Abstract
The anti-inflammatory effect of phellodendrine (PHE), derived from Phellodendri Chinensis Cortex, has been verified in previous studies. Major depressive disorder (MDD) is associated with immune dysregulation and inflammatory processes. This study aimed to explore the therapeutic effects of PHE on MDD through network pharmacology and experimental validation. Multiple databases were used to predict the targets of PHE and MDD. The intersection targets between PHE and MDD were obtained to identify as targets for PHE against MDD, followed by protein–protein interaction network, Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathway analyses. Molecular docking was applied to further confirm the anti-MDD effects of PHE. The mitochondrial DNA (mtDNA) copy number, inflammatory cytokines and pathway-related mRNA expressions in PC12 cell were determined via quantitative PCR (qPCR) and enzyme-linked immunosorbent assay to verify our finding. Thirty-eight intersection targets were obtained between PHE and MDD. PHE exerted an anti-MDD effect by regulating SLC6A4, SLC6A3, SLC6A2, MAOA and other targets through serotonergic synapse, salivary secretion, dopaminergic synapse, and cAMP signalling pathway. In vitro, PHE induced an increment in mtDNA copy number compared with the CORT group. PHE affected the levels of IL6 and IL1β with different concentrations. The mRNA levels of CHRM1, HTR1A and key targets of the PI3K/Akt signalling pathway were also influenced. Our research reveals novel mechanisms underlying the anti-MDD effects of PHE through network pharmacology and experiments, which provides a new direction for the development of antidepressants.
Similar content being viewed by others
Introduction
Depressive disorders, a prominent category within neuropsychiatric disorders, is characterised by persistent sadness, loss of interest or pleasure, disturbed sleep or appetite and poor concentration1,2. According to a systematic analysis for the Global Burden of Disease, depressive disorders were responsible for the largest proportion of mental disability-adjusted life years in 20193. Depressive disorders, including major depressive disorder (MDD) and dysthymia, are ranked as the single largest contributor to nonfatal health loss2. The pathogenesis of depression involves multiple factors, such as the nervous, immune and endocrine systems, with a high complication of various correlated pathways1,4. The complex and unclear pathogenesis of MDD complicates the treatment options. Available treatments for depression clinically remain relatively inefficient with only 30% reaching complete remission after receiving an adequate pharmacological treatment5. Therefore, based on the present situation, the discovery of novel antidepressants is urgently needed and will be of great significance in the future, offering potential alternative options for target individuals with MDD.
Traditional Chinese medicine (TCM), renowned for its clinical efficacy, minimal side effects and multiple targets and pathways, has garnered significant attention for its distinctive advantages in antidepression. For example, Kaixinsan, SiNiSan, Xiaoyaosan and Chaihu-Shugan-San, classic prescriptions of TCM, have demonstrated impressive antidepressant efficacy6,7,8,9.
Certain components of Chinese medicinal herbs also play an important role in the prevention of depression. Previous studies have reported that quercetin (from Radix Bupleuri), icariin (from Epimedium brevicornum Maxim.), baicalin (from Scutellaria baicalensis Georgi.) and rehmannioside A (from Rehmanniae Radix) exhibited antidepressant effects10,11,12,13. Various studies have found that MDD is associated with immune dysregulation and inflammatory processes. The hypothesis regarding cytokines in depression proposes that external and internal stressors can induce depressive symptoms by boosting the generation of proinflammatory cytokines and decreasing anti-inflammatory cytokines1,14. The inhibitory effect of phellodendrine (PHE, from Cortex Phellodendri) on inflammation has been confirmed in our previous research15. However, whether PHE is beneficial for MDD remains unknown. Therefore, the main objective of the present study is to observe the therapeutic potential and modulatory mechanisms of PHE in MDD.
The hypothalamic-pituitary-adrenal (HPA) axis and the glucocorticoids (corticosterone in rodents, cortisol in humans) known as the stress hormone play important roles in the pathophysiology of depression. Hyperactivity of the HPA axis and high level of corticosterone(CORT) ( the final products of the HPA axis) are biological features in severe depression individuals16. Chronic high levels of glucocorticoids activate glucocorticoid receptors and affect the activity, proliferation, neurogenesis of neuronal cell in the brain, finally result in depression symptoms17. Therefore, CORT is commonly used to construct depression models in vitro and in vivo. Many studies have confirmed that mice exposed to CORT displayed dysfunction of the HPA axis and depressive-like behaviors, including increased immobility time in forced swimming test, decrease in sucrose preference, and so on18,19.
