Abstract
The coexistence of multiple resistance genes within a single bacterial strain presents a significant public health challenge, as it complicates treatment options and accelerates the spread of multidrug resistance. While the co-occurrence of blaNDM-1 and mcr-1 genes in Klebsiella pneumoniae (KP) is uncommon, this study reports the biological characterization of a K. pneumoniae isolate, L5151, derived from a patient with diarrhea. This strain carries blaNDM-1, mcr-1, and blaCTX-M-199 resistance genes simultaneously. Multilocus sequence typing (MLST) analysis identified the L5151 strain as an ST499 type. Antimicrobial susceptibility testing (AST) was conducted via agar dilution and the broth microdilution procedure. The AST results revealed that L5151 is resistant to a variety of antibiotics. Whole-genome sequencing (WGS) and bioinformatics analysis were performed to determine the genetic composition of the strain, including the presence and characteristics of resistance genes. The results of S1 nuclease-pulsed field gel electrophoresis (S1-PFGE) and Southern blotting confirmed that the L5151 strain harbors three plasmids and that the plasmid carrying mcr-1 (pL5151_MCR_CTX) is of the IncI2(Delta) type, whereas the plasmid carrying blaNDM-1 (pL5151_NDM) is of the IncN type. The combination of these plasmids in recipient strains conferred enhanced resistance to carbapenems and colistin, highlighting the potential for increased treatment challenges. This study emphasizes the importance of ongoing surveillance of multidrug-resistant strains to prevent their spread and outbreaks. These findings provide critical insights for clinical treatment strategies and infection prevention and control measures. Enhanced surveillance and targeted interventions are essential to manage the public health risks posed by multidrug-resistant strains.
Similar content being viewed by others
Introduction
Carbapenem-resistant Klebsiella pneumoniae (CRKP) is considered one of the most serious global health threats today1 because of the high morbidity and mortality linked to infections caused by such bacteria2,3. The increasing resistance of K. pneumoniae to a wide range of antibiotics has become a major clinical challenge, largely driven by the widespread use of antibiotics and the dissemination of resistance genes via plasmids capable of horizontal transfer4,5.
The gene blaNDM is responsible for the production of New Delhi metallo-β-lactamase (NDM), an enzyme that breaks down the majority of β-lactam antibiotics, which are typically considered first-line drugs to treat infections6,7. The emergence of NDM-producing bacteria severely restricts the availability of clinical therapeutic alternatives, exacerbating the complexity of medical interventions against the infectious diseases induced by these pathogens. In addition to NDM, the mcr-1 gene represents another critical resistance factor encoding a transferase that confers resistance to colistin. Colistin is usually considered the last resort of defense against gram-negative bacterial infections8,9,10. The clinical effectiveness of colistin has been compromised by the emergence and prevalence of mcr resistance genes, which has consequently led to significant treatment challenges11,12. Furthermore, among Enterobacteriaceae worldwide, CTX-M-type β-lactamases have emerged as the predominant form of extended-spectrum β-lactamases (ESBLs)13. Recent studies have shown that the blaCTX-M-199 gene, in particular, has gained attention because it originates from a mutation of blaCTX-M-64, resulting in a variant that demonstrates emerging resistance to cephalosporins and tazobactam-sulbactam combinations, which poses a new challenge for therapeutic selection14.
Previous studies have indicated that the coexistence of blaNDM-1 and mcr-1 is rare in K. pneumoniae15,16. Our study is the first to report the co-occurrence of blaCTX-M-199 alongside blaNDM-1 and mcr-1 within the same strain. The genetic background of the isolate was examined through whole-genome sequencing. Our research aims to evaluate the phenotypes of K. pneumoniae strains harboring blaNDM-1, mcr-1, and blaCTX-M-199 and to establish a scientific foundation for the formulation of efficacious treatment and control strategies during the ensuing period. Our results highlight the necessity of additional monitoring of K. pneumoniae strains that carry both β-lactamase and colistin resistance genes. This ongoing monitoring is crucial for deepening our understanding of these resistant strains and guiding future therapeutic and control measures to combat multidrug-resistant pathogens.
