. & Kang, S. C. Free radical scavenging activity from different extracts of leaves of Bauhinia vahlii Wight & Arn. Saudi J. Biol. Sci. 20, 319–325 (2013).
Ullah, F. et al. Sodium nitroprusside and melatonin improve physiological vitality and drought acclimation via synergistically enhancing antioxidant response in dryland maize. J. Plant Growth Regul. https://doi.org/10.1007/s00344-024-11498-2 (2024).
Keshari, A. K., Srivastava, A., Verma, A. K. & Srivastava, R. Free Radicals Scavenging and Protein Protective Property of Ocimum sanctum (L). J. Pharmaceut. Res. Int. 1–10. https://doi.org/10.9734/BJPR/2016/31445 (2016).
Kavimani, S., Vetrichelvan, T. & Nagarajan, N. S. Possible mechanism of anti-inflammatory activity of biochanin-A isolated from Dalbergia sissoides. Indian Drugs 39, 161–162 (2002).
Morsy, M. A. et al. Screening and molecular docking of novel benzothiazole derivatives as potential antimicrobial agents. Antibiotics (Basel) 9, 221 (2020).
Muzammil, S. et al. Aluminium oxide nanoparticles inhibit EPS production, adhesion and biofilm formation by multidrug resistant Acinetobacter baumannii. Biofouling 36, 492–504 (2020).
D’Andrea, M. M., Fraziano, M., Thaller, M. C. & Rossolini, G. M. The urgent need for novel antimicrobial agents and strategies to fight antibiotic resistance. Antibiotics (Basel) 8, 254 (2019).
ALRashdi, B.M., Germoush, M.O., Sani, S. S., Ayub, I., Bashir, W., Hussain, B., Mazhar, M., Ali, S., Zahid, Z., Ayesha, S., & Rafique, A. Biosynthesis of Salvia hispanica based Silver nanoparticles and evaluation of their antibacterial activity in-vitro and rat model. Pak. Vet. J. 43(2), 283–289 (2023).
G, S. et al. Green and chemical synthesis of CuO nanoparticles: A comparative study for several in vitro bioactivities and in vivo toxicity in zebrafish embryos. J. King Saud Univ. Sci. 34, 102092 (2022).
Alhalili, Z. Green synthesis of copper oxide nanoparticles CuO NPs from Eucalyptus Globoulus leaf extract: Adsorption and design of experiments. Arab. J. Chem. 15, 103739 (2022).
Upadhyay, S. K. et al. Efficient removal of total arsenic (As3+/5+) from contaminated water by novel strategies mediated iron and plant extract activated waste flowers of marigold. Chemosphere 313, 137551 (2023).
Tshireletso, P., Ateba, C. N. & Fayemi, O. E. Spectroscopic and antibacterial properties of CuONPs from orange, lemon and tangerine peel extracts: Potential for combating bacterial resistance. Molecules 26, 586 (2021).
Shankar, S. S., Ahmad, A., Pasricha, R. & Sastry, M. Bioreduction of chloroaurate ions by geranium leaves and its endophytic fungus yields gold nanoparticles of different shapes. J. Mater. Chem. 13, 1822–1826 (2003).
Ramzan, M. et al. Green synthesis of copper oxide nanoparticles using Cedrus deodara aqueous extract for antibacterial activity. Mater. Today Proc. 36, (2020).
Rana, A., Yadav, K. & Jagadevan, S. A comprehensive review on green synthesis of nature-inspired metal nanoparticles: Mechanism, application and toxicity. J. Clean. Prod. 272, 122880 (2020).
Atri, A. et al. Green synthesis of copper oxide nanoparticles using Ephedra Alata plant extract and a study of their antifungal, antibacterial activity and photocatalytic performance under sunlight. Heliyon 9, e13484 (2023).
Alhalili, Z. Metal oxides nanoparticles: general structural description, chemical, physical, and biological synthesis methods, role in pesticides and heavy metal removal through wastewater treatment. Molecules 28, 3086 (2023).
Ali, M. et al. Biogenic synthesis, characterization and antibacterial potential evaluation of copper oxide nanoparticles against Escherichia coli. Nanoscale Res. Lett. 16, 148 (2021).
Gamedze, N. P., Mthiyane, D. M. N., Babalola, O. O., Singh, M., & Onwudiwe, D. C. Physico-chemical characteristics and cytotoxicity evaluation of CuO and TiO2 nanoparticles biosynthesized using extracts of Mucuna pruriens utilis seeds. Heliyon 8(8) (2022).
Bhavyasree, P. G. & Xavier, T. S. Green synthesised copper and copper oxide based nanomaterials using plant extracts and their application in antimicrobial activity: Review. Curr. Res. Green Sustain. Chem. 5, 100249 (2022).
Singh, S., Singh, M., Agrawal, V. & Kumar, A. An attempt to develop surface plasmon resonance based immunosensor for Karnal bunt (Tilletia indica) diagnosis based on the experience of nano-gold based lateral flow immuno-dipstick test. Thin Solid Films 519, 1156–1159 (2010).
Wang, L., Hu, C. & Shao, L. The antimicrobial activity of nanoparticles: Present situation and prospects for the future. Int. J. Nanomed. 12, 1227–1249 (2017).