PC12 cells, a type of cell lineage, are derived from a pheochromocytoma of the adrenal gland in an irradiated rat20. PC12 cells show typical neuronal characteristics, have the general characteristics of neuroendocrine cell, and express high levels of glucocorticoid receptors21,22. CORT-stimulated PC12 cells were widely used as an model of depression in vitro for evaluating the antidepressant effect and underlying molecular mechanisms of drugs21.
Materials and methods
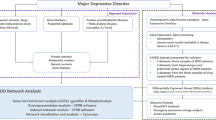
Acquisition of PHE and depression targets
TCMSP (https://old.tcmsp-e.com/tcmsp.php), PharmMapper (http://lilab-ecust.cn/pharmmapper/index.html) and SwissTargetPrediction (http://www.swisstargetprediction.ch/index.php) were used to collect the targets of PHE. Depression targets were selected from the OMIM (http://www.omim.org), DrugBank (https://go.drugbank.com/), DisGeNET (http://www.disgenet.org/) and GeneCards (https://www.genecards.org/) databases by searching ‘Major Depressive Disorder’ as a key word.
The intersection targets of PHE and depression were obtained using the Venn 2.1.0 platform.
Construction of a protein–protein interaction (PPI) network
Intersection targets of PHE and depression were uploaded to the STRING database (https://cn.string-db.org/) to appraise key regulatory targets and identify potential relationships23. The minimum required interaction score was set to 0.4 (medium confidence). In addition, betweenness centrality, closeness centrality and degree centrality were calculated to assess the topological characteristics of the nodes.
Function enrichment analysis
The intersection targets were analysed for Gene ontology (GO) function and Kyoto.
Encyclopedia of Genes and Genomes (KEGG) enrichment using the database for annotation, visualization and integrated discovery (DAVID, https://david.ncifcrf.gov/, version 6.8)24,25. p < 0.05 was considered significant in enrichment results.
Molecular docking
Molecular docking was conducted via Schrödinger to confirm the connection and underlying mechanisms of action between candidate proteins and PHE. We obtained the crystal structures of candidate proteins from AlphaFold2 or RCSB Protein Data Bank (RCSB PDB, https://www.rcsb.org/). PDB ID of the selected targets are as follows DRD2 (PDB ID: 7DFP, https://www.rcsb.org/structure/7DFP), ADRB1 (PDB ID:7BVQ,https://www.rcsb.org/structure/7BVQ), SLC6A4 (PDB ID: 5I6X, https://www.rcsb.org/structure/5I6X), DRD1 (PDB ID: 7JVP, https://www.rcsb.org/structure/7JVP). Then, the structure of SLC6A3 was predicted by AlphaFold2. The specific steps for this process were identical to those described in the previous literature26.
Cell culture and treatments
PC12 cells were purchased from Haixing Biosciences (Suzhou, Jiangsu, China). Cells were cultured in Dulbecco’s modified eagle’s medium (HyClone, New York, NY, USA) containing 10% foetal bovine serum (Gibco, New York, NY, USA) and incubated with 5% CO2 at 37 °C. Cells were seeded in various culture plates at appropriate density for subsequent experiments. For PC12 cells, corticosterone(CORT, CAS:50-22-6, Shanghai Aladdin Biochemical Technology Co., Ltd., Shanghai, China) (400µM) pretreatment was performed for 2 h, followed by stimulation with definite-concentration PHE for 24–48 h.
Cell viability assay
Cell viability was measured by Cell Counting Kit-8 (CCK-8 Beyotime Biotechnology, China) in accordance with the manufacturer’s instructions. The brief operation steps were as follows: 20 µL of CCK-8 solution was added to each well, incubated for 2.5 h at 37 °C and measured at 450 nm using a microplate reader.
Enzyme-linked immunosorbent assay (ELISA)
The media of three wells were collected from each group in 96-well plates. They were centrifuged at 3000 rpm for 10 min to obtain samples for ELISA kits (Shanghai Jining Shiye Co., Ltd., Shanghai, China). The measurement of data followed the instructions in the manual.