Methods
This study was approved by the Institutional Review Board (IRB) of the Research Ethics Committee of the First Affiliated Hospital, College of Medicine, Zhejiang University (Approval No. 2018-752). As this was a retrospective study, the requirement for informed consent was waived in accordance with the Ethical Review Measures for Life Science and Medical Research Involving Human Subjects, issued by the National Science and Technology Ethics Committee of China.
A carbapenem-resistant K. pneumoniae strain, designated strain L5151, was isolated from fecal samples of a patient who presented with diarrhea. The isolate was subjected to species identification, along with the determination of carbapenemase-encoding genes and the colistin resistance gene, via matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) (Bruker, Bremen, Germany) and PCR amplification following previously described methods. 17,18
The minimum inhibitory concentration (MIC) values of the strains against commonly used antibiotics were determined via the agar dilution method and broth microdilution method for antimicrobial susceptibility testing (AST). Escherichia coli ATCC 25,922 served as the reference for quality assurance. The interpretation of colistin was carried out following the guidelines of the European Committee for Accreditation of Pharmacovigilance Tests (EUCAST) (https://www.eucast.org/). The assessment of tigecycline, omadacycline, and eclacycline adhered to the definitions provided by the U.S. Food and Drug Administration (FDA) (https://www.fda.gov), which can be retrieved from the FDA’s official website (Tigecycline-Injectable Products | U.S. Food and Drug Administration; Omadacycline-Injectable and Oral Products |U.S. Food and Drug Administration; Elacycline-Injectable Products |U.S. Food and Drug Administration).19 The remaining antibiotics were evaluated based on the standards set by the Clinical and Laboratory Standards Institute (CLSI), which are accessible at CLSI (https://clsi.org). Virulence-associated genes were identified using the Virulence Factor Database (VFDB) (http://www.mgc.ac.cn/VFs/).20
The S1 nuclease pulsed-field gel electrophoresis (S1-PFGE) method was employed to validate the quantity and dimensions of the plasmids within the K. pneumoniae strain L5151. DIG-labeled probes were subsequently adopted to conduct Southern blotting and hybridization procedures, and mcr-1 and blaNDM-1-specific probes were employed to identify the locations of the mcr-1 and blaNDM genes. 21.To evaluate the transferability of plasmid pL5151_MCR, we carried out conjugation experiments. Using rifampicin-resistant E. coli EC600 as a recipient strain, we co-cultured donors and recipients and inoculated them on MH medium supplemented with 2 μg/mL colistin and 200 μg/mL rifampicin for selection of transconjugants. Resistance genes such as mcr-1 and blaCTX-M-199 were confirmed by PCR in the transconjugants. The transfer efficiency was calculated by dividing the number of transconjugants (CFU/mL) by the number of donor bacteria (CFU/mL)22.
The genomic DNA of K. pneumoniae L5151 was extracted using a Bacterial DNA Kit (QIAGEN, Hilden, Germany) for both Illumina and Nanopore sequencing. It was then sequenced on the Illumina NovaSeq 6000 (Illumina, San Diego, CA, USA) to generate high-accuracy short reads and on the Oxford Nanopore platform (Oxford Nanopore Technologies, Oxford, UK) to obtain long reads. Hybrid genome assembly was performed using Unicycler v0.4.7(https://github.com/rrwick/Unicycler), with Nanopore long reads used for preliminary assembly and Illumina short reads for error correction to improve accuracy.23
The genome was annotated using Proksee (https://proksee.ca/). The ResFinder24 (https://cge.food.dtu.dk/services/ResFinder/) and PlasmidFinder25 (https://cge.food.dtu.dk/services/PlasmidFinder/) databases were utilized to ascertain the antibiotic resistance genes (ARGs) and plasmid incompatibility types that were obtained. The multilocus sequence typing (MLST) method was established via pubMLST (http://pubmlst.org/ecloacae). Finally, BLAST Ring Image Generator (BRIG) version 0.95 (https://brig.sourceforge.net/) was used to plot the rings of the plasmids pL5151_NDM and pL5151_MCR_CTX plasmids 26. Plasmids obtained for antimicrobial resistance genes and replicon types were identified using an online tool (http://www.genomicepidemiology.org/). Transposons and insertion sequence (IS) elements were characterized using the ISFinder database (http://www-is.biotoul.fr/).