Thekkae Padil, V. V. & Černík, M. Green synthesis of copper oxide nanoparticles using gum karaya as a biotemplate and their antibacterial application. Int. J. Nanomed. 8, 889–898 (2013).
Sharma, G. Pterocarpus marsupium Derived Phyto-Synthesis of Copper Oxide Nanoparticles and their Antimicrobial Activities. J. Microbial Biochem. Technol. 07, (2015).
Azam, Z. et al. Microbial synthesized cadmium oxide nanoparticles induce oxidative stress and protein leakage in bacterial cells. Microb. Pathog. 144, 104188 (2020).
Abbaszadegan, A. et al. The effect of charge at the surface of silver nanoparticles on antimicrobial activity against gram-positive and gram-negative bacteria: A preliminary study. J. Nanomater. 2015, e720654 (2015).
Pardesi, K., Pable, A., Bhagat, D. & Satpute, S. Applications of metal nanoparticles to combat biofilm forming ESKAPE pathogens. in 1–40 (2019).
Shehabeldine, A. M. et al. Potential antimicrobial and antibiofilm properties of copper oxide nanoparticles: Time-kill kinetic essay and ultrastructure of pathogenic bacterial cells. Appl. Biochem. Biotechnol. 195, 467–485 (2023).
Chari, N., Felix, L., Davoodbasha, M., Sulaiman Ali, A. & Nooruddin, T. In vitro and in vivo antibiofilm effect of copper nanoparticles against aquaculture pathogens. Biocatal. Agric. Biotechnol. 10, 336–341 (2017).
Pemovska, T., Bigenzahn, J. W. & Superti-Furga, G. Recent advances in combinatorial drug screening and synergy scoring. Curr. Opin. Pharmacol. 42, 102–110 (2018).
Alazavi, F., Molavi, F.- & Tehranipoor, M. Synergistic effect of copper oxide nanoparticles and chloramphenicol antibiotic on MexA gene expression of pump efflux system in drug-resistant Pseudomonas aeruginosa isolates. Med. J. Tabriz Univ. Med. Sci. 44, 127–138 (2022).
El-Sherbiny, G. M. et al. Biogenic synthesis of CuO-NPs as nanotherapeutics approaches to overcome multidrug-resistant Staphylococcus aureus (MDRSA). Artif. Cells Nanomed. Biotechnol. 50, 260–274 (2022).
Kaur, H. et al. A novel and one-pot synthesis of Punica granatum mediated copper oxide having flower-like morphology as an efficient visible-light driven photocatalyst for degradation of textile dyes in waste water. J. Mol. Liquids 355, 118966 (2022).
Brooks, B. & Brooks, A. Therapeutic strategies to combat antibiotic resistance. Adv. Drug Deliv. Rev. 78, (2014).
Liu, Z. et al. Role of ROS and nutritional antioxidants in human diseases. Front. Physiol. 9(477), 477 (2018).
Yasin, A. et al. Fabrication of copper oxide nanoparticles using Passiflora edulis extract for the estimation of antioxidant potential and photocatalytic methylene blue dye degradation. Agronomy 12, 2315 (2022).
Sarfraz, M. H., Muzammil, S., Hayat, S., Khurshid, M. & Sayyid, A. H. Fabrication of chitosan and Trianthema portulacastrum mediated copper oxide nanoparticles: Antimicrobial potential against MDR bacteria and biological efficacy for antioxidant, antidiabetic and photocatalytic activities. Int. J. Biol. Macromol. 242, 124954 (2023).
Rehana, D., Mahendiran, D., Kumar, R. S. & Rahiman, A. K. Evaluation of antioxidant and anticancer activity of copper oxide nanoparticles synthesized using medicinally important plant extracts. Biomed. Pharmacother. 89, 1067–1077 (2017).
Muthuvel, A. & Manoharan, C. Synthesis of copper oxide nanoparticles by chemical and biogenic methods: photocatalytic degradation and in vitro antioxidant activity. Nanotechnol. Environ. Eng. 5 (2020).
Acknowledgements
The authors would like to extend their sincere appreciation to the Researchers Supporting Project Number (RSP2025R134), King Saud University, Riyadh, Saudi Arabia.
Funding
The authors would like to extend their sincere appreciation to the Researchers Supporting Project Number (RSP2025R134), King Saud University, Riyadh, Saudi Arabia.
Author information
Authors and Affiliations
Contributions
Conceptualization, S.H. and S.M.; field research, M.S., and M.S.; software, S.H., A.A., and M.S.; formal analysis, M.H.S., S.H., A.M.K. and S.M.; data curation, A.M.K., M.H.S., and A.A.; Resources, S.M., M.H.S., and A.M.K.; Validation, S.M., S.H.,; investigation, M.S.; writing-original draft preparation, M.S., A.H., G.D.A.Q., E.F.A. and S.H.; writing-review and editing, S.M., A.H., G.D.A.Q. and E.F.A.; funding acquisition: A.H. and E.F.A. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Siddique, M.H., Sadia, M., Muzammil, S. et al. Biofabrication of copper oxide nanoparticles using Dalbergia sisso leaf extract for antibacterial, antibiofilm and antioxidant activities. Sci Rep 14, 31867 (2024). https://doi.org/10.1038/s41598-024-83199-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-83199-5