Quantitative real-time PCR analysis
Total RNAs were extracted from each group using TRIzol reagent (Thermo Fisher Scientific) and reverse transcribed into cDNA using PrimeScript™ RT Reagent Kit (Takara Biomedical Technology, Beijing, China). Next, quantitative real-time PCR was conducted using TB Green®Premix Ex Taq™ (Takara Biomedical Technology, Beijing, China) following the instructions in the manual. The primers of β-actin were provided by Sangon Biotech (Shanghai, China). The 2−ΔΔCT method was used to estimate the relative expression levels of genes. The primer sequences are shown in Table 1.
Statistical analysis
All data were expressed as mean ± SEM and analysed for statistical significance using SPSS 17.0. One-way ANOVA and Tukey’s post-hoc test were selected for the analysis of statistical differences (a significance level of p < 0.05).
Results
Potential targets of PHE and depression
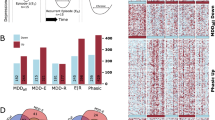
During the preliminary screening, 380 potential targets of PHE were collected from the databases, including 17 from TCMSP, 275 from PharmMapper and 88 from SwissTargetPrediction (Probability > 0.097). After target names were standardised and unified and duplicates were removed, a total of 369 targets were obtained (Fig. 1A).
Furthermore, 566 targets of depression were identified, including 108 from DrugBank, 30 from OMIM, 163 from GeneCards (Relevance score > 27.5398) and 265.
from DisGeNET (Score_gda > 0.3). After the duplicates were deleted, 463 targets remained (Fig. 1B). The intersection of PHE and depression targets resulted in 38 common targets (Fig. 1C).
PPI network
The 38 intersection targets were imported to the STRING database, and interactions with medium confidence were chosen. The PPI network consisted of 38 nodes, 130 edges and 6.84 average node degree. The network was visualised in Cytoscape 3.7.2, where the size and colour of the nodes represented by the circular shape varied on the basis of the value of their degree (Fig. 2A). In addition, 18 core targets were obtained and ranked by degree values (Fig. 2B). The top 10 targets were SLC6A4, SLC6A3, SLC6A2, MAOA, ALB, DRD2, HTR2A, ABCB1, DRD4 and ADRB2.
GO and KEGG pathway enrichment analyses
The 38 antidepressant targets of PHE were imported into DAVID to perform GO and KEGG pathway enrichment analyses. The GO analysis showed that the numbers of biological processes, cellular components and molecular functions were 132, 26 and 38, respectively(p < 0.05), and the top 10 significant entries are shown in Fig. 3A. Seventeen items were selected in the KEGG enrichment analysis (p < 0.05). The significantly enriched pathways were the neuroactive ligand–receptor interaction, calcium signalling pathway, cGMP-PKG signalling pathway, serotonergic synapse, salivary secretion, dopaminergic synapse, cAMP signalling pathway, gap junction, cocaine addiction and regulation of lipolysis in adipocytes. The 10 top KEGG pathway enrichments and the involved targets are shown in Fig. 3B.
Molecular docking
The docking results of PHE with five key targets (DRD1, SLC6A4, SLC6A3, ADRB1 and DRD2), including their XP Gscore and MM-GBSA dG Bind values, are presented in Table 2. The docking score and MM-GBSA result were less than − 6 and − 30 kcal/mol respectively, indicating effective binding to PHE. The docking score between PHE and DRD1 was − 6.910, with an MM-GBSA analysis result of − 31.42 kcal/mol (Fig. 4A). The docking score between PHE and SLC6A4 was − 6.304, with an MM-GBSA analysis result of − 33.42 kcal/mol (Fig. 4B). The docking scores of PHE with SLC6A3, ADRB1 and DRD2 were − 4.917, − 4.876 and − 3.858, respectively. The MM-GBSA analysis results were − 43.84, − 35.30 and − 43.60 kcal/mol (Fig. 4C–E).
