Abstract
The aim of this study was to investigate the changes in the gut microbiota and proteins related to metabolism and immunity caused by childhood obesity and insulin resistance (IR) and to assess the underlying relationship between the gut microbiota and IR in children. Nineteen children with obesity and sixteen healthy children were recruited. Children with obesity were divided into two groups: obese with IR and obese without IR. All of the obese children participated in a 1-month lifestyle-based weight loss program. Faecal metagenomics and serum Olink proteomics combined with clinical parameters were used to identify the metabolic alterations associated with childhood obesity and IR. The gut microbiota and serum protein were significantly altered in obese children with IR. The relative abundances of Akkermansia muciniphila, IGFBP1 and GP6 were decreased in obese children with IR. Butyricicoccus sp. AM29-23AC, DLK1, CD163, CCL16 and CTSD levels were recovered after IR improved. TNFR1 had a significant indirect effect on the interaction between s-Citrobacter.freundii and fasting insulin. In conclusion, obese children with IR have abnormal gut microbiota and serum proteins related to metabolism and immunity, which can be improved after weight loss. The gut microbiome of children with obesity may contribute to the development of IR by triggering metabolic inflammation.
Clinical Trial Registration: This study was registered with the Chinese Clinical Trial Registry (Registration number: ChiCTR2300072179).
Similar content being viewed by others
Introduction
The dramatic increase in childhood obesity prevalence has become a public health problem worldwide1,2. Insulin resistance (IR) is hypothesized to be the important pathophysiologic link between adiposity and the future development of type 2 diabetes and cardiovascular disease, which is characterized by a decrease in the efficiency of insulin in glucose uptake and utilization. Insulin resistance (IR) serves not only as a fundamental mechanism underlying the pathogenesis of obesity and a central contributor to metabolic dysfunction-associated fatty liver disease (MAFLD) but also as a significant risk factor for conditions such as polycystic ovarian syndrome and hyperuricaemia3. The underlying pathological mechanisms have not been fully investigated. Identifying effective and feasible targets for the treatment and prevention of childhood obesity is imperative.
Accumulating evidence has demonstrated that the gut microbiota is the primary endogenous factor affecting obesity and IR4. A metagenome-wide association study revealed that gut microbial carbohydrate metabolism was associated with the development of obesity4. Studies focusing on the gut microbiota during childhood IR are scarce5,6. It is essential to obtain data on gut microbiota changes in obese children with IR before and after treatment to characterise the underlying mechanism of insulin resistance comprehensively. Proteomics has emerged as an effective method for exploring the pathological mechanism of diseases7. Olink provides a new perspective for studying the pathological mechanisms of disease using minimal sample volume. Proteomic data may serve as a bridge to investigate the relationships between the gut microbiota and disease8. However, studies to elucidate the role of the gut microbiota through protein analysis in obese children are scarce, especially in children with IR. Integrated analyses of the gut microbiome, host metabolic proteins and phenotype may represent promising strategies to explore these multiomics data and yield mechanistic insight into the development of childhood obesity and IR.
In the present study, we aimed to compare the changes in the gut microbiome and serum proteins related to metabolism and immunity caused by childhood obesity and IR and to identify biomarkers after lifestyle-based weight loss intervention. Additionally, we investigated how altered gut microbiota modulates host metabolism via serum proteins in children with obesity and IR by performing multiomics data analysis. Our findings are important for the comprehensive prevention and intervention of childhood obesity and IR through the gut microbiota.
Methods
Study cohort
Nineteen obese children (13 boys and 6 girls) aged 6–14 years who visited the clinic of the Paediatric Endocrinology Department at the Second Affiliated Hospital of Xi’an Jiaotong University were recruited for this study. The inclusion criterion was an age- and sex-specific BMI ≥ 95th percentile. Patients were excluded if they had endocrine or genetic obesity syndromes, infectious disease, or autoimmune disease or if they consumed any medicine. Sixteen healthy children (8 boys and 8 girls) who visited a child health care clinic were included as controls. All the children were divided into a prepubertal group (Tanner’s stage I) and a pubertal group (Tanner’s stage II–V) according to the Tanner stage. According to the homeostasis model assessment of IR (HOMA-IR) index (prepubertal > 2.5, pubertal > 4.0), children with obesity were divided into two groups: obese with IR and obese without IR9. This study was approved by the Second Affiliated Hospital of Xi’an Jiaotong University (No. 2022245) and was registered with the Chinese Clinical Trial Registry (06/06/2023, ChiCTR2300072179). All parents agreed to participate in the study and provided written informed consent. This research was performed in accordance with the Declaration of Helsinki.