Results and discussion
The K. pneumoniae strain L5151 was isolated from fecal samples, it was classified as ST499 via a search within the K. pneumoniae motif/sequence definition database (https://cge.cbs.dtu.dk/services/MLST/). Based on the WGS data, one chromosome and two plasmids were found in strain L5151. Among them, the chromosome has a length of 5,300,774 bp and its GC content was 57.4%. The resulting plasmid pL5151_NDM, which carried blaNDM-1 was 58,867 bp in length and had a GC content of 52.0%. The plasmid pL5151_MCR_CTX carrying mcr-1 and blaCTX-M-199 was 64,247 bp long and its GC content was 42.6%.
AST results showed that K. pneumoniae strain L5151 exhibited resistance to multiple antibiotics, including carbapenems, cephalosporins, fluoroquinolones, and β-lactam/β-lactamase inhibitor combinations, while remaining susceptible to aztreonam, aminoglycosides, fosfomycin, chloramphenicol, trimethoprim-sulfamethoxazole, and tigecycline. Detailed MIC values are provided in Table 1. A total of 13 ARGs were identified in strain L5151 based on ResFinder results (Table 2). These results corresponded to the resistance phenotype.
The presence of an ~ 64.2 kb IncI2 (Delta) plasmid (pL5151_MCR_CTX) and an ~ 58.9 kb IncN plasmid (pL5151_NDM) in isolate L5151 was verified by S1-PFGE and Southern blot analyses (Figs. 1 and 2). The detailed results of the S1 nuclease PFGE and subsequent Southern blot hybridization are presented in Supplementary Fig. 2(A) (for blaNDM-1) and Supplementary Fig. 2(B) (for mcr-1).The transconjugant L5151-EC, which carried the ~ 64.2 kb IncI2 pL5151_MCR_CTX plasmid, had an MIC of colistin of 4 mg/L. Additionally, the transconjugant L5151-EC, which acquired the ~ 54.4 kb IncN plasmid pL5151_NDM, exhibited an MIC of 8 mg/L for imipenem, indicating successful transfer of carbapenem resistance. Conjugation experiments also confirmed that the IncI2 plasmid pL5151_MCR_CTX was successfully transferred into the rifampicin-resistant recipient strain E. coli EC600, with a conjugation efficiency of 4.08 × 10–4. AST results of the transconjugants demonstrated that both pL5151_MCR_CTX and pL5151_NDM were transferable and conferred resistance to colistin and carbapenems, respectively, leading to elevated MIC values (Table 1).
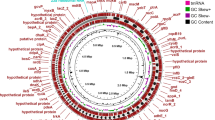
Comparison of plasmids carrying mcr-1/blaNDM-1 detected in this study. (A) Circumplex genome comparison of pL5151-NDM with four plasmids carrying blaNDM-1 (pNDM-1, pNDM-Cf7308, pNDM2963, pNDM-BTR). (B) Genomic mapping of mcr-1 and blaCTX-M-199 co-carrying IncI2 pL5151_MCR_CTX plasmid with four closely related plasmids (pEC16-50-MCR, pM-199-232, pM-199-C35, pSLDY13). Circular maps were generated by BLAST Ring Image Generator (BRIG) software.
NCBI BLAST analysis of the complete sequence of pL5151_MCR_CTX revealed that it has the greatest genetic similarity with the pEC16-50-MCR (MG515249.1), pM-199–232 (MT773664.1), pM-199-C35 (MW218147.1), and pSLDY13 (CP149969.1) plasmids. These similarities were marked by a query coverage exceeding 95% and an identity exceeding 99%. Similarly, the complete sequence of pL5151_NDM shows the highest genetic similarity to plasmids pNDM-1 (CP160626.1), pNDM-Cf7308 (CP092465.1), pNDM2963 (CP138536.1), and pNDM-BTR (KF534788.2), with both query coverage and identity exceeding 99% (Supplementary Table S3). Genetic background analysis revealed that multiple mobile elements (IS26, ISKpn19, and IS3000) flank NDM-1, which facilitates the clustering and integration with resistance genes, thereby contributing to the transfer of plasmids. These findings highlight the urgent necessity of developing and implementing effective prevention strategies to restrain the further spread of plasmids harboring mcr-1 and blaCTX-M-199.