Effects of PHE on cell viability
To assess the anti-MDD effect, we first detected the viability of PC12 cells after incubation with PHE solutions at different concentrations. For 24 h, as shown in Fig. 5A, the viability of PC12 cells showed no significant change with increasing concentrations of PHE, except for an increase caused by 5 mg/L of PHE (p < 0.01). The viability of PC12 cells dipped to 57.36 ± 1.67% when coincubated with CORT (p < 0.001), and different concentrations of PHE (40, 10, 5 and 2.5 mg/L) reversed the decline (p < 0.05, p < 0.001, p < 0.001 and p < 0.01, respectively; Fig. 5C). For 48 h, PC12 cells’ viability were reduced by all concentrations of PHE (Fig. 5B). PHE at 10 mg/L elevated the cell viability, which then decreased because of CORT treatment (Fig. 5D).
Effects of PHE on mitochondrial DNA (mtDNA) copy number
As indicated in Fig. 6, mtDNA copy number was significantly decreased in the CORT group (p < 0.01) compared with that in the control group. By contrast, PHE (20 mg/L) induced an increase compared with the CORT group (p < 0.05).
Effects of PHE on IL-6 and IL-1β
We investigated the effects of PHE on the proinflammatory cytokines (IL-6 and IL-1β) of PC12 cells induced by CORT. As indicated in Fig. 7A, for 24 h, the level of IL-6 was significantly elevated in the CORT group (p < 0.05) compared with that in the control group. Significant decreases were observed in 80 and 20 mg/L PHE groups (p < 0.01 and p < 0.05, respectively). IL-6 levels were markedly decreased after exposure to 400 µM CORT for 48 h (p < 0.01) but were increased in the 20 mg/L PHE group (p < 0.05; Fig. 7B).
Figure 7C demonstrates that the CORT-stimulated secretion of IL-1β for 24 h significantly decreased compared with that in the control group (p < 0.01). The IL-1β levels continuously declined in the 10 mg/L PHE group compared with that in the CORT group (p < 0.05). Figure 7D illustrates an opposite result for 48 h to the result for 24 h in the CORT group (p < 0.01). Compared with the CORT group, the 80 and 40 mg/L PHE groups significantly decreased the IL-1β production of PC12 cells (p < 0.05 and p < 0.01, respectively).
Effects of PHE on the mRNA levels of the PI3K/Akt/mammalian target of rapamycin (mTOR) signalling pathway
The mRNA levels of PHE on CHRM1, HTR1A and key targets of the PI3K/Akt signalling pathway (PI3K, Akt, mTOR) in CORT-stimulated PC12 for 24 and 48 h are shown in Fig. 8. The mRNA levels of CHRM1 significantly increased in the CORT group (p < 0.001), whereas PHE treatment inhibited the expression (p < 0.01; Fig. 8A). The expression of HTR1A mRNA for 24 and 48 h (p < 0.05 and p < 0.01, respectively) was significantly higher in the CORT group than that in the control group, whereas a different trend was found in PHE groups, indicating different functions with varying concentrations.
The mRNA level of PI3K was markedly elevated in the CORT group compared with that in the control group (p < 0.001), whereas it was significantly suppressed in PHE groups compared with that in the control group for 24 h (p < 0.01, p < 0.05 and p < 0.01; Fig. 8C,D).The result for 48 h was consistent with that for 24 h but insignificant.
As shown in Fig. 8E, the result for 48 h revealed that CORT treatment significantly increased the mRNA expression of AKT (p < 0.001). However, the PHE (5 and 20 mg/L) treatment partially reversed the effect of CORT (p < 0.001 and p < 0.001, respectively). The result for 24 h displayed a reduced level in the CORT group, whereas PHE showed no significant effect.
CORT and PHE had no significant effect on the expression of mTOR (Fig. 8F).
Discussion
MDD is a highly pervasive and recurrent neuropsychiatric illness causing work/social dysfunction and affecting millions of people worldwide and is a significant financial burden5. The pathogenesis of MDD is complex and obscure, involving inflammatory cytokines, the hypothalamic–pituitary–adrenal (HPA) axis, neurotrophins, the neurotransmitter system, mitochondrial dysfunction and epigenetics. At present, MDD still lacks safe and efficient therapeutic drugs. The main purpose of our study is to explore the potential molecular targets and signalling pathways of PHE in MDD via network pharmacology, molecular docking simulation and cell experiment.