Study design
The baseline anthropometric characteristics and biological samples of the cohort were obtained. Nineteen children with obesity participated in a lifestyle intervention program under the guidance of professional doctors. Each parent received an appropriate and strict diet sheet after nutritional evaluation. The participants were required to exercise for 30 min every day. Sedentary activities were limited to less than 2 h per day. In addition, the children were rewarded in stages based on weight loss. At the end of the 1-month intervention, the physical examination was repeated. Blood and stool samples were collected.
Clinical and biochemical assessment
Stand height and weight without shoes were measured using a calibrated scale and a stadiometer. The BMI data were converted into a body mass index standard deviation score (BMI-SDS) according to the Chinese children’s and adolescents’ age- and sex-specific percentile standards10. Overnight fasting blood samples and stool samples were collected at baseline and after intervention from all the children. The blood samples were stored at − 80 ℃ after centrifugation. The collected stool samples were quickly placed into liquid nitrogen cold raffinate and stored at − 80 ℃ until the experiments.
Fasting glucose, total triglyceride (TG), cholesterol (TC), high-density lipoprotein cholesterol (HDL-C), low-density lipoprotein cholesterol (LDL-C), very low-density lipoprotein cholesterol (VLDL-C), alanine aminotransferase (ALT) and aspartate aminotransferase (AST) levels were tested using an autoanalyzer (Hitachi 747, Japan). Serum insulin levels were measured using radioimmunoassay (BeiFang systems, Beijing, China). The HOMA-IR was used to estimate insulin resistance.
Olink proteomics
Olink proteomics was performed by iomics (Beijing, China). In brief, protein samples were prepared from serum and incubated with paired antibodies conjugated to specific DNA barcodes. After binding the paired antibody to the target protein and extending and amplifying the conjugated oligonucleotide strands, the DNA barcodes were detected using qPCR to quantify the protein.
Faecal whole-genome shotgun sequencing
Total DNA was isolated from the faecal samples using a QIAamp Fast DNA Stool Mini Kit (Qiagen, USA). Paired-end 2 × 150 bp sequencing was performed using the Illumina NovaSeq 6000 high-throughput sequencing platform. The effective sequences were annotated using Kraken2, and the data from the RefSeq genome database of NCBI were used as references to construct a database with an adjusted confidence level of 0.5. A species abundance table was obtained after the species abundance was normalized. MetaGeneMark software was used to analyse the contigs. The filtered protein sequence sets were aligned with common protein databases to annotate gene functions in all the samples.
The α diversity indices were individually determined using QIIME software. Composition and abundance distribution tables were obtained for each sample at the six taxonomic levels. The contrasts between the groups were performed using the “microeco” R package11. Enriched taxa between the groups were obtained by linear discriminant analysis effect size (LEfSe) analysis.
Statistical analysis
Differences in the clinical data, relative abundance of proteins and gut microbiota species, or functional pathways were analysed via SPSS 23.0. Normal distributions were assessed by the Kolmogorov–Smirnov test. The data are expressed as the means ± standard errors of the means or counts (percentages). Comparisons between groups were performed using t tests or chi-square tests at baseline. A paired t test was used to compare the follow-up parameters between obese children with IR before and after the intervention.
Partial least square differential analysis (PLS-DA) was run using the R package “mixOmics”12. Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) analyses were performed using the R package “clusterProfiler”13. Protein–protein interactions (PPIs) were calculated using the STRING database (https://string-db.org) and visualized via Cytoscape.
A random forest regression model was constructed on the basis of the relative abundance of gut species and proteins using the “randomForest” R package. The “Boruta” R package was used to perform the Boruta algorithm14. Correlation analysis (Spearman’s correlation coefficients) between proteins, metagenomics, and clinical indices was performed using the “corr.test” function. Mediation analyses were performed using the “mediation” R package15. The “ggplot2” R package was used to visualize the results. FDR was controlled using the Benjamini–Hochberg method, and a p value < 0.05 was considered to indicate statistical significance.
