Abstract
The seasonality and epidemiology of viral acute respiratory infections (ARIs) have changed since the coronavirus disease 2019 pandemic. However, molecular-based ARI surveillance has not been conducted in Japan. We developed a regional surveillance program to define the local epidemiology of ARIs. Between December 2023 and March 2024, 2,992 upper respiratory samples collected from patients suspected of having ARIs at five facilities in Kyoto City, Japan, were tested for SARS-CoV-2, influenza virus, and respiratory syncytial virus (RSV) using RT‒PCR. Samples negative for these viruses were randomly selected for testing with the FilmArray Respiratory Panel, and the detection rates of other viruses were estimated. SARS-CoV-2, influenza virus, and RSV were detected in 598 (20.3%), 165 (5.6%), and 40 (1.4%) of the 2,949 samples with valid RT‒PCR results, respectively. The most prevalent viruses in the < 6, 6–17, 18–64, and ≥ 65 year age groups were rhinovirus/enterovirus, RSV, and SARS-CoV-2; influenza virus, seasonal coronavirus, and rhinovirus/enterovirus; SARS-CoV-2, seasonal coronavirus, and influenza virus; and SARS-CoV-2, seasonal coronavirus, and influenza virus, respectively. Significant differences in the detection rates of these viruses were detected between the age groups. This study highlights the importance of age-stratified molecular-based surveillance for a comprehensive understanding of the epidemiology of ARIs.
Similar content being viewed by others
Introduction
The seasonality and epidemiology of viral acute respiratory infections (ARIs) have changed since the beginning of the coronavirus disease 2019 (COVID-19) pandemic in 20201,2. For example, the seasonal peak of human respiratory syncytial virus (RSV) infection in Japan shifted from autumn to summer after no peak was observed in 20203. The COVID-19 pandemic has highlighted the importance of reliable molecular detection tests (nucleic acid amplification tests) for the diagnosis and control of this disease4. Although molecular testing platforms for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) have been introduced in many Japanese hospitals, rapid antigen tests that have lower detection sensitivity than do molecular tests5,6 are the mainstay of diagnostic methods for respiratory viral infections, including COVID-19 and influenza7. Rapid antigen tests for RSV, adenovirus, and human metapneumovirus are available for in vitro diagnostics but are not used for adults due to a lack of reimbursement. Recently, multiplexed molecular assays that can detect SARS-CoV-2, influenza virus, and RSV (SARS-CoV-2/Flu/RSV) simultaneously and the BioFire FilmArray molecular panel assay, which can additionally detect adenovirus, seasonal human coronavirus, human metapneumovirus, human parainfluenza virus, Bordetella pertussis, Bordetella parapertussis, Chlamydia pneumoniae, and Mycoplasma pneumoniae, have become available8,9. Despite increasing recognition of the importance of molecular assays for clinical management, antimicrobial stewardship, and infection control10,11, these tests are usually performed for a limited number of patients because of their high cost and lack of availability in the majority of hospitals and clinics.
Japan has a case-based national sentinel surveillance (National Epidemiological Surveillance of Infectious Diseases, NESID) program, which is based on the Infectious Diseases Control Law12, for only SARS-CoV-2/Flu/RSV among respiratory viral infections. Surveillance for COVID-19 transitioned from all-case reports to sentinel surveillance in May 2024. Currently, COVID-19 cases are defined based on clinical diagnosis and positive rapid antigen or molecular detection tests. COVID-19 cases can also be diagnosed based on clinical diagnosis if they have confirmed COVID-19 in their family members. Influenza and RSV cases are defined based on a clinical diagnosis and positive rapid antigen tests. Clinical diagnosis was made according to clinicians’ discretion without defined criteria. In Kyoto city, which has a population of approximately 1 million, 69 medical institutions (43 pediatric sites, 22 clinics, and 4 hospitals) report the number of influenza and COVID-19 cases, and 43 pediatric sentinel sites report the number of RSV cases weekly. This surveillance program may not accurately represent epidemiology because the spectrum of pathogens, targeted populations, and diagnostic methods are limited. For example, RSV surveillance does not target adults or elderly individuals with RSV, who have more severe disease than do those with COVID-19 or influenza13.