In this study, the core targets, including SLC6A4, SLC6A3, SLC6A2, MAOA, ALB, DRD2, HTR2A, ABCB1, DRD4 and ADRB2, were identified through PPI network analysis. The SLC6A4 (solute carrier family 6 member 4) gene encodes the serotonin transporter (integral membrane protein) that transports the neurotransmitter serotonin from synaptic spaces into presynaptic neurons27. Several studies have indicated that SLC6A4 polymorphisms are associated with the efficacy of antidepressant drugs, and SLC6A4 is the major pharmacodynamic gene identified for selective serotonin reuptake inhibitors (commonly used antidepressant drugs)28,29,30. In addition, elevated SLC6A4 methylation is considered a significant predictor of blunted HPA axis reactivity in the MDD group and blunted cortisol reactivity to stress31. Elevated SLC6A4 methylation is further associated with an increased risk of MDD, and the methylation status is linked to the diagnosis of depression32.
The SLC6A3 gene is responsible for encoding the dopamine (DA) transporter, which is a membrane-spanning protein and primarily serves to eliminate DA from synapses33. The SLC6A2 gene encodes the noradrenaline transporter or the norepinephrine transporter and is a key target of several antidepressant agents33.
Monoamine oxidases (MAOs, with two isoforms: MAOA and MAOB) are involved in regulating the levels of neurotransmitters, including serotonin and DA, located in mitochondrial outer membranes34. Specifically, MAOA metabolises serotonin, noradrenaline and adrenaline. It primarily influences neurotransmitters that are deemed crucial in depression, as a lasting antidepressant target35. Therefore, monoamine oxidase inhibitors have been generally used for antidepression treatment since the early 1950s, although they are rarely used today because of their serious side effects36. Our results are consistent with those reported in the literature.
β-adrenergic receptors are a group of G protein-coupled receptors responsible for transducing cellular signalling from the sympathetic nervous system37. β2-adrenergic receptor (ADRB2) is the primary subtype of β-adrenergic receptors and contributes to many mental disorders (such as MDD). Previous animal studies have indicated that activating central ADRB2 can counteract depressive-like behaviours, reversed by ADRB2 inhibition eliciting antidepressant-like responses38. Consistent with previous research, decreased expression of ADRB2 has also been detected in myeloid cells harvested from the spleen, after repeated social stress39. In addition, the ADRB2-c-Myc-sirt1 pathway inhibits the development of depression influenced by fat mass and obesity40. ADRB2 may show an antidepressant effect by regulating the expression of GR and promoting hippocampal neuroplasticity and neurogenesis41. In sum, our findings indicated that PHE might exert its anti-MDD effects by targeting dopaminergic synapses (DRD2, DRD4, MAOA, SLC6A3), neuroactive ligand–receptor interactions (HTR2A, ADRB2, DRD2, DRD4), serotonergic synapses (HTR2A, MAOA, SLC6A4) and synaptic vesicle cycles (SLC6A2, SLC6A3, SLC6A4). These findings are consistent with the monoamine neurotransmitter and receptor hypothesis of MDD.
DA, a pivotal catecholamine neurotransmitter in the central nervous system, is involved in psychiatric disorders such as schizophrenia, bipolar disorder, depression and attention-deficit disorder42. A frequent approach to treating these disorders requires the use of medications that target DA receptors (DRD1, DRD2, DRD3, DRD4 and DRD5). Several neuropsychiatric diseases are associated with DA D2 receptor (DRD2), making it a key target for antipsychotic drugs43. Studies have revealed that DRD2 alteration is implicated in the onset of depression42. Through the activation of mTOR, DRD2 modulates synaptic pruning and long-term depression43. Furthermore, stepholidine (a specific agonist of DRD1) exerts antidepressant effects and upregulates synaptogenesis-related protein expression through activating the PKA/mTOR pathway44.
The monoamine neurotransmitter 5-HT associated with the PI3K/Akt/mTOR signalling pathway may serve as a mechanism for Chaigui granules to reverse depression-like behaviour in rats45. The PI3K/Akt/mTOR pathway plays an important role in angiogenesis, cell proliferation, differentiation, migration, apoptosis and other processes46. Recent studies have demonstrated a close association between the PI3K/Akt/mTOR signalling pathway and depression. Plant-derived natural products have been proven to be important modulators of the PI3K/Akt/mTOR signalling pathway in depression. For example, schisantherin B (an active lignan separated from Schisandra chinensis) showed antidepressant activities through regulating the PI3K/Akt/mTOR pathway47. Therefore, this study indicates that PHE may exert antidepressant effects by influencing the PI3K/Akt/mTOR signalling pathway through DRD1, which is consistent with previous research reports.