Results
Baseline demographic and clinical data
The clinical trial process is illustrated in Fig. 1a. The clinical characteristics are shown in Table 1. No significant differences in sex or age were observed among the obese children with IR, obese children without IR and control groups. Compared with the control group, the obesity groups presented significantly higher BMI-SDS, fasting insulin, HOMA-IR, ALT, AST, TG, TC, and LDL-C levels, whereas their fasting glucose levels were similar (t test).
Study design and gut microbiota changes at baseline. (a) Workflow of the clinical research in this study. (b) Composition of the phyla in different groups. (c) The top 10 relative abundance classes of the gut microbiota. The genera whose average relative abundance in all participants was less than 0.5% were integrated as “Other genera”. (d) The top 6 most abundant species of the gut microbiota. (e) Differences in the gut microbiota between obese individuals with and without IR according to LEfSe analysis (linear discriminant analysis (LDA) threshold of 2).
Faecal metagenomic analyses
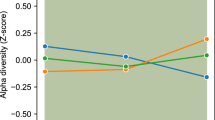
For the baseline analysis, the faecal metagenomes of the obese children were compared with those of the controls. No significant differences were observed in the Shannon, Simpson, ACE, or CHAO indices among healthy controls, obese children with IR and obese children without IR. There were no significant differences in the α diversity indices after IR improvement (Figure S1). Wilcoxon’s test with Benjamini–Hochberg correction revealed significant changes in the relative abundances of gut microbiota phyla, genera, and species among the control, obesity with IR, and obesity without IR groups. The proportion of Firmicutes was highest, and the proportion of Bacteroidetes was lowest in obese individuals without IR. The obesity with IR group presented the lowest proportion of Actinobacteria (Fig. 1b). The top 10 genera in terms of overall abundance are shown in Fig. 1c. The Bacteroides was the most enriched genus in our cohort. The 6 most abundant species are shown in Fig. 1d. The relative abundance of s-Akkermansia muciniphila was lowest in the IR-positive obesity group, whereas the relative abundance of s-Megamonas funiformis was lowest in the control group. LEfSe analysis was used to compare differences in the gut microbiota composition among the three groups. In obese individuals with IR, increased levels of s-Rothia kristinae were observed. In obese individuals without IR, s-Weissella cibaria, s-Weissella confusa, g-Weissella and f-Leuconostocaceae were enriched (Fig. 1e).
Olink proteomics and correlation analyses between obese children with and without IR
We compared the relative abundance of 92 proteins spanning metabolic and inflammatory pathways by Olink proteomics. There were 27 proteins significantly changed in children with obesity compared with controls (Figure S2a). PLS-DA revealed that Olink proteomics effectively distinguished the three groups (Fig. 2a). There were four proteins whose expression significantly changed in obese individuals with IR compared with obese individuals without IR (Fig. 2b). The upregulated proteins included CHI3L1 (log2FC = 0.500, p = 0.047) and GDF15 (log2FC = 0.736, p = 0.012). The downregulated proteins included GP6 (log2FC = -0.615, p = 0.023) and IGFBP1 (log2FC = -0.618, p = 0.046). GO and KEGG enrichment analyses were performed to compare obese children with and without IR. Significantly different proteins were enriched in leukocyte migration, regulation of cell‒cell adhesion, endopeptidase activity, secretory granule lumen and diabetic cardiomyopathy (Fig. 2c). On the basis of degree and betweenness centrality, the PPI network identified EPCAM, COL1A1 and EGFR as hub modules (Fig. 2d).
Comparisons of serum Olink proteomics profiles and protein functions between obese individuals with IR and obese individuals without IR. (a) Partial least square differential analysis (PLS-DA) of Olink proteomics in the control, obesity without IR and obesity with IR groups. (b) Volcano plot of different serum protein levels in obese individuals with IR compared with obese individuals without IR, as determined using t tests. (c) Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses of the differentially expressed proteins. (d) Protein‒protein interaction network of different serum proteins, ranked by betweenness.
We combined the gut microbiota and serum proteins to establish a random forest model to distinguish obesity with IR from obesity without IR. Paired with the feature selection and error rates, the number of random forest classification trees stabilized after 400 iterations (Figure S2b). In terms of the mean decrease in accuracy, s-Megasphaera.micronuciformis, s-Veillonella.sp.T11011.6, s-Veillonella.dispar and PGLYRP1 were included in the equation (Fig. 3a). The Boruta algorithm was used to compare the correlation between features and random detection. Figure S2c shows the change in the importance score of each variable. A total of eight variables were screened, including s-Veillonella sp. T11011.6, s-Megasphaera micronuciformis and s-Veillonella dispar (Fig. 3b).