We developed a local surveillance framework based on molecular testing for COVID-19 (CovPCRnet) in Kyoto and its neighboring regions. After the withdrawal of the COVID-19 containment strategy by the government in May 2023, we enhanced this framework to include other common viral pathogens to conduct molecular-based regional surveillance that can fill knowledge gaps in NSEID. In this study, we report the age-stratified epidemiology of viral ARIs in the 2023–24 winter season in Kyoto city, Japan.
Results
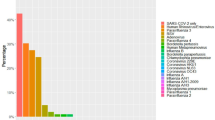
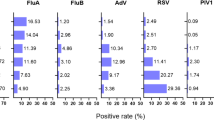
A total of 2,992 upper respiratory samples were obtained, and 2,949 samples that yielded valid CDC RT‒PCR results were included in the analysis (Table 1 and Supplementary Table S1). SARS-CoV-2, influenza virus, and RSV were detected in 598 (20.3%), 165 (5.6%), and 40 (1.4%) of the samples, respectively. Among the remaining 2,155 SARS-CoV-2/Flu/RSV-negative samples, 928 (43.1%) were selected for FilmArray testing. Random selection was applied to samples from three facilities that provided > 200 samples and was not applied to those from the other two facilities that had fewer samples from the ≥ 65 year age group (53.5–83.5% vs. 0–12.2%; Supplementary Fig. S1). This selection resulted in a relatively lower proportion of samples from the ≥ 65 year age group for FilmArray testing (Table 1). FilmArray detected seasonal coronavirus, rhinovirus/enterovirus, metapneumovirus, parainfluenza virus, and adenovirus in 86 (9.3%), 82 (8.8%), 26 (2.8%), 12 (1.4%), and 11 (1.2%) of the 928 samples, respectively. FilmArray detected SARS-CoV-2 in 4 samples (0.4%), whereas influenza virus and RSV were not detected. Based on these FilmArray results, the detection rates of viruses other than SARS-CoV-2/Flu/RSV were estimated as follows: seasonal coronavirus, 6.8%; rhinovirus/enterovirus, 6.5%; metapneumovirus, 2.0%; parainfluenza virus, 1.0%; and adenovirus, 0.9% (Figs. 1 and 2). The three most prevalent viruses were rhinovirus/enterovirus (32.6%), RSV (15.4%), and SARS-CoV-2 (9.8%) in the < 6 year age group; influenza virus (15.9%), seasonal coronavirus (14.3%), and rhinovirus/enterovirus (9.5%) in the 6–17 year age group; and SARS-CoV-2 (24.4% and 18.6%), seasonal coronavirus (8.6% and 3.6%), and influenza virus (8.2% and 2.8%) in the 18–64 and ≥ 65 year age groups, respectively. SARS-CoV-2, seasonal coronavirus, rhinovirus/enterovirus, and influenza virus were most commonly detected in the 18–64, 6–17, < 6, and 6–17 year age groups, respectively (Fig. 2). RSV was most commonly detected in the < 6 year age group (15.4%), followed by the 6–17 year age group (2.3%). The 18–64 and ≥ 65 year age groups had lower detection rates (0.8% and 0.6%, respectively). Adenovirus and parainfluenza virus were most commonly detected in the < 6 year age group (Fig. 2). Metapneumovirus was most commonly detected in the 6–17 year age group. Influenza virus A, RSV A, coronavirus OC43, and parainfluenza virus 1 were the most common subtypes across all age groups, except for the higher detection rate of influenza virus B than influenza virus A in the 6–17 year age group (Supplementary Fig. S2). Only one sample was positive for bacterial pathogens (Mycoplasma pneumoniae in the 6–17 year age group; Supplementary Fig. S2). Multiple pathogens (combinations of SARS-CoV-2, influenza virus, or RSV) were detected in 0.3–1.1% of the samples in each age group (Supplementary Fig. S3a). Among the 928 SARS-CoV-2/Flu/RSV-negative samples, multiple pathogens were detected most frequently in the < 6 year age group (13.9%) but were not common in the other groups (0–1.4%; Supplementary Fig. S3b).