VFDB analysis revealed that L5151 harbors multiple virulence-associated genes, including those related to adhesion (fimABCDEFGHIK, mrkABCDFHIJ), iron acquisition (enterobactin: entABCDEFS, iroEN; aerobactin: iutA), and other factors (Supplementary Table S1). These findings suggest that these genes may enhance the strain’s ability to colonize and persist in the host environment.
In this study, we characterized a multidrug-resistant K. pneumoniae strain (L5151) that carries the blaNDM-1, mcr-1, and blaCTX-M-199 genes, conferring resistance to carbapenems, colistin, and β-lactamase inhibitors (tazobactam and sulbactam), respectively. This strain is the first clinical isolate of K. pneumoniae simultaneously harboring these three resistance genes in China, highlighting the increasing challenge posed by such multi-drug resistant pathogens in clinical settings.
Virulence factor analysis revealed that strain L5151 carries a variety of genes associated with pathogenicity, including adhesion factors (fim and mrk gene clusters), iron uptake systems, and biofilm formation factors (Supplementary Table S1). These factors may enhance the strain’s ability to colonize and persist within the host, suggesting that the strain not only exhibits high resistance but also possesses pathogenic potential.
BLAST alignment of the IncI2 (Delta) plasmid in strain L5151 revealed significant sequence similarity with plasmids found in multiple E. coli isolates, as well as a K. pneumoniae strain and a Salmonella isolate, all showing greater than 95% coverage and identity. Most of these plasmids originated from China (Supplementary Table S2). IncI2-type plasmids are primarily found in E. coli isolates, suggesting that the horizontal gene transfer of the IncI2 plasmid in L5151 may have originated from E. coli.
Notably, these plasmids are frequently detected in various clinical strains in China, suggesting that they may have a certain level of prevalence in the region. Recent studies in China have reported a high detection rate of E. coli strains harboring the IncI2 plasmid in hospital settings, with some also found in community environments (as reported by Cai et al.14, Shen et al.27 and Chen et al.28). The widespread presence of these plasmids may exacerbate the transmission of antibiotic resistance genes across species, further complicating public health management. As the transfer of resistance genes across species increases, future studies should focus on the transmission dynamics and epidemiological characteristics of the IncI2 plasmid in China, in order to provide scientific evidence for controlling the spread of resistant pathogens.
Conjugation experiments confirmed the transferability of the IncI2 plasmid. The results showed that the plasmid could successfully transfer from L5151 to rifampicin-resistant E. coli EC600, with a transfer frequency of 4.08 × 10–4. This significantly increased the MIC values of various antibiotics, including colistin, in the transconjugants (Table 1), demonstrating the ability of the plasmid to spread resistance across bacterial species.
From a clinical perspective, the significance of strain L5151 lies in its high-level resistance to multiple classes of antibiotics and its potential pathogenicity. The spread of the mcr-1 gene, particularly in the context of carbapenem resistance, reduces the options available for effective treatment, and the presence of various virulence factors may lead to severe infections, treatment failure, and prolonged hospital stays. Moreover, the strain’s origin from a hospitalized patient raises concerns about the potential spread of high-risk clones in hospital environments. The transferable plasmids it carries further amplify the risk of cross-species transmission of resistance genes, which could lead to more widespread outbreaks.
In conclusion, this study provides molecular insights of horizontal plasmid transmission and highlights the integration of drug resistance and virulence in K. pneumoniae. It emphasizes the need for continuous monitoring of such high-risk pathogens in both clinical and community settings. Furthermore, the findings suggest that effective infection control measures must be implemented to prevent the spread of these multidrug-resistant pathogens and the plasmids that facilitate the transmission of resistance genes.
Although a phylogenetic analysis was not performed in this study, we fully acknowledge the importance of such an approach. In future studies, phylogenetic analysis could play a pivotal role in providing deeper insights into the evolutionary relationships among multidrug-resistant K.pneumoniae strains, their spread within local and national environments, and their genetic connections to other circulating clones. Such analysis may help elucidate the origins and dissemination of resistant strains and provide additional insights into the transmission dynamics of resistance plasmids.
