Abstract
Idiopathic pulmonary fibrosis (IPF) is an age-related disease with an unclear pathogenesis. The senescence and insufficient regeneration of alveolar epithelial cells are significant factors in the development and progression of IPF. Currently, effective treatment methods are lacking. The aim of this study is to explore the mechanism of action of uridine in delaying the aging of AECs and intervening in IPF. In vitro, Western blot and qRT-PCR analyzed uridine’s effects on bleomycin-induced senescence, EMT, cell viability, and cell cycle. In vivo, uridine’s impact on lung aging and fibrosis in BLM-induced mice was assessed by weight, staining, Ashcroft scoring, and Western blot. Uridine reduced senescence markers in A549 cells, suppressed epithelial-mesenchymal transition, improved antioxidant capacity, and delayed pulmonary fibrosis and lung aging in mice. The effects of uridine were mediated through the NRF2 signaling pathway, which regulates antioxidant defense and autophagy. Uridine enhanced autophagic degradation of Keap1, possibly through p62/SQSTM1-mediated autophagy. These findings suggest that uridine inhibits AEC senescence via the NRF2 pathway, mitigating IPF progression and offering a potential strategy for treating age-related pulmonary fibrosis by targeting oxidative stress.
Similar content being viewed by others
Introduction
Idiopathic Pulmonary Fibrosis (IPF) is a chronic, progressive lung disease, with an increasing incidence, particularly among individuals over 65 years old. The global prevalence ranges from 3 to 45 cases per 100,000 people1, with a median survival time of approximately 3–5 years. The exact cause of IPF remains unknown, but alveolar epithelial cells (AECs) senescence is considered a crucial factor in its progression2,3.
In IPF, senescent cells accumulate in aging tissues and contribute to the development of age-related diseases4. These cells not only lose their normal function but also secrete a variety of pro-inflammatory cytokines, chemokines, and growth factors, collectively known as the senescence-associated secretory phenotype (SASP)4,5. SASP factors such as interleukin-6 (IL-6), interleukin-8 (IL-8), matrix metalloproteinases (MMPs), and others can activate and recruit lung fibroblasts, promoting collagen deposition and exacerbating inflammation6. Furthermore, SASP factors disrupt normal tissue repair mechanisms, leading to chronic inflammation and accelerating the fibrotic process7.
The existing treatments for IPF include rehabilitation treatment, oxygen therapy, lung transplantation, and immunomodulatory therapy8. Currently, the main treatment is nintedanib, which has been shown to have gastrointestinal toxicity, hepatotoxicity, nephrotoxicity, and increased bleeding risk in some patients9. Moreover, nintedanib typically only slows disease progression and does not cure IPF10. Notably, recent studies suggest that targeting and clearing senescent epithelial cells in lung tissue can effectively alleviate the progression of pulmonary fibrosis11. Therefore, investigating a drug that can effectively improve AECs senescence is a potential target for treating pulmonary fibrosis.
For senescent epithelial cell clearance, (nuclear factor E2-related factor 2) Nrf2 signaling pathway is a key regulator of intracellular antioxidant responses, playing a critical role in the development of IPF12. Nrf2 combats oxidative stress by inducing the transcription of genes encoding antioxidant enzymes. Its regulation is tightly controlled by Kelch ECH-associated protein 1 (Keap1), which normally promotes Nrf2 ubiquitination and degradation13. Interestingly, the selective autophagy receptor p62/SQSTM1 enhances Nrf2 activation by promoting Keap1 degradation through autophagy14. Phosphorylated p62 binds to Keap1 with higher affinity, facilitating its sequestration into autophagosomes and subsequent degradation, thereby lifting the inhibition of Nrf2 and maintaining the continuous activation of the Nrf2 signaling pathway, which strengthens the cell’s anti-oxidative and anti-aging capabilities15,16.
In view of this, this study focused on the small molecule compound Uridine, which, with its unique anti-inflammatory, anti-aging and tissue regeneration properties, is a potential effective candidate for intervention in IPF17,18,19. Previous studies have shown that uridine can effectively improve the aging state of mesenchymal stem cells and intestinal epithelial cells, and has shown potential to reduce lung inflammatory response in mouse models20,21,22. However, it remains to be determined whether and how uridine inhibits the IPF process by regulating Nrf2 signaling to reduce oxidative stress-induced AECs senescence. Therefore, this study aims to reveal the effect of uridine on AECs aging and epithelial-mesenchymal transition (EMT) through in vivo and in vitro experiments, and explore the molecular mechanism of uridine protecting alveolar structure through activation of Nrf2-p62 axis-mediated autophagy pathway, thus inhibiting the pulmonary fibrosis process. It provides new insights into the pathophysiology of IPF and opens new avenues for the clinical development of targeted anti-oxidative stress therapies, especially for the treatment of age-related pulmonary fibrosis.
Results
Uridine inhibits bleomycin (BLM)- induced senescence and Epithelial-Mesenchymal transition (EMT) in A549 cells
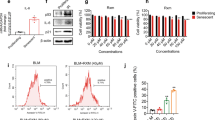
We established the BLM model with reference to the Jeengar, M.K., et al. and Della Latta, V., et al. study23,24 and selected the concentration according to the experimental results of the effects of BLM and uridine on the activity of A549 cells (Supplementary Fig. 1), so as to ensure that the activity of cells was in a state suitable for the experiment.
Cell cycle-assays and cell proliferation assays were used to assess cell senescence25. Uridine treatment alleviated BLM-induced S phase cell cycle arrest (Fig. 1a). We then measured SA-β-gal activity in A549 cells, a widely used senescence marker. BLM treatment significantly increased the number of positive cells, while uridine treatment significantly reduced the number of SA-β-gal positive cells in a concentration-dependent manner (Fig. 1b). Similarly, uridine treatment effectively reduced the expression levels of senescence markers p16, p21, and p53 compared to the BLM group (Fig. 1c). Moreover, RT-qPCR showed that uridine treatment significantly reduced the expression of SASP-related genes IL-6, IL-1β, TNF-α, Mmp12, TGF-β1, and Wisp1 in BLM-treated A549 cells (Fig. 1d).