Detection rates of respiratory viruses according to age group, 2023–24 winter season, Kyoto, Japan. The detection rates of SARS-CoV-2/Flu/RSV were calculated for 2,949 samples, and those of the other pathogens were estimated from a subset of 928 SARS-CoV-2/Flu/RSV-negative samples that underwent FilmArray testing. FilmArray results for SARS-CoV-2/Flu/RSV were not considered. Subtypes of influenza virus (A and B), RSV (A and B), seasonal human coronavirus (229E, HKU1, NL63, and OC43), and human parainfluenza virus (1–4) are shown in Supplementary Fig. S2.
Comparison of the detection rates of respiratory viruses among different age groups, 2023–24 winter season, Kyoto, Japan. The detection rates of SARS-CoV-2/Flu/RSV were calculated for 2,949 samples (panel a), and those of the other pathogens were calculated for a subset of 928 SARS-CoV-2/Flu/RSV-negative samples that underwent FilmArray testing (panel b). The data in panel b are not estimated for 2,966 samples. Asterisks indicate a P value of < 0.05.
The mean monthly number of COVID-19 cases per sentinel site reported by the NESID program peaked between January and February (Fig. 3). The monthly mean number of positive samples per site for SARS-CoV-2 showed a similar trend, which was largely influenced by the samples obtained from the 18–64 and ≥ 65 year age groups. For influenza cases, the NESID data revealed high numbers in December–February and a lower number in March. A gradual monthly decrease was observed in the 18–64 and ≥ 65 year age groups, whereas the 6–17 year age group peaked in January, and the < 6 year age group had a second peak in February. For RSV, the NESID data showed nearly zero cases in December–February and an increase in March. The total number of positive samples per site from all age groups tended to increase. Positive samples from the 18–64 year age group were present in all months and increased over time. The samples from < 6 year age group, followed by ≥ 65 and 18–64 year age groups contributed to a large increase in March. The monthly trends for seasonal coronavirus, rhinovirus/enterovirus, metapneumovirus, parainfluenza virus, and adenovirus detections are shown in Supplementary Fig. S4.
Monthly changes in the mean number of positive samples per site of SARS-CoV-2 (panel a), influenza virus (panel b), and RSV (panel c) among different age groups and the mean number of cases per sentinel site according to the National Epidemiological Surveillance of Infectious Diseases (NESID) program in Kyoto city, Japan.
Discussion
This molecular-based regional surveillance for viral ARIs was designed to address several limitations of the current surveillance, NESID. First, the spectrum of pathogens (only SARS-CoV-2/Flu/RSV) in the NESID is limited. Second, sampling and reporting biases are present. The participating sites are concentrated in pediatric clinics. In particular, RSV cases have been limited to children. Cases are determined at the clinician’s discretion without defined criteria, and microbiological diagnosis is mostly dependent on rapid antigen tests. Third, only the mean number of cases per site without age information was reported, without denominators. In contrast, our surveillance targets a broader spectrum of pathogens from all age groups and can generate age-stratified positive sample counts and rates. Our laboratory-based surveillance performed centralized molecular testing, which can provide highly accurate detection of pathogens compared with rapid antigen tests at each site. The collected clinical samples can also be used for further testing (e.g., genomic analysis and viral culture) and research.