PHE may emerge as a candidate component for a new antidepressant drug, which can offer more opportunities for for patients. PHE or its derivatives could potentially be used in relieving depression in further. PHE can complement existing antidepressants by regulating PI3K/Akt signalling pathway and other molecular mechanism, thereby enhancing antidepressant efficacy. This complementary effect may help improve the treatment efficiency of depression, reduce adverse effects of drugs, and thus improve the quality of life of patients. Future research can explore the comprehensive mechanism of PHE in antidepressant treatment, as well as how to improve its antidepressant effect by optimizing its chemical structure.
Conclusion
Our study preliminarily proved that PHE has an anti-MDD effect by regulating the mRNA levels of CHRM1, HTR1A and key targets of the PI3K/Akt signalling pathway (PI3K, Akt, mTOR). Network pharmacology analysis revealed the multitarget and multipathway mechanism of PHE. The key targets of PHE in treating MDD probably were SLC6A4, SLC6A3, SLC6A2, MAOA, ALB, DRD2, HTR2A, ABCB1, DRD4 and ADRB2, and the underlying mechanism would be associated with the cGMP-PKG signalling pathway, serotonergic synapse, salivary secretion, dopaminergic synapse and cAMP signalling pathway. In conclusion, our research provides compelling evidence for PHE in MDD therapy through network pharmacology and experiments and a theoretical basis for the clinical application of PHE in the future.
Data availability
The datasets used and analyzed in this study are available in this article.
Change history
17 June 2025
A Correction to this paper has been published: https://doi.org/10.1038/s41598-025-03862-3
References
Bai, Y. et al. Immunotherapy for depression: recent insights and future targets. Pharmacol. Ther. J. Pharmther. 257, 108624 (2024).
World Health Organization. Depression and other common mental disorders: global health estimates. https://apps.who.int/iris/bitstream/handle/10665/254610/WHO-MSD-MER-2017.2-eng.pdf;jsessionid= (2017).
GBD 2019 Mental Disorders Collaborators. Global, regional, and national burden of 12 mental disorders in 204 countries and territories, 1990–2019: a systematic analysis for the global burden of Disease Study 2019. Lancet Psychiatry 9 (2), 137–150 (2022).
Fries, G. R. et al. Molecular pathways of major depressive disorder converge on the synapse. Mol. Psychiatry. 28 (1), 284–297 (2023).
Caldiroli, A., Capuzzi, E., Tagliabue, I. & Cet Augmentative pharmacological strategies in treatment-resistant major depression: a comprehensive review. Int. J. Mol. Sci. 22, 13070 (2021).
Bo, M. et al. Systematic review of Kaixinsan in treating depression: efficacy and pharmacological mechanisms. Front. Behav. Neurosci. 16, 1061877 (2022). Dec 6;.
Ye, L. et al. Si-Ni-San alleviates early life stress-induced depression-like behaviors in adolescence via modulating Rac1 activity and associated spine plasticity in the nucleus accumbens. Front. Pharmacol. 14, 1274121 (2023).
Jiao, H. et al. Xiaoyaosan ameliorates CUMS-induced depressive-like and anorexia behaviors in mice via necroptosis related cellular senescence in hypothalamus. J. Ethnopharmacol. 318(Pt A), 116938 (2024).
Wang, Y., Fan, R. & Huang, X. Meta-analysis of the clinical effectiveness of traditional Chinese medicine formula Chaihu-Shugan-San in depression. J. Ethnopharmacol. 141 (2), 571–577 (2012).
Ge, C. et al. Quercetin attenuates brain apoptosis in mice with chronic unpredictable mild stress-induced depression. Behav. Brain Res. 2465, 114934. https://doi.org/10.1016/j.bbr.2024.114934 (2024).