Uridine Inhibits Bleomycin (BLM)- Induced Senescence and Epithelial-Mesenchymal Transition (EMT) in A549 Cells. (a) Cell cycle analysis of A549 cells treated with uridine and BLM. (b) Representative images and quantification of SA-β-gal staining in A549 cells treated with uridine and BLM. (c) Western blot analysis of senescence markers p16, p21, and p53 expression levels in A549 cells treated with uridine and BLM. (d) qRT-PCR analysis of SASP (senescence-associated secretory phenotype) markers IL-6, IL-1β, TNF-α, Mmp12, TGF-β1, and Wisp1 mRNA expression in A549 cells treated with uridine and BLM. (e) qRT-PCR analysis of fibrotic markers Collagen1a1, Collagen5a3, and Fibronectin mRNA expression in A549 cells treated with uridine and BLM. (f) Western blot analysis of fibrotic markers Collagen1a1, Collagen5a3, and Fibronectin expression levels in A549 cells treated with uridine and BLM. (g) qRT-PCR analysis of lung epithelial markers Sftpc, Sftpa1, and T1α mRNA expression in A549 cells treated with uridine and BLM. (h) Western blot analysis of lung epithelial markers Sftpc, Sftpa1, and T1α expression levels in A549 cells treated with uridine and BLM. Bars in (b) represent 100 μm. Data in (a, b, d, e, g) are presented as mean ± SD (n = 3, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; ns indicates no significance; student’s t-test).
Due to the important role of EMT in IPF, we detected EMT related characterization. We found that uridine treatment significantly reduced the gene and protein expression of key EMT markers Collagen1a1, Collagen5a3, and Fibronectin (Fig. 1e-f). Additionally, the gene and protein expression of alveolar epithelial cell markers Sftpc, Sftpa1, and T1α significantly increased compared to the untreated group (Fig. 1g-h).
Uridine improves BLM-Induced pulmonary fibrosis in mice
Given the significant effects of uridine on improving A549 cell senescence in vitro, our next step was to test whether uridine could improve BLM-induced pulmonary fibrosis in vivo. We used intratracheal instillation of BLM to create a mouse model of pulmonary fibrosis. Considering that the inflammation phase occurs 7 days after BLM instillation (2 mg/kg) and the fibrosis phase occurs from days 8–1026, we started oral administration of nintedanib (100 mg/kg) and uridine (20, 40 mg/kg) on the 10 th day post-operation, maintaining the treatment for 10 days. Lung tissues were collected for further analysis on the 20 th day post-operation. Initial observations of the collected lung tissues revealed hemorrhagic necrosis and focal scarring in BLM-treated mice, which were improved to varying degrees in the nintedanib and uridine treatment groups (Fig. 2a).
Uridine Improves BLM-Induced Pulmonary Fibrosis in Mice. (a) Macroscopic view of lung tissues from mice treated with uridine and nintedanib following intratracheal instillation of 2 mg/kg BLM for 10 days, followed by 10 days of oral gavage treatment with uridine (20, 40 mg/kg) and nintedanib (100 mg/kg). (b) Lung weight/body weight ratio in mice treated with uridine and nintedanib. (c) Representative images of lung tissues stained with H&E from mice treated with uridine and nintedanib. (d) Ashcroft score of lung fibrosis in histopathological sections from mice. (e) Representative images of lung tissues stained with Masson’s trichrome and Sirius Red from mice treated with uridine and nintedanib. (f) Hydroxyproline levels in lung tissues from mice treated with uridine and nintedanib. (g) Representative images of immunofluorescence staining for epithelial markers Ki67 and E-cadherin in lung tissues from mice treated with uridine and nintedanib. (h) Western blot analysis of epithelial markers Ki67 and E-cadherin expression levels in lung tissues from mice treated with uridine and nintedanib. (i) Representative images of immunofluorescence staining for fibrotic markers α-SMA and Collagen1a1 in lung tissues from mice treated with uridine and nintedanib. (j) Western blot analysis of fibrotic markers α-SMA and Collagen1a1 expression levels in lung tissues from mice treated with uridine and nintedanib. Bars in (c, e, g, i) represent 100 μm. Data in (b, d, f) are presented as mean ± SD (n = 3, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; ns indicates no significance; student’s t-test).
By measuring lung weight and body weight and calculating the lung-to-body weight ratio, we found that uridine reduced the lung-to-body weight ratio in a concentration-dependent manner in mice with pulmonary fibrosis (Fig. 2b), indicating that uridine effectively ameliorated lung consolidation. Additionally, HE staining and Ashcroft scoring showed that uridine treatment effectively improved BLM-induced alveolar structural damage and inflammatory infiltration in mice (Fig. 2c-d).
Next, we examined the effect of uridine on improving pulmonary fibrosis in mice. Masson’s trichrome staining and Sirius red staining revealed that uridine significantly reversed BLM-induced pulmonary fibrotic connective tissue formation and reduced the levels of type I and type III collagen in lung tissue compared to the PBS group (Fig. 2e). Similarly, uridine treatment reduced hydroxyproline levels in lung tissue, an important indicator of early pulmonary fibrosis (Fig. 2f). By observing alveolar epithelial cell proliferation markers E-cadherin and Ki67, we found that uridine treatment effectively improved the reduction in alveolar epithelial cell regeneration induced by BLM (Fig. 2g-h).
Furthermore, uridine treatment significantly decreased the deposition of fibrosis markers α-SMA and Collagen1a1 in the lungs (Fig. 2i-j). Given that the occurrence of IPF is closely related to insufficient regeneration of alveolar epithelial cells and the progression of EMT, our study found that uridine treatment effectively reduced BLM-induced loss of alveolar epithelial phenotype and mesenchymal transition, preventing the progression of EMT and significantly improving lung tissue structural remodeling and fibrosis development.
Uridine alleviates pulmonary senescence in mice
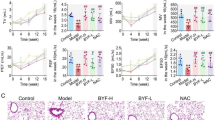
p21 and p53 are significant marker of cellular senescence27. Uridine treatment reduced the levels of the cellular aging markers p21 and p53 in the lung tissue of mice with pulmonary fibrosis (Fig. 3a-b). Similarly, uridine treatment effectively decreased the expression of genes related to SASP in mouse lung tissue (Fig. 3c).