According to our surveillance program, we estimated the prevalence of viral ARIs for each age group. The detection rates of respiratory viruses, except for that of metapneumovirus, significantly varied according to age group (Fig. 2). These results are concordant with those of previous studies that reported different prevalences of pathogens according to age, period, and region14,15,16. The discrepancies in the monthly trends and the number of cases or positive samples between the NESID and our data (Fig. 3) can be largely explained by the sampling bias of NESID toward children, although differences in the sampling sites might also have affected the data. These observations confirmed the importance of age-stratified local surveillance.
The viruses detected in this study are all considered causes of hospital admission17 and pneumonia18,19. Among these, SARS-CoV-2/Flu/RSV are especially important because of their prevalence and the availability of vaccines and therapeutics. Following recent approval of vaccines for elderly individuals and maternal immunization20, our surveillance provides valuable data regarding adult RSV infections. Data for viruses other than SARS-CoV-2/Flu/RSV are of public health importance because these viruses can cause endemics14,15,16,17 and outbreaks within facilities21,22. Previous reports indicate that the detection of viruses other than SARS-CoV-2/Flu/RSV has led to the identification of causative organisms in ARI outbreaks due to unknown etiology, the initiation and cessation of isolation and transmission-based precautions in outbreak settings or routine screening activities for symptomatic and asymptomatic admissions, and the avoidance of mixing patients with different organisms11,21,22,23,24,25. In a clinical setting, the detection of these viruses may facilitate subsequent modifications of empiric broad-spectrum antimicrobials, even in the absence of specific interventions.
There are no surveillance systems that target a broad range of respiratory viruses in Japan. Only several retrospective studies from a single institution using FilmArray tests have been reported. One study conducted in Yamanashi that compared the COVID-19 pandemic and post-COVID-19 (May–September 2023) periods revealed an increase in overall pathogen detection rates (specifically, metapneumovirus, rhinovirus/enterovirus, and RSV) in all age groups15. In children ≤ 10 years old, the positivity rates of adenovirus, Bordetella pertussis, and parainfluenza viruses 2 and 4 also increased, whereas the positivity rates of SARS-CoV-2, seasonal coronaviruses HKU1 and OC43, and parainfluenza virus 1 decreased. During the COVID-19 pandemic (2022–2022), a study in Nara detected a higher positivity rate for rhinovirus/enterovirus than SARS-CoV-2 throughout most of the study period and peaks of RSV and parainfluenza virus 3 detection in the summer of 202116. A large-scale study that examined > 50,000 samples obtained from inpatients or children for whom admission was planned in the USA reported the resurgence of viruses other than SARS-CoV-2 during 202217. Among pediatric patients aged < 18 years, rhinovirus/enterovirus had the highest incidence in almost all months, with distinct seasonal increases in the incidence of different viruses in the following order: seasonal coronavirus, influenza virus, metapneumovirus, parainfluenza virus, and RSV. Our results indicate that viruses other than SARS-CoV-2/Flu/RSV cause ARIs more frequently. Rhinovirus/enterovirus was the most prevalently detected in the < 6 year age group; seasonal coronavirus and rhinovirus/enterovirus were more prevalent than SARS-CoV-2 and influenza virus were in the 6–17 year age group; and seasonal coronavirus was more prevalent than influenza virus was in the ≥ 18 year age group. These epidemiological data highlight the importance of a real-time local surveillance program with molecular diagnostics that cover a broad range of viruses.
Multiplexed pathogen detection assays can detect viral coinfections14,26. A recent study in the USA revealed that viral coinfection occurred more frequently in children (21% vs. 4% in those < 18 and ≥ 18 years, respectively), and coinfection rates were much lower than expected on the basis of the incidence of each virus, suggesting the presence of viral exclusionary effects17. Our data also revealed a relatively high rate of multiple viruses in children, which was in line with these observations.