Data availability
The complete genome sequence of Klebsiella pneumoniae L5151 has been deposited into GenBank, and its accession numbers are CP175678-CP175681.
References
Bonomo, R. A. et al. Carbapenemase-producing organisms: a global scourge. Clin. Infect. Dis. 66(8), 1290–1297. https://doi.org/10.1093/cid/cix893 (2018).
Wu, C., Zheng, L. & Yao, J. Analysis of risk factors and mortality of patients with carbapenem-resistant Klebsiella pneumoniae Infection. Infect. Drug Resist. 15, 2383–2391. https://doi.org/10.2147/idr.S362723 (2022).
Carlos, C. C. et al. Genome sequencing identifies previously unrecognized klebsiella pneumoniae outbreaks in neonatal intensive care units in the Philippines. Clin. Infect. Dis. 73(4), S316–S324. https://doi.org/10.1093/cid/ciab776 (2021).
Ji, J. et al. Co-colonization of different species harboring KPC or NDM carbapenemase in the same host gut: insight of resistance evolution by horizontal gene transfer. Front. Microbiol. 15, 1416454. https://doi.org/10.3389/fmicb.2024.1416454 (2024).
Wu, W. et al. Emergence of carbapenem-resistant Enterobacter hormaechei ST93 plasmids co-harbouring bla(NDM-1), bla(KPC-2), and mcr-9 in bloodstream infection. J. Glob. Antimicrob. Resist. 34, 67–73. https://doi.org/10.1016/j.jgar.2023.06.009 (2023).
Durante-Mangoni, E., Andini, R. & Zampino, R. Management of carbapenem-resistant Enterobacteriaceae infections. Clin. Microbiol. Infect. 25(8), 943–950. https://doi.org/10.1016/j.cmi.2019.04.013 (2019).
Yong, D. et al. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53(12), 5046–5054. https://doi.org/10.1128/aac.00774-09 (2009).
Wang, Y. et al. Prevalence, risk factors, outcomes, and molecular epidemiology of mcr-1-positive Enterobacteriaceae in patients and healthy adults from China: an epidemiological and clinical study. Lancet Infect. Dis. 17(4), 390–399. https://doi.org/10.1016/s1473-3099(16)30527-8 (2017).
Ling, Z. et al. Epidemiology of mobile colistin resistance genes mcr-1 to mcr-9. J. Antimicrob. Chemother. 75(11), 3087–3095. https://doi.org/10.1093/jac/dkaa205 (2020).
Hu, Y. & Coates, A. Mefloquine enhances the activity of colistin against antibiotic-resistant Enterobacterales in vitro and in an in vivo animal study. Int. J. Antimicrob Agents. 57(4), 106309. https://doi.org/10.1016/j.ijantimicag.2021.106309 (2021).
Liu, Y. Y. et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16(2), 161–168. https://doi.org/10.1016/s1473-3099(15)00424-7 (2016).
Jia, Y. et al. Melatonin protects against colistin-induced intestinal inflammation and microbiota dysbiosis. J. Pineal Res. 76(5), e12989. https://doi.org/10.1111/jpi.12989 (2024).
Riccobono, E. et al. Characterization of IncI1 sequence type 71 epidemic plasmid lineage responsible for the recent dissemination of CTX-M-65 extended-spectrum β-lactamase in the Bolivian Chaco region. Antimicrob. Agents Chemother. 59(9), 5340–5347. https://doi.org/10.1128/aac.00589-15 (2015).
Cai, J. et al. Genetic and functional characterization of bla(CTX-M-199), a novel tazobactam and sulbactam resistance-encoding gene located in a conjugative mcr-1-bearing IncI2 Plasmid. Antimicrob. Agents Chemother. 61, 7. https://doi.org/10.1128/aac.00562-17 (2017).
Zheng, B. et al. Low prevalence of MCR-1-producing Klebsiella pneumoniae in bloodstream infections in China. Clin. Microbiol. Infect. 24(2), 205–206. https://doi.org/10.1016/j.cmi.2017.08.004 (2018).