Uridine Alleviates Pulmonary Senescence in Mice. (a) Representative immunofluorescence images of p21 and p53 in lung tissues from uridine-treated mice with lung fibrosis. (b) Western blot analysis of p21 and p53 expression in lung tissues from uridine-treated mice with lung fibrosis. (d) qRT-PCR analysis of mRNA expression levels of SASPs (IL-6, IL-1β, TNF-α, Mmp12, TGF-β1, and Wisp1) in lung tissues from uridine-treated mice. Bars in (a) represent 100 μm. Data in (c) are presented as mean ± SD (n = 3, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; ns indicates no significance; student’s t-test).
These results suggest that uridine treatment mitigates BLM-induced damage and senescence of alveolar epithelial cells, reduces SASP levels, and influences the EMT process induced by alveolar epithelial cell senescence, thereby treating pulmonary fibrosis. Overall, uridine treatment can effectively reverse BLM-induced alveolar epithelial cell senescence. This outcome is consistent with the antifibrotic effects observed in previous experiments. Therefore, we believe that the therapeutic effect of uridine on IPF is achieved by ameliorating alveolar epithelial cell senescence.
Uridine activates the Nrf2 signaling pathway
In order to verify the specific mechanisms involved in aging in vivo and in vitro, we used ROS level detection methods for verification, and the results showed that uridine treatment can reduce the ROS level of A549 cells (Fig. 4a).
Uridine Activates the Nrf2 Signaling Pathway. (a) Representative images of ROS fluorescence in A549 cells induced by BLM with uridine treatment. (b) Representative immunofluorescence images of Nrf2 in uridine-treated A549 cells. (c) qRT-PCR analysis of Nrf2 gene expression levels in uridine-treated A549 cells. (d) Western blot analysis of Nrf2 protein expression levels in uridine-treated A549 cells. (e) Western blot analysis of downstream genes of Nrf2 (NQO1, HO-1, GCLM, GCLC) expression levels in uridine-treated A549 cells. (f) qRT-PCR analysis of downstream gene expression levels of Nrf2 in uridine-treated A549 cells. (g) Treatment of A549 cells with CHX (300 µM) followed by Western blot analysis of Nrf2 protein levels at 0, 8, 16, and 24 h. (h) Treatment of A549 cells with CHX (300 µM) and MG132 (10 µM) followed by Western blot analysis of Nrf2 protein levels at 0, 8, 16, and 24 h. (i) Treatment of A549 cells with CHX (300 µM) and uridine (300 µM) followed by Western blot analysis of Nrf2 protein levels at 0, 8, 16, and 24 h. Bars in (a) represent 200 μm, and in (b) represent 50 μm. Data in (c) and (f) are presented as mean ± SD (n = 3, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; ns indicates no significance; student’s t-test).
Our next experiment explored whether the protective effect of uridine against cellular senescence depends on the Nrf2 signaling pathway. Immunofluorescence and Western blot results showed that uridine treatment increases Nrf2 accumulation in A549 cells (Fig. 4b, Supplementary Fig. 2). Additionally, Western blot results indicated that uridine treatment significantly increases Nrf2 protein expression in A549 cells (Fig. 4d), although no increase in Nrf2 mRNA transcription was observed (Fig. 4c).
Next, we evaluated the transcription levels of major downstream target genes of activated Nrf2, such as NQO1, HO-1, GCLM, and GCLC, using qRT-PCR. We found that the expression levels of Nrf2 downstream regulatory genes were significantly elevated in uridine-treated A549 cells compared to the control group. Similarly, the protein levels of NQO1, HO-1, GCLM, and GCLC also increased markedly after uridine treatment (Fig. 4e-f).
Then we treated A549 cells with the protein synthesis inhibitor cycloheximide (CHX, 300 µM), the proteasome inhibitor MG132 (10 µM), and uridine, and measured Nrf2 protein levels at different time points. The results showed that the degradation level of Nrf2 can be significantly decreased by MG132 (Fig. 4g-h), and uridine treatment can markedly extend the half-life of intracellular Nrf2 (Fig. 4i).
Based on these results, we hypothesize that the increased intracellular levels of Nrf2 protein may not be due to changes in DNA transcription but rather through enhanced stability of the Nrf2 protein. Therefore, we conclude that uridine treatment reduces the ubiquitination of Nrf2, thereby enhancing its stability.
Uridine exerts Anti-aging and EMT-suppressing effects depend on the Nrf2 signaling pathway
To further demonstrate that uridine protects against alveolar epithelial cell senescence and improves pulmonary fibrosis via the Nrf2 signaling pathway, we conducted additional in vitro and in vivo experiments using the Nrf2 signaling pathway inhibitor ML38528. The experimental results confirmed that pretreatment with the Nrf2 inhibitor ML385 (5 µM) resulted in a significant limitation of uridine-induced reduction in ROS levels in A549 cells (Fig. 5a).
Uridine Exerts Anti-aging and EMT-suppressing Effects Depend on the Nrf2 Signaling Pathway. (a) Representative images of ROS fluorescence in A549 cells. (b) Western blot analysis of the expression levels of aging marker proteins p16, p21, and p53 in A549 cells. (c) Representative images of SA-β gal staining in A549 cells. (d) qRT-PCR analysis of SASP (IL-6, IL-1β, TNF-α, Mmp12, TGF-β1, and Wisp1) gene expression levels in lung tissues of mice treated with ML385 and uridine. (e) qRT-PCR analysis of fibrosis markers (Collagen1a1, Collagen5a3, and Fibronectin) DNA expression levels in lung tissues of mice. (f) qRT-PCR analysis of epithelial markers (Sftpc, Sftpa1, and T1α) gene expression levels in lung tissues of mice. (g) Western blot analysis of fibrosis markers (Collagen1a1, Collagen5a3, Fibronectin) expression levels in lung tissues of mice. (h) Western blot analysis of epithelial markers (Sftpc, Sftpa1, and T1α) expression levels in lung tissues of mice. (i) Representative images of HE staining in lung tissues of mice. (j) Representative immunofluorescence images of γ-H2 AX, α-SMA and Collagen1a1 in lung tissues of mice. (k) Western blot analysis of γ-H2 AX, α-SMA and Collagen1a1 expression levels in lung tissues of mice. Bars in (a) represent 200 μm, and in (c), (i) and (j) represent 100 μm. Data in (c), (d), (e) and (f) are presented as mean ± SD (n = 3, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; ns indicates no significance; student’s t-test).