This study has several limitations. We probably underestimated the detection rates of viruses other than SARS-CoV-2/Flu/RSV because of the presence of coinfections in SARS-CoV-2/Flu/RSV-positive samples. This bias may not have a large impact because SARS-CoV-2, influenza virus, and RSV are associated with the lowest probability of coinfection17. Sampling bias could be present due to the relatively low numbers of participating facilities and samples from children and the use of a random sampling strategy for FilmArray testing. Notably, this surveillance is based on the test positivity of symptomatic patients and may include recovered patients with prolonged viral shedding from prior infection episodes.
We demonstrated the differences in the detection rates and trends of respiratory viruses among age groups using the developed local molecular surveillance program. This surveillance is unique in terms of age stratification, the molecular detection basis for SARS-CoV-2/Flu/RSV, and the inclusion of a broad range of viruses. Our data will help accurately elucidate the epidemiology of viral ARIs at different ages and may help clinicians and public health professionals plan infection control and prevention strategies. Further detailed analysis, including viral genomics, will enhance our knowledge of the spread of respiratory viruses.
Methods
Clinical specimens
Between December 2023 and March 2024, all nasopharyngeal or nasal swabs in viral transport medium that were collected for molecular testing for SARS-CoV-2 or SARS-CoV-2/Flu/RSV because of the clinical suspicion of ARIs, including COVID-19, at five acute care hospitals or PCR testing centers were sent to the reference laboratory at Kyoto University (Supplementary Table S1). The collected patient information included age and outpatient/inpatient status. The definitions of outpatient and inpatient status were determined based on the patient’s ___location at the time of specimen collection.
Molecular diagnosis
RNA was extracted from 200 μL samples using a MagNA Pure 96 DNA and Viral NA Small Volume Kit and a MagNA Pure 96 Instrument (Roche, Basel, Switzerland) and was eluted in a final volume of 50 μL. The CDC Influenza SARS-CoV-2 Multiplex assay was performed with TaqPath 1-Step Multiplex Master Mix (Thermo Fisher Scientific) following the instructions (https://www.cdc.gov/coronavirus/2019-ncov/lab/multiplex.html). Samples that yielded invalid results due to negative human RNase P internal control results were excluded from the analysis. The CDC RSV duplex assay was performed following the original protocol27. RT‒PCR was performed on a QuantStudio5 Real-Time PCR System (Thermo Fisher Scientific). Samples that tested negative for SARS-CoV-2/Flu/RSV by these two assays were randomly selected (if > 200 samples were collected from a facility) for FilmArray® Respiratory Panel 2.1 (bioMérieux Japan, Tokyo, Japan) testing. A systematic random sampling approach was applied to select samples in the order in which they were received. Every third sample was picked starting from the first sample in the sequence. The detection rates of respiratory viruses other than SARS-CoV-2/Flu/RSV (seasonal coronavirus, rhinovirus/enterovirus, metapneumovirus, parainfluenza virus, and adenovirus) were estimated via the following formula: the rate that tested negative for SARS-CoV-2/Flu/RSV multiplied by the positive rate of the FilmArray results. The FilmArray results for SARS-CoV-2/Flu/RSV were not used. Surveillance data were made public on the laboratory website and updated regularly.
Statistical analysis
The detection rates of pathogens were calculated according to four age groups: < 6 (infants and preschoolers), 6–17 (school-aged), 18–64 (adults), and ≥ 65 (elderly) years. Fisher’s exact test with Bonferroni correction (two-tailed) was applied for multiple comparisons to compare the positive rates among the age groups. A p value < 0.05 was considered to indicate statistical significance. All the statistical analyses were performed using the R software (https://cran.r-project.org).
NESID data for the number of cases of COVID-19, influenza, and RSV infection were obtained from the official webpage of the Kyoto City Institute of Public Health (https://www.city.kyoto.lg.jp/menu3/category/41-6-3-0-0-0-0-0-0-0.html).
Data availability
Data is provided within the manuscript or supplementary information files.