Wu, X. et al. Icariin prevents depression-like behaviors in chronic unpredictable mild stress-induced rats through Bax/cytoplasm C/caspase-3 axis to alleviate neuronal apoptosis. Cell Mol. Biol. (Noisy-le-grand) 69 (7), 196–204 (2023).
Ma, Z. et al. Baicalin attenuates chronic unpredictable mild stress-induced hippocampal neuronal apoptosis through regulating SIRT1/PARP1 signaling pathway. Behav. Brain Res. 441, 114299 (2023).
Yuan, M. & Yuan, B. Antidepressant-like effects of Rehmannioside A on rats induced by chronic unpredictable mild stress through inhibition of endoplasmic reticulum stress and apoptosis of hippocampus. J. Chem. Neuroanat. 125, 102157 (2022).
Gao, J. et al. Probiotics for the treatment of depression and its comorbidities: a systemic review. Front. Cell. Infect. Microbiol. 13, 1167116 (2023).
Hu, L. et al. Utilizing network pharmacology and experimental validation to investigate the underlying mechanism of phellodendrine on inflammation. PeerJ 10, e13852 (2022).
Keller, J. et al. HPA axis in major depression: cortisol, clinical symptomatology and genetic variation predict cognition. Mol. Psychiatry 22, 527–536 (2017).
Murray, F., Smith, D. W. & Hutson, P. H. Chronic low dose corticosterone exposure decreased hippocampal cell proliferation, volume and induced anxiety and depression like behaviours in mice. Eur. J. Pharmacol. 583(1), 115–127 (2008).
Huang, J. et al. Dihydromyricetin attenuates depressive-like behaviors in mice by inhibiting the AGE-RAGE signaling pathway. Cells 11 (23), 3730 (2022).
Sun, J-Y. et al. Essential oil from the roots of Paeonia lactiflora pall.has protective effect against corticosterone-induced depression in mice via modulation of PI3K/Akt signaling pathway. Front. Pharmacol. 13, 999712 (2022).
Tian, J. S. et al. Metabolomics studies on corticosterone-induced PC12 cells: a strategy for evaluating an in vitro depression model and revealing the metabolic regulation mechanism. Neurotoxicol. Teratol. 69, 27–38 (2013).
Zhang, L. et al. Erxian decoction, a famous Chinese medicine formula, antagonizes corticosterone-induced injury in PC12 cells, and improves depression-like behaviours in mice. Pharm. Biol. 58 (1), 498–509 (2020).
Terada, K. et al. Inhibition of nerve growth factor-induced neurite outgrowth from PC12 cells by dexamethasone: signaling pathways through the glucocorticoid receptor and phosphorylated Akt and ERK1/2. PLoS One 9(3), e93223 (2014).
Szklarczyk, D. et al. The STRING database in 2021: customizable pro-tein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 49, D605–D612 (2021).
Sherman, B. T. et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 50 (W1), W216–W221 (2022).
Kanehisa, M. et al. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 51, D587–D592 (2023).
Hu, L. et al. Network pharmacology combined with bioinformatics analysis to TEXPLORE the potential mechanism of phellodendri chinensis cortex against bladder cancer. Cell Biochem. Biophys. https://doi.org/10.1007/s12013-024-01414-6 (2024).
Li, Y. et al. Integrated network pharmacology and GC-MS-based metabolomics to investigate the effect of Xiang-Su volatile oil against menopausal depression. Front. Pharmacol. 12, 765638 (2021).
Bonvicini C, et al. Serotonin transporter gene polymorphisms and treatment-resistant depression. Prog. Neuropsychopharmacol. Biol. Psychiatry. 34 (6), 934-939 (2010)
Dong C, Wong ML, & Licinio J. Sequence variations of ABCB1, SLC6A2, SLC6A3, SLC6A4, CREB1, CRHR1 and NTRK2: association with major depression and antidepressant response in Mexican-Americans. Mol. Psychiatry. 14 (12), 1105-1118 (2009)
Bi, Y. et al. Influence and interaction of genetic, cognitive, neuroendocrine and personalistic markers to antidepressant response in Chinese patients with major depression. Prog. Neuropsychopharmacol. Biol. Psychiatry 104, 110036 (2021).
Bakusic, J. et al. Increased methylation of NR3C1 and SLC6A4 is associated with blunted cortisol reactivity to stress in major depression. Neurobiol. Stress 13, 100272 (2020).