Correspondingly, A549 cells treated with the Nrf2 inhibitor showed a pronounced increase in levels of senescence marker proteins p16, p21, p53, and SA-β gal-positive cells (Fig. 5b-c). Given that uridine treatment improves the transcription levels of SASP-related genes in A549 cells, this effect was significantly reversed upon addition of the Nrf2 inhibitor (Fig. 5d). Furthermore, we assessed the impact of ML385 on EMT levels in A549 cells. The results indicated a significant increase in mRNA and protein levels of fibrotic markers Collagen1a1, Collagen5a3, and Fibronectin in the ML385-treated cell group (Fig. 5e, g). Similarly, the upward trend in gene transcription levels of alveolar epithelial cell markers Sftpc, Sftpa1, and T1α was reversed (Fig. 5f, h).
In vivo experiments also confirmed that ML385 treatment (5 mg/kg) significantly reversed the therapeutic effects of uridine treatment on pulmonary fibrosis in mice. HE staining showed that lung structural improvements observed in the uridine treatment group were significantly reversed with ML385 treatment (Fig. 5i), accompanied by a significant increase in Ashcroft scores (Supplementary Fig. 3a). Mice treated with ML385 also exhibited more severe fibrotic characteristics, as pathological staining indicated marked proliferation of fibrotic tissue and increased collagen positivity in lung tissues (Supplementary Fig. 3b).
Moreover, we found that ML385 effectively countered uridine’s anti-EMT and anti-DNA damage function, with the ML385 group showing higher levels of fibrotic markers Collagen1a1, α-SMA and DNA damage marker γ-H2 AX (Fig. 5j-k).
Previous results indicated that uridine could elevate levels of lung epithelial cell markers in the pulmonary fibrosis mouse model, a trend that was attenuated by the Nrf2 inhibitor ML385 (Supplementary Fig. 3c). Collectively, these findings suggest that uridine’s protective effect against alveolar epithelial cell senescence is mediated through the Nrf2 signaling pathway.
Uridine promotes p62-Mediated Keap1 autophagy
We determined the regulatory effect of uridine on Nrf2 by detecting Keap1 levels, and the results showed that uridine treatment effectively reduces Keap1 protein levels (Fig. 6a). Similar conclusions were drawn from in vivo experiments (Supplementary Fig. 4).
Uridine Promotes p62-Mediated Keap1 Autophagy. (a) Western blot analysis of Keap1 levels in uridine-treated A549 cells. (b) Western blot analysis of Nrf2 and Keap1 levels in A549 cells treated with uridine and 3-MA. (c) Transmission electron microscopy observation of autophagy in uridine-treated A549 cells. (d) Representative immunofluorescence images of LC3 and Keap1 in A549 cells treated with uridine and BLM. (e) Western blot analysis of LC3, Beclin1, and p62 levels in A549 cells treated with uridine and 3-MA. (f) Western blot analysis of p62 protein expression in A549 cells transfected with control and p62 siRNA after 72 h. (g) Western blot analysis of p16, p21, and p53 expression levels in uridine-treated A549 cells transfected with control and p62 siRNA. (h) Western blot analysis of Collagen1a1, Collagen5a3, and Fibronectin expression levels in uridine-treated A549 cells transfected with control and p62 siRNA. (i) Co-immunoprecipitation (co-IP) analysis of the interaction levels between Keap1 and p62 in A549 cells treated with uridine and the p62 inhibitor XRK3 F2 (5 µM). Bars represent 20 μm in (d).
To further confirm the induction of autophagy by uridine, we performed transmission electron microscopy (TEM) on A549 cells treated with BLM alone, uridine alone, and a combination of both BLM and uridine. TEM images revealed a significant increase in the number of autophagic vesicles in uridine-treated cells compared to the control group (Fig. 6c). Next, we observed dual fluorescence autophagy in A549 cells infected with lentivirus. Based on the co-localization of red/green fluorescence, we found that the addition of uridine significantly increased the yellow fluorescence dots, indicating that uridine can induce an increase in intracellular autophagy levels (Fig. 6d).
In order to further verify the influence of autophagy on the Keap1-Nrf2 mechanism, we conducted a western-blot experiment, and the results showed that uridine treatment could significantly increase the level of Nrf2 and decrease the level of Keap1, but this phenomenon disappeared when the autophagy process was inhibited (Fig. 6b). We further found that uridine’s reduction of Keap1 and increase in Nrf2 levels correlated with the degradation of autophagic substrate p62 and the upregulation of autophagy markers LC3 and Beclin1. Similarly, this process was inhibited by 3-MA (Fig. 6e).
To further validate the role of p62 in uridine-mediated improvement of senescence and EMT in A549 cells, we transfected A549 cells with p62-siRNA to silence p62 expression (Fig. 6f). Following uridine treatment, cells transfected with p62-siRNA exhibited higher expression levels of senescence markers p16, p21, p53, and EMT markers Collagen1a1, Collagen5a3, and Fibronectin compared to cells transfected with negative control siRNA (Fig. 6g-h).
To confirm the connection between uridine-induced Keap1 reduction and p62, we added a p62 inhibitor, XRK3 F2. Co-immunoprecipitation experiments showed that uridine enhanced the interaction between p62 and Keap1, promoting Keap1 degradation. Adding XRK3 F2 reversed this trend, resulting in low p62 levels and high Keap1 levels (Fig. 6i).
These results show that uridine-induced autophagy leads to selective degradation of Keap1 via p62, which correlates with promoted Nrf2 activation and improvements in cellular senescence and EMT in A549 cells.
Discussion
Despite the ongoing deepening of our understanding of the pathogenesis of IPF, traditional drugs primarily function to slow down disease progression rather than achieve complete cure29.This limitation arises largely because current therapies for IPF focus on improving lung inflammation and inhibiting fibrotic pathways30. However, recent research suggests that a subset of lung epithelial stem cells, primarily composed of type II alveolar epithelial cells, plays a significant role in the progression of IPF through cellular senescence. Therefore, traditional drug treatments for IPF are inherently limited.