Zheng, B. et al. Complete genome sequencing and genomic characterization of two Escherichia coli strains co-producing MCR-1 and NDM-1 from bloodstream infection. Sci. Rep. 7(1), 17885. https://doi.org/10.1038/s41598-017-18273-2 (2017).
Liu, R. et al. Genomic characterization of two Escherichia fergusonii isolates harboring mcr-1 gene from farm environment. Front. Cell Infect Microbiol. 12, 774494. https://doi.org/10.3389/fcimb.2022.774494 (2022).
Chen, C. et al. Co-existence of KPC-2, LAP-2, and CTX-M-65 in an ST1469 multidrug-resistant Klebsiella pneumoniae strain in China. Infect. Drug Resist. 15, 6731–6737. https://doi.org/10.2147/idr.S392063 (2022).
Liu, X. et al. Emergence of plasmid-borne tet(X4) resistance gene in clinical isolate of eravacycline- and omadacycline-resistant Klebsiella pneumoniae ST485. Microbiol. Spectr. 12(9), e0049624. https://doi.org/10.1128/spectrum.00496-24 (2024).
Liu, B., Zheng, D., Zhou, S., Chen, L. & Yang, J. VFDB 2022: a general classification scheme for bacterial virulence factors. Nucleic Acids Res. 50(D1), D912–D917. https://doi.org/10.1093/nar/gkab1107 (2022).
Ge, H. et al. Emergence and clonal dissemination of KPC-3-producing Pseudomonas aeruginosa in China with an IncP-2 megaplasmid. Ann. Clin. Microbiol. Antimicrob. 22(1), 31. https://doi.org/10.1186/s12941-023-00577-z (2023).
Zhang, X. et al. Emergence of a clinical isolate of E. coil ST297 co-carrying bla(NDM-13) and mcr-11 in China. J. Infect. Public Health 16(11), 1813–1820. https://doi.org/10.1016/j.jiph.2023.09.007 (2023).
Wick, R. R., Judd, L. M., Gorrie, C. L. & Holt, K. E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13(6), e1005595. https://doi.org/10.1371/journal.pcbi.1005595 (2017).
Zankari, E. et al. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67(11), 2640–2644. https://doi.org/10.1093/jac/dks261 (2012).
Carattoli, A. et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58(7), 3895–3903. https://doi.org/10.1128/aac.02412-14 (2014).
Alikhan, N. F., Petty, N. K., Ben Zakour, N. L. & Beatson, S. A. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12, 402. https://doi.org/10.1186/1471-2164-12-402 (2011).
Shen, Y. et al. Heterogeneous and flexible transmission of mcr-1 in hospital-associated Escherichia coli. MBio 9, 4. https://doi.org/10.1128/mBio.00943-18 (2018).
Chen, J., Chen, S., Jiang, Y., Zhang, R. & Cai, J. Fecal carriage and genetic characterization of CTX-M-1/9/1-producing Escherichia coli from healthy humans in Hangzhou, China. Front. Microbiol. 12, 616687. https://doi.org/10.3389/fmicb.2021.616687 (2021).
Funding
This study was sponsored by Science and Technology Research Program of Henan Province (No. 212102310191).
Author information
Authors and Affiliations
Contributions
Xinjun Hu and Bingyou Yin wrote the main manuscript text, and Ruishan Liu prepared Figs. 1–2. Lu Gong, Xiaolu Yang, Zhenghao Lou, and Haowei Ye: methodology, data acquisition. Bingyang Shang, Yibing Shang: data acquisition. Yingjian Zhang: Funding acquisition, conceptualization. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics statement
The research protocol was reviewed and approved by the Research Ethics Review Committee of the First Affiliated Hospital of Zhejiang University Medical School. All methods are carried out in accordance with relevant guidelines and regulations. Due to the retrospective nature of the study, the institutional review board waived the need for informed consent. All patient data were anonymized prior to analysis (2018–752).
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Hu, X., Yin, B., Liu, R. et al. Coexistence of blaNDM-1, mcr-1 and blaCTX-M-199 in an ST499 multidrug resistant Klebsiella pneumoniae Isolate. Sci Rep 15, 19132 (2025). https://doi.org/10.1038/s41598-025-03759-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-03759-1