Okada, S. et al. The potential of SLC6A4 gene methylation analysis for the diagnosis and treatment of major depression. J. Psychiatr Res. 53, 47–53 (2014).
Yin, L. et al. SLC6A2, SLC6A3, DRD2 and DRD4 polymorphisms and neuroendocrine factors predict SSRIs treatment outcome in the Chinese population with major depression. Pharmacopsychiatry 48 (3), 95–103 (2015).
Mathew, B. et al. Enzyme inhibition assays for monoamine oxidase. Methods Mol. Biol. 2761, 329–336 (2024).
Shi, Q. et al. Cardiac MAO-A inhibition protects against catecholamine-induced ventricular arrhythmias via enhanced diastolic calcium control. Cardiovasc. Res. 10, cvae012. https://doi.org/10.1093/cvr/cvae012 (2024).
Thase, M. E., Trivedi, M. H. & Rush, A. J. MAOIs in the contemporary treatment of depression. Neuropsychopharmacology 12, 185–219 (1995).
Huang, T. et al. Association of ovarian tumor β2-adrenergic receptor status with ovarian cancer risk factors and survival. Cancer Epidemiol. Biomark. Prev. 25 (12), 1587–1594 (2016).
Liu, S. et al. Fat mass and obesity-associated protein regulates RNA methylation associated with depression-like behavior in mice. Nat. Commun. 26 (1), 6937 (2021).
Rajalingam, D. et al. Repeated social defeat promotes persistent inflammatory changes in splenic myeloid cells; decreased expression of β-arrestin-2 (ARRB2) and increased expression of interleukin-6 (IL-6). BMC Neurosci. 21 (1), 25 (2020).
Li, C. et al. RNA methylations in depression, from pathological mechanism to therapeutic potential. Biochem. Pharmacol. 215, 115750. https://doi.org/10.1016/j.bcp.2023.115750 (2023).
Bortolotto, V. et al. Salmeterol, a β2 adrenergic agonist, promotes adult hippocampal neurogenesis in a region-specific manner. Front. Pharmacol. 10, 1000. https://doi.org/10.3389/fphar.2019.01000 (2019).
Graham, D. L. et al. Loss of dopamine D2 receptors increases parvalbumin-positive interneurons in the anterior cingulate cortex. ACS Chem. Neurosci. 6 (2), 297–305 (2015).
Zhang, Y. Q. et al. Dopamine D2 receptor regulates cortical synaptic pruning in rodents. Nat. Commun. 12 (1), 6444 (2021).
Zhao, Y. et al. The pharmacological mechanism of chaihu-jia-longgu-muli-Tang for treating depression: integrated meta-analysis and network pharmacology analysis. Front. Pharmacol. 14, 1257617 (2023).
Tian, J. S. et al. Combining network pharmacology and experimental verification to reveal the mechanism of Chaigui granules in the treatment of depression through PI3K/Akt/mTOR signaling pathways. Metab. Brain Dis. 38 (8), 2849–2864 (2023).
Fakhri, S. et al. Natural products attenuate PI3K/Akt/mTOR signaling pathway: a promising strategy in regulating neurodegeneration. Phytomedicine 91, 153664 (2021).
Xu, M. et al. Schisantherin B improves the pathological manifestations of mice caused by behavior desperation in different ages-depression with cognitive impairment. Biomol. Ther. 27, 160–167 (2019).
Acknowledgements
This work was supported by Fundamental Research Program of Shanxi Province (No. 202203021222278).
Author information
Authors and Affiliations
Contributions
Lili Hu designed the project. Lili Hu, Na Wu, and Jue Wang performed the experiment. Na Wu and Jue Wang collected and analyzed the data. Lili Hu, Donghui Cai and Na Wu wrote the manuscript. All authors have approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this Article was revised: The original version of the Article contained an error in Figure 7, where an image in Figure 7D was a duplicate of Figure 7C.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Hu, L., Wu, N., Wang, J. et al. Network pharmacology combined with experimental verification for exploring the potential mechanism of phellodendrine against depression. Sci Rep 15, 1958 (2025). https://doi.org/10.1038/s41598-024-84771-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-84771-9