In this study, we demonstrated that uridine mitigates oxidative stress-induced damage to alveolar epithelial cells through the Nrf2 signaling pathway, effectively reducing cellular senescence and EMT processes, thus improving IPF. Specifically, the observed reduction in senescence markers and EMT markers in A549 cells treated with uridine suggests that uridine may play a role in restoring the regenerative capacity of damaged AECs. This finding aligns with previous studies demonstrating uridine’s ability to rejuvenate aged human stem cells and promote tissue regeneration20. Our study demonstrated that uridine can reduce the levels of aging and EMT markers in A549 cells. Uridine has been shown in previous studies to rejuvenate aged human stem cells and promote regeneration of various tissues in vivo20. However, the effect of uridine has not been reported in BLM-Induced Pulmonary Fibrosis Animal Model and IPF. We have demonstrated for the first time that uridine inhibits BLM-induced senescence and EMT in A549 cells, and this effect is not limited to in vitro cell models but is also observed in an in vivo mouse model.
The mechanism underlying the anti-aging and anti-fibrotic effects of uridine involves the activation of the Nrf2 signaling pathway, a key player in cellular defense against oxidative stress and inflammation31,32. By promoting the degradation of Keap1 through autophagy, uridine enhances the stabilization and nuclear translocation of Nrf2, leading to the induction of antioxidant and cytoprotective genes33. We demonstrate that uridine could improve BLM-induced pulmonary senescence and fibrosis in vivo. The therapeutic effect of uridine on IPF is achieved by ameliorating alveolar epithelial cell senescence. Uridine could alleviate chondrocyte and MSCs senescence in vitro by evaluating a series of aging markers and furthermore, uridine could also relieve OA in vivo21. We demonstrate for the first time that uridine inhibits IPF in vivo and that this effect is caused by delaying senescence of lung epithelial cells.
For the first time, we propose a novel mechanism by which uridine treats IPF: uridine-induced autophagy selectively degrades Keap1 via p62, thereby promoting Nrf2 activation and improving cellular senescence and EMT in A549 cells. (Nrf2 activation is a core event in attenuating oxidative stress-associated aging34. Under normal conditions, Nrf2 is sequestered in the cytoplasm by its repressor protein, Keap1, which promotes Nrf2 ubiquitination and degradation35. However, when cells are exposed to oxidative stress or treated with uridine, Keap1 is selectively degraded via autophagy, a process that is mediated by p6236. Phosphorylated p62 increases its binding affinity for Keap1 and competes with Nrf2 to bind Keap1, thereby preventing Keap1-mediated Nrf2 ubiquitination and promoting the sustained activation of Nrf237. The degradation of Keap1 by p62-mediated autophagy leads to the stabilization and nuclear translocation of Nrf2, which then binds to antioxidant response elements (AREs) in the promoter regions of genes encoding antioxidant enzymes and other cytoprotective proteins38. Importantly, the selective degradation of Keap1 by p62-mediated autophagy is a novel mechanism that has not been previously reported in the context of IPF36. This mechanism represents a significant advancement in understanding how uridine can mitigate the oxidative stress and senescence associated with IPF39. Moreover, the role of p62 in this process is indispensable. p62 acts as an adaptor protein that binds both Keap1 and LC3, facilitating the sequestration of Keap1 into autophagosomes and its subsequent degradation. The downregulation of p62 expression by siRNA negates the beneficial effects of uridine, supporting the hypothesis that p62 is essential for the protective effects of uridine in A549 cells.
Our findings not only support the known mechanism of Nrf2 activation but also introduce new therapeutic targets and avenues for the treatment of IPF. The strong correlation between uridine treatment and improved cellular senescence and EMT markers highlights the potential of uridine as a therapeutic agent for IPF. However, additional research is required to explore uridine’s protective effects on alveolar epithelial cells in lung fibrosis: Experimental evidence indicates that uridine supplementation enhances p62-mediated Keap1 autophagy. Future experiments will focus on the interaction between uridine and p62, as well as the direct impact on the p62-Keap1 interaction; Our research demonstrates that uridine upregulates the autophagic receptor protein p62 to exert anti-senescence effects. Further investigation is warranted to understand the upstream regulatory mechanisms of autophagy, including HIF-1α, Rubicon, mTOR, AMPK, TFEB, and ATG740[,41; while we have evaluated cell viability in vitro, the potential in vivo toxicities of uridine remain uncertain. Future experiments will include comprehensive assessments of uridine’s safety, including peripheral blood counts, organ function tests, and histopathological analyses.
Conclusion
In conclusion, our study demonstrates that uridine effectively alleviates senescence and EMT in pulmonary epithelial cells, both in vitro and in vivo, through the modulation of the Nrf2 signaling pathway. Uridine enhances p62-mediated Keap1 autophagy, leading to sustained Nrf2 activation and upregulation of downstream antioxidant genes. These findings suggest that uridine may serve as a promising therapeutic agent for IPF by inhibiting alveolar epithelial cell senescence and deficiency. Further research is warranted to explore the full therapeutic potential of uridine and to evaluate its safety profile in vivo.
Materials and methods
Cell culture
Alveolar epithelial cells (A549) were obtained from ATCC and maintained in DMEM high glucose medium (C7076-500 ml, Bioss) supplemented with 10% fetal bovine serum and 1% streptomycin/penicillin. The cells were cultured at 37 °C in a 5% CO2 atmosphere. Cells were seeded at a density of 2.5 × 10^4 cells and regularly passaged. Bleomycin (11-B608166, Boer) was dissolved in PBS, and uridine (11-B612407, Boer) was dissolved in PBS for cell treatment and stored at −20 °C before use.
Cell viability assay
The Cell Counting Kit-8 (CCK-8) assay (AKCE001-1, BOXBIO) was used to evaluate the effect of uridine on A549 cell proliferation. Briefly, A549 cells were seeded at an initial density of 5 × 10^3 cells/well in 96-well plates and cultured with different concentrations of uridine (100, 200, 300, and 400 µM) for 24, 48, and 72 h. At each time point, 10 µl of CCK-8 solution and 90 µl of culture medium were added to each well and incubated at 37 °C for 2 h. Absorbance was measured at 450 nm.
SA-β-Galactosidase staining
Cells were seeded in six-well plates and treated with 100, 200, 300, and 400 µM uridine for 24 h. Then, the cells were further treated with BLM for 72 h. SA-β-gal staining was performed using a staining kit (G1580-100 T, Solarbio) according to the manufacturer’s instructions. The staining working solution was prepared accordingly. The six-well plates were sealed with plastic wrap to prevent evaporation and incubated overnight at 37 °C. Senescence-positive cells were identified as blue-stained cells under a light microscope.