References
Schuz, M. L. et al. Global prevalence of respiratory virus infections in adults and adolescents during the COVID-19 pandemic: A systematic review and meta-analysis. Int. J. Infect. Dis. 137, 16–24 (2023).
Rios-Guzman, E. et al. Deviations in RSV epidemiological patterns and population structures in the United States following the COVID-19 pandemic. Nat. Commun. 15, 3374 (2024).
National Institute of Infectious Diseases. Infectious Diseases Weekly Report (IDWR) https://www.niid.go.jp/niid/en/idwr-e.html.
Mardian, Y., Kosasih, H., Karyana, M., Neal, A. & Lau, C. Y. Review of current COVID-19 diagnostics and opportunities for further development. Front. Med. (Lausanne). 8, 615099 (2021).
Azar, M. M. & Landry, M. L. Detection of influenza A and B viruses and respiratory syncytial virus by use of Clinical Laboratory Improvement Amendments of 1988 (CLIA)-waived point-of-care assays: A paradigm shift to molecular tests. J. Clin. Microbiol. 56 (2018).
Matsumura, Y., Yamazaki, W., Noguchi, T., Yamamoto, M. & Nagao, M. Analytical and clinical performances of seven direct detection assays for SARS-CoV-2. J. Clin. Virol. Plus 3, 100138 (2023).
Kyo, H. et al. A population-based study of the trend in SARS-CoV-2 diagnostic modalities from the beginning of the pandemic to the Omicron surge in Kyoto City, Kyoto, Japan. BMC Public Health 23, 2551 (2023).
Jensen, C. B. et al. Evaluation of the analytical and clinical performance of two RT-PCR based point-of-care tests; Cepheid Xpert(R) Xpress CoV-2/Flu/RSV plus and SD BioSensor STANDARD M10 Flu/RSV/SARS-CoV-2. J. Clin. Virol. 172, 105674 (2024).
Huang, H. S. et al. Multiplex PCR system for the rapid diagnosis of respiratory virus infection: Systematic review and meta-analysis. Clin. Microbiol. Infect. 24, 1055–1063 (2018).
Hanson, K. E. et al. Molecular testing for acute respiratory tract infections: Clinical and diagnostic recommendations from the IDSA’s diagnostics committee. Clin. Infect. Dis. 71, 2744–2751 (2020).
Wils, J., Saegeman, V. & Schuermans, A. Impact of multiplexed respiratory viral panels on infection control measures and antimicrobial stewardship: A review of the literature. Eur. J. Clin. Microbiol. Infect. Dis. 41, 187–202 (2022).
Zaraket, H. & Saito, R. Japanese surveillance systems and treatment for influenza. Curr. Treat. Options Infect. Dis. 8, 311–328 (2016).
Surie, D. et al. Disease severity of respiratory syncytial virus compared with COVID-19 and influenza among hospitalized adults aged >/=60 Years - IVY Network, 20 U.S. States, February 2022-May 2023. MMWR Morb. Mortal. Wkly. Rep. 72, 1083–1088 (2023).
Tsagarakis, N. J. et al. Age-related prevalence of common upper respiratory pathogens, based on the application of the FilmArray Respiratory panel in a tertiary hospital in Greece. Medicine (Baltimore) 97, e10903 (2018).
Hirotsu, Y. et al. Changes in viral dynamics following the legal relaxation of COVID-19 mitigation measures in Japan from children to adults: A single center study, 2020–2023. Influenza Other Respir. Viruses 18, e13278 (2024).
Kitagawa, D. et al. Epidemiology of respiratory tract infections using multiplex PCR in a Japanese acute care hospital during the COVID19 pandemic. Heliyon 9, e14424 (2023).
Weidmann, M. D., Green, D. A., Berry, G. J. & Wu, F. Assessing respiratory viral exclusion and affinity interactions through co-infection incidence in a pediatric population during the 2022 resurgence of influenza and RSV. Front. Cell. Infect. Microbiol. 13, 1208235 (2023).