ROS analysis
Cells were seeded in 24-well plates and treated with uridine and BLM. DCFH-DA (50101ES01, Yeasen) was diluted 1:1000 in serum-free medium and added to the cells. The cells were incubated in the dark at 37 °C for 30 min. After washing three times with serum-free medium, the cells were observed under a fluorescence microscope.
Cell cycle detection
Cells were treated with uridine and BLM, trypsinized with EDTA at 37 °C, and the reaction was stopped with FBS-containing DMEM. Following centrifugation and PBS washes, cells were fixed in 70% cold ethanol, then stained with propidium iodide and assessed using a flow cytometer (Beckman CytoFlex).
Quantitative Real-Time PCR analysis
Total RNA was extracted from A549 cells and lung tissues using TRIzol reagent (YZ-15596018, Acmec). RNA concentration was measured with a NanoDrop One spectrophotometer (Thermo). cDNA was synthesized using Hifair® II Reverse Transcriptase (11110ES92*, Yeasen) under the following conditions: 25 °C for 5 min, 42 °C for 40 min, and 85 °C for 5 min. Amplification was performed using Hieff® qPCR SYBR Green Master Mix (11203ES08, Yeasen) with the following cycling conditions: 95 °C for 5 min, followed by 40 cycles of 95 °C for 10 s, 60 °C for 20 s, and 72 °C for 20 s. β-Actin was used as the internal control, and all data were normalized to the control. Primer sequences are listed in Supplementary Table 1.
Western blot analysis
Total protein was extracted using cold RIPA buffer with protease inhibitors. Protein concentration was measured using a BCA kit (BX-2142728, Pierce). Proteins were separated on 8–15% Tris-glycine SDS-PAGE gels at 80 V, then 120 V, and transferred to PVDF membranes (bsp0161, PALL). Membranes were blocked with 5% non-fat milk and probed with primary antibodies overnight at 4 °C. After washing, membranes were incubated with secondary antibodies for 90 min at room temperature. Bands were visualized using a chemiluminescence imaging system (Biorad). Primary antibodies used included P16, P21, P53, γ-H2 AX, Ki67, E-cadherin, α-SMA, Collagen1a1, Collagen5a3, Fibronectin, Sftpc, Sftpa 1, T1α, Nrf2, NQO1, HO-1, GCLM, GCLC, Keap1, Beclin1, p62, and LC3 (1:2000, Proteintech).
BLM-Induced pulmonary fibrosis animal model
Pulmonary fibrosis was induced in male C57BL/6 mice (6 weeks old). The mice were obtained from Xiamen University Laboratory Animal Center and housed under specific pathogen-free (SPF) conditions with a 12-hour light/dark cycle and free access to food and water. All procedures involving animals were approved by the Institutional Animal Care and Use Committee (IACUC) of Xiamen University (ethics approval number: XMULAC20240137). The animals used in this study were handled following the ARRIVE guidelines. All methods were carried out in accordance with relevant guidelines and regulations.
Briefly, mice were anesthetized with 1% sodium pentobarbital (60 mg/kg), and bleomycin (2 mg/kg in 40 µl sterile saline) was administered intratracheally. Control mice received the same volume of sterile saline. Starting on day 10 post-BLM injection, nintedanib (11-B618731, Boer) and uridine (11-B612407, Boer) dissolved in saline were administered by gavage daily to anesthetized mice. Mice were sacrificed by cervical dislocation on day 20 post-BLM injection, and lung tissue samples were collected. On day 28 of uridine administration, mice were sacrificed after anesthesia. Body weight (BW) was recorded, and the whole lung was excised and weighed (lung weight, PW). The lung weight index was calculated as PI = PW/BW × 100.
Immunofluorescence staining
Cell Immunofluorescence: A549 cells were cultured on glass coverslips, pretreated with BLM (25 µM) for 24 h, and then treated with uridine (300 µM) for 72 h. After washing, the cells were fixed with 4% paraformaldehyde for 15 min and permeabilized with 0.5% Triton X-100 in PBS for 20 min at room temperature. After blocking with normal goat serum, primary antibodies diluted 1:60 were added and incubated in a humidified chamber at 4 °C for 12 h. After washing, the cells were incubated with fluorescent secondary antibodies (1:400) for 1 h and counterstained with DAPI for 5 min. The slides were mounted with anti-fade mounting medium and observed using a high-sensitivity laser confocal microscope (Zeiss LSM980).
Tissue Immunofluorescence: Fresh lung tissues were cryosectioned and fixed with acetone. After blocking with goat serum for 20 min, primary antibodies diluted 1:60 were added and incubated in a humidified chamber at 4 °C for 12 h. After warming to room temperature for 45 min, the sections were incubated with fluorescent secondary antibodies (1:400) for 1 h, counterstained with DAPI, and mounted with anti-fade mounting medium. Images were captured using a high-sensitivity laser confocal microscope (Zeiss LSM980). Primary antibodies used included γ-H2 AX, Ki67, E-cadherin, α-SMA, Collagen1a1, Nrf2, LC3 and Keap1, with Alexa Fluor® 594 (red) or 488 (green)-conjugated secondary antibodies (1:400, Invitrogen).
HE stain, Masson’s trichrome stain and modified Sirius red stain
Lung tissues were collected from male C57BL/6 mice (6 weeks old) on day 28 after BLM or PBS administration. The lungs were fixed in 4% paraformaldehyde, embedded in paraffin, and sectioned at 5 μm. Sections were stained with Hematoxylin-Eosin (HE) Stain Kit (G1120, Solarbio), Masson’s Trichrome Stain Kit (G1340, Solarbio), and Modified Sirius Red Stain Kit (G1472, Solarbio). The degree of lung fibrosis was assessed using the Ashcroft scoring system: grade 0, normal lung tissue; grade 1, mild fibrosis with minimal thickening of alveolar or bronchiolar walls; grades 2–3, moderate thickening of alveolar walls without obvious damage to lung architecture; grades 4–5, increased fibrosis with significant lung structure damage, forming fibrous bands or small fibrous masses; grades 6–7, severe lung structure destruction with large fibrous areas, potentially accompanied by honeycomb lung formation; grade 8, total fibrosis with all lung tissue occluded and necrotic.
Hydroxyproline content measurement
Hydroxyproline levels in the lungs were measured using a hydroxyproline assay kit (Nanjing Jiancheng Bioengineering Institute, China) at 550 nm, with results expressed as micrograms of hydroxyproline per milligram of lung tissue.
Transfection
p62 and control siRNA were purchased from Cell Signaling Technology (Danvers, MA, USA). The RFP-GFP-LC3 adenovirus vector, ATG5 siRNA, and control siRNA were designed and synthesized by Jikai (Guangzhou, China). Transfection of THP-1 cells was performed according to the manufacturer’s protocol (The primer sequences for transfection are provided in Supplementary Table 2). Autophagic flux was observed using a fluorescence microscope (Olympus).
Transmission Electron microscopy
Cells treated with 300 µM uridine were harvested and immediately fixed in 2.5% glutaraldehyde overnight at 4 °C, followed by post-fixation with 2% osmium tetroxide for 1 h at 37 °C. Cells were then embedded, stained with uranyl acetate/lead citrate, and imaged using a transmission electron microscope (TEM) (ThermoFisher Helios 5 UC).
Co-immunoprecipitation analysis
For co-immunoprecipitation (Co-IP), 500 µg of total protein was extracted from each cell group and incubated with an anti-P62 antibody. Protein A + G agarose beads were used to remove nonspecific binding. The antibody-antigen complex was allowed to react overnight, and then protein A + G agarose beads were added to capture the complex. After washing and centrifugation, the pellet was resuspended, boiled, and used for Western blot analysis.
Statistical analysis
Statistical analysis was performed using SPSS 20.0 software (SPSS Inc., Chicago, IL, USA). Data are presented as mean ± SD. Statistical significance was determined using the Student’s t-test and one-way analysis of variance. Asterisks indicate statistical significance (* P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001, ns indicates no significance).
Data availability
The datasets used and analysed during the current study available from the corresponding author on reasonable request.
References
Anna, J. P. et al. Idiopathic pulmonary fibrosis: state of the Art for 2023. Eur. Respir. J. https://doi.org/10.1183/13993003.00957-2022 (2023).
Paul, J. W. et al. Time for a change: is idiopathic pulmonary fibrosis still idiopathic and only fibrotic? The Lancet Respiratory Medicine (2018). https://doi.org/10.1016/s2213-2600(18)30007-9
Luca, R., Harold, R. C. & Mark, G. J. Idiopathic pulmonary fibrosis. Lancet https://doi.org/10.1016/s0140-6736(17)30866-8 (2017).
Taylor, A. et al. Single-cell RNA-seq reveals ectopic and aberrant lung-resident cell populations in idiopathic pulmonary fibrosis. Sci. Adv. https://doi.org/10.1126/sciadv.aba1983 (2020).
La Tonya, J. H. et al. Senolytics decrease senescent cells in humans: preliminary report from a clinical trial of dasatinib plus Quercetin in individuals with diabetic kidney disease. EBioMedicine https://doi.org/10.1016/j.ebiom.2019.08.069 (2019).
Lulu, S. et al. Potential role of senescent macrophages in radiation-induced pulmonary fibrosis. Cell Death Dis. https://doi.org/10.1038/s41419-021-03811-8 (2021).
Chin, C. et al. Loss of IGFBP2 mediates alveolar type 2 cell senescence and promotes lung fibrosis. Cell. Rep. Med. https://doi.org/10.1016/j.xcrm.2023.100945 (2023).
Vivien, S. et al. The therapy of idiopathic pulmonary fibrosis: what is next? Eur. Respiratory Rev. https://doi.org/10.1183/16000617.0021-2019 (2019).
John, S. et al. (ed, K.) Comparison of Pirfenidone and Nintedanib: Post-Hoc analysis of the CleanUP-IPF study. Chest https://doi.org/10.1016/j.chest.2023.11.035 (2023).
Yvette, N. L. & Nintedanib A Review in Fibrotic Interstitial Lung Diseases. Drugs (2021). https://doi.org/10.1007/s40265-021-01487-0
Yi, Z. et al. Orally-active, clinically-translatable senolytics restore α-Klotho in mice and humans. EBioMedicine (2022). https://doi.org/10.1016/j.ebiom.2022.103912
Solomon, K. et al. Reciprocal REGγ-Nrf2 regulation promotes long period ROS scavenging in oxidative Stress-Induced cell aging. Oxidative Med. Cell. Longev. https://doi.org/10.1155/2023/4743885 (2023).
Shengnan, L., Jingbo, P. & Qiang, Z. Signal amplification in the KEAP1-NRF2-ARE antioxidant response pathway. Redox Biol. https://doi.org/10.1016/j.redox.2022.102389 (2022).
Vijay, A. et al. Carvacrol abates NLRP3 inflammasome activation by augmenting Keap1/Nrf-2/p62 directed autophagy and mitochondrial quality control in neuropathic pain. Nutr. Neurosci. https://doi.org/10.1080/1028415x.2021.1892985 (2021).
Shifeng, C. et al. Up-regulation of Nrf2/P62/Keap1 involves in the anti-fibrotic effect of combination of monoammonium glycyrrhizinate and cysteine hydrochloride induced by CCl4. Eur. J. Pharmacol. https://doi.org/10.1016/j.ejphar.2021.174628 (2021).
Emma, T., John, A. C., Prabhakaran, G., Susan, C. & Richard, F. L. Age and oxidative stress regulate Nrf2 homeostasis in human articular chondrocytes. Osteoarthr. Cartil. https://doi.org/10.1016/j.joca.2023.05.004 (2023).
Zunpeng, L. et al. Cross-species metabolomic analysis identifies uridine as a potent regeneration promoting factor. Cell. Discovery. https://doi.org/10.1038/s41421-021-00361-3 (2022).
Isabelle, A. et al. Pyruvate and uridine rescue the metabolic profile of OXPHOS dysfunction. Mol. Metabolism. https://doi.org/10.1016/j.molmet.2022.101537 (2022).
Ruth, H. et al. Circulating uridine dynamically and adaptively regulates food intake in humans. Cell. Rep. Med. https://doi.org/10.1016/j.xcrm.2022.100897 (2023).
Sanja, C. et al. Uridine supplementation exerts anti-inflammatory and anti-fibrotic effects in an animal model of pulmonary fibrosis. Respir. Res. https://doi.org/10.1186/s12931-015-0264-9 (2015).