Ruuskanen, O., Lahti, E., Jennings, L. C. & Murdoch, D. R. Viral pneumonia. Lancet 377, 1264–1275 (2011).
Jain, S. et al. Community-acquired pneumonia requiring hospitalization among U.S. children. N. Engl. J. Med. 372, 835–845 (2015).
Zeng, B. et al. Efficacy and safety of vaccines to prevent respiratory syncytial virus infection in infants and older adults: A systematic review and meta-analysis. Int. J. Infect. Dis. 146, 107118 (2024).
Ohyama, K. et al. Resurgence of human coronavirus OC43 at a long-term care facility during the coronavirus disease 2019 (COVID-19) pandemic: Outbreak investigation. Antimicrob. Steward. Healthc. Epidemiol. 3, e97 (2023).
Karimata, Y. et al. Clinical features of human metapneumovirus pneumonia in non-immunocompromised patients: An investigation of three long-term care facility outbreaks. J. Infect. Dis. 218, 868–875 (2018).
Kuribayashi, M. et al. Clinical influence of multiplex polymerase chain reaction routine uses in urgent pediatric admissions. Pediatr. Int. 65, e15525 (2023).
Babady, N. E. et al. Comparison of the Luminex xTAG RVP Fast assay and the Idaho Technology FilmArray RP assay for detection of respiratory viruses in pediatric patients at a cancer hospital. J. Clin. Microbiol. 50, 2282–2288 (2012).
Cunha, B. A., Connolly, J. J., Musta, A. C. & Abruzzo, E. Infection control implications of protracted lengths of stay with noninfluenza viral influenza-like illnesses in hospitalized adults during the 2015 influenza A (H3N2) epidemic. Infect. Control Hosp. Epidemiol. 36, 1368–1370 (2015).
Lustrek, M. et al. Influenza A, Influenza B, human respiratory syncytial virus and SARSCoV-2 molecular diagnostics and epidemiology in the post COVID-19 era. Respir. Res. 25, 234 (2024).
Wang, L. et al. Duplex real-time RT-PCR assay for detection and subgroup-specific identification of human respiratory syncytial virus. J. Virol. Methods 271, 113676 (2019).
Acknowledgements
We thank Tsunehiro Shimizu, Junko Akeyama, Akihiko Hayashi (Kyoto City Hospital), Keita Ninomiya (Kyoto Katsura Hospital), Yoshinori Sanada (Kyoto Kujo Hospital), Yukiko Okamoto, Yuko Nishikawa, and Masami Tada (PSS-Kyoto University Laboratory) for sample collection. We also thank Ayumi Sakaguchi, Nana Imai, Azusa Asai, Kaori Ishizaki, Isao Nakamoto, and Kazuki Kitamura (Department of Clinical Laboratory Medicine, Kyoto University Graduate School of Medicine) for their technical assistance.
Author information
Authors and Affiliations
Contributions
YM designed the study and the experiments. YM, MY and MN contributed materials and data collection. YM, MY, Y.Tsuda, KS, Y.Tsuchido, SY, and TN conducted the experiments. KT and MN acquired the funding and supervised the study. YM analyzed the data and wrote the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
YM received research funds from Beckman Coulter, Precision System Science, and Toyobo. MN received research funds from Beckman Coulter and Precision System Science. MY, Y.Tsuda, KS, Y.Tsuchido, SY, TN, and KT declare no potential conflict of interest.
Ethical approval
This study was performed in accordance with the principles of the Declaration of Helsinki. The Ethics Committee of Kyoto University Graduate School and the Faculty of Medicine approved this study (R4195), and the need to obtain informed consent from each patient was waived.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Matsumura, Y., Yamamoto, M., Tsuda, Y. et al. Epidemiology of respiratory viruses according to age group, 2023–24 winter season, Kyoto, Japan. Sci Rep 15, 924 (2025). https://doi.org/10.1038/s41598-024-85068-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-85068-7