Yali, J., Zhihui, J., Sen, C. & Weichun, G. Uridine relieves MSCs and chondrocyte senescence in Vitvo and exhibits the potential to treat osteoarthritis in vivo. Cell. Cycle. https://doi.org/10.1080/15384101.2021.2010170 (2022).
Manish Kumar, J., Dinesh, T., Mattias, M., Naidu, V. G. M. & Srinivas, U. Uridine ameliorates dextran sulfate sodium (DSS)-Induced colitis in mice. Sci. Rep. https://doi.org/10.1038/s41598-017-04041-9 (2017).
Veronica Della, L. et al. Lung inflammation after bleomycin treatment in mice: selection of an accurate normalization strategy for gene expression analysis in an ex-vivo and in-vitro model. Int. J. Biochem. Cell Biol. https://doi.org/10.1016/j.biocel.2017.05.016 (2017).
Veronica Della, L., Antonella, C., Silvia Del, R. & Maria-Aurora, M. Bleomycin in the setting of lung fibrosis induction: from biological mechanisms to counteractions. Pharmacol. Res. https://doi.org/10.1016/j.phrs.2015.04.012 (2015).
Colin, A. H., Wei, S., Fangwu, W., Margarita, W., Connie, J. E. & M. & Aging alters the cell cycle control and mitogenic signaling responses of human hematopoietic stem cells. Blood https://doi.org/10.1182/blood.2022017174 (2023).
Aswin, S., Ying, C., Lauren, D. B., Bree, B. A. & David, L. K. Engineered cell and tissue models of pulmonary fibrosis. Adv. Drug Deliv. Rev. https://doi.org/10.1016/j.addr.2017.12.013 (2018).
Gabriele, S. & Raffaella Di, M. Fabrizio D’Adda Di, F. Crosstalk between chromatin state and DNA damage response in cellular senescence and cancer. Nat. Rev. Cancer. https://doi.org/10.1038/nrc3344 (2012).
Jing, H. et al. Polydopamine-loaded Prunetin nanomaterials activate DRD2 to reduce UV-induced inflammation by stabilizing and promoting Nrf2 nuclear translocation. Acta Biomater. https://doi.org/10.1016/j.actbio.2023.07.050 (2023).
Pinheiro, P. M., José, F., Roberto de Oliveira, F. & Gardênia Carmen Gadelha, M. & Daniel Pereira, B. Chloroquine and hydroxychloroquine in antitumor therapies based on autophagy-related mechanisms. Pharmacological Research (2021). https://doi.org/10.1016/j.phrs.2021.105582
Katie, K. InterMune and Boehringer Blaze trails for idiopathic pulmonary fibrosis drugs. Nat. Rev. Drug Discovery. https://doi.org/10.1038/nrd4382 (2014).
Safaa, A. F., Noha, M. S., Reem, N. E. N., Iriny, M. A. & Samar, S. A. Hepatoprotective effect of cranberry nutraceutical extract in Non-alcoholic fatty liver model in rats: impact on insulin resistance and Nrf-2 expression. Front. Pharmacol. https://doi.org/10.3389/fphar.2020.00218 (2020).
Guihe, Y. et al. Astaxanthin suppresses oxidative stress and calcification in vertebral cartilage endplate via activating Nrf-2/HO-1 signaling pathway. Int. Immunopharmacol. https://doi.org/10.1016/j.intimp.2023.110159 (2023).
Huankai, Y. et al. Swinhoeic acid from Potentilla fragarioides ameliorates high glucose-induced oxidative stress and accumulation of ECM in mesangial cells via Keap1-dependent activation of Nrf2. Redox Rep. https://doi.org/10.1080/13510002.2022.2134755 (2022).
Klaudia, J. et al. Reactive oxygen species, toxicity, oxidative stress, and antioxidants: chronic diseases and aging. Arch. Toxicol. https://doi.org/10.1007/s00204-023-03562-9 (2023).
Ilaria, B., Ileana, G., Alba, M. & Rosario, D. Nrf2-Keap1 signaling in oxidative and reductive stress. Biochimica et biophysica acta (BBA) - Molecular Cell Research (2018). https://doi.org/10.1016/j.bbamcr.2018.02.010
Lingming, K. et al. Sitagliptin activates the p62–Keap1–Nrf2 signalling pathway to alleviate oxidative stress and excessive autophagy in severe acute pancreatitis-related acute lung injury. Cell Death Dis. https://doi.org/10.1038/s41419-021-04227-0 (2021).
Xin, W. et al. DDRGK1 enhances osteosarcoma chemoresistance via inhibiting KEAP1-Mediated NRF2 ubiquitination. Adv. Sci. https://doi.org/10.1002/advs.202204438 (2023).
Tao, J. et al. p62 links autophagy and Nrf2 signaling. Free Radic. Biol. Med. https://doi.org/10.1016/j.freeradbiomed.2015.06.014 (2015).
Diptiman, C. et al. Developmental pathways in the pathogenesis of lung fibrosis. Mol. Aspects Med. https://doi.org/10.1016/j.mam.2018.08.004 (2019).
Junlan, Z. et al. NF-κB1 p50 stabilizes HIF-1α protein through suppression of ATG7-dependent autophagy. Cell Death Dis. https://doi.org/10.1038/s41419-022-05521-1 (2022).
Clément, B., Mercedes, T., Piedad del Socorro, M., Raúl, V. D. & Glutamine MTOR and autophagy: a multiconnection relationship. Autophagy https://doi.org/10.1080/15548627.2022.2062875 (2022).
Acknowledgements
This research was funded by National Natural Science Foundation of China, grant number 81870388.
Author information
Authors and Affiliations
Contributions
DCC carried out the studies, participated in collecting data, and drafted the manuscript. ZRJ and LQ performed the statistical analysis and participated in its design. GZZ,HJJ and YZW participated in acquisition, analysis, or interpretation of data and draft the manuscript. LGY: funding acquisition, writing–review and editing, and project administration. All authors read and approved the final manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
All methods were carried out in accordance with relevant guidelines and regulations.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ding, C., Zuo, R., Liao, Q. et al. Uridine alleviates the aging of alveolar epithelial cells in idiopathic pulmonary fibrosis through the Keap1-Nrf2 signaling pathway. Sci Rep 15, 22952 (2025). https://doi.org/10.1038/s41598-025-04053-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-04053-w