Important features and correlations of parameters between obese children with and without IR were identified. Ranking of variable importance of features identified by random forest (a) and burota (b). (c) Heatmap of Spearman’s correlation coefficients between clinical parameters with significantly changed proteins and the gut microbiota in obese children with and without IR. (d) Mediation model of TNFR1 between s-Citrobacter freundii and fast insulin. * p < 0.05, ** p < 0.01.
Finally, correlations between clinical parameters, significantly altered gut microbiota and serum proteins were assessed by Spearman correlation analysis. The relative abundance of s-Citrobacter.freundiii was positively associated with fasting insulin and GP6 (Fig. 3c). For these significant gut microbiota and protein associations, we performed a mediation analysis to test whether the proteins related to metabolism and immunity can mediate the gut microbiota and clinical characteristics. TNFR1 had a significant indirect effect on the interaction between s-Citrobacter.freundii and fasting insulin (effect ratio = 32.5%, p = 0.04) (Fig. 3d).
Proteome and gut Microbiome after weight loss
After 1 month of lifestyle-based weight loss, 16 children successfully lost more than 0.5% of their body weight. Among the subset of seven obese children with baseline insulin resistance, a 4-week lifestyle intervention resulted in statistically significant improvements in metabolic parameters: HOMA-IR decreased from 4.85 ± 2.32 (baseline) to 1.87 ± 0.5 (posttreatment) (p < 0.05, paired t test), with six of seven participants achieving levels within the normal range. The fasting glucose levels also decreased, although the difference was not statistically significant. Additionally, VLDL-C levels decreased significantly after the intervention (paired t test, Fig. 4a). The proportions of Firmicutes and Bacteroidetes increased, the proportion of Bacteroidetes decreased after weight loss, and IR improved. (Fig. 4b). At the species level, the s-Enterobacter cloacae and s-Actinomyces johnsonii levels decreased, whereas the s-Butyricicoccus sp. AM29-23AC level increased (Fig. 4c). The results of the KEGG analysis revealed that lipoarabinomannan biosynthesis, ferroptosis, the thyroid hormone signalling pathway, fatty acid degradation and oxidative phosphorylation were significantly changed (Fig. 4d). In terms of gut CAZyme enrichment, the differentially expressed CAZymes were mainly glycoside hydrolases (GH37, GH53, GH113, and GH135), polysaccharide lyase (PL30) and carbohydrate esterase (CE4) (Fig. 4e).
Alterations in clinical parameters and the gut microbiota after IR improvement in obese children with IR. (a) Comparison of BMI-SDS, fasting insulin, fasting glucose, HOMA-IR and serum lipid contents before and after IR improvement in obese children with IR. (b) Composition of the phyla before and after IR improvement in obese children with IR. (c) The significantly changed species of the gut microbiota after IR improvement. (d) KEGG pathway enrichment analysis before and after IR improvement in obese children with IR. (e) Ranking of significantly changed CAyZE after IR improvement. *p < 0.05, **p < 0.01.
Finally, we compared the metabolic protein profiles before and after the intervention in obese children with IR. The levels of CCL16, CD163, CTSD and DLK1 were significantly decreased, whereas those of FABP4 and ICAM2 were significantly increased (Fig. 5a). In the correlation analysis matrix, we found that HOMA-IR was negatively correlated with FABP4 and s-Butyricicoccus sp. AM29-23AC but positively correlated with s-Enterobacter cloacae and s-Actinomyces johnsonii. The correlations of fasting insulin with other measures remained consistent with those of the HOMA-IR. In addition, fasting insulin was positively associated with CD163 (Fig. 5b).
Alterations in serum proteins related to metabolism and immunity and relationships of parameters after IR improvement in obese children with IR. (a) The significantly changed serum proteins related to metabolism and immunity after IR improvement. (b) Heatmap of Spearman’s correlation coefficients between clinical parameters with significantly changed proteins and the gut microbiota after IR improvement. *p < 0.05, **p < 0.01.
Discussion
Several previous studies reported an association between altered gut microbial composition and clinical traits of IR in children with obesity5,6. Nonetheless, the unique mechanism underlying the involvement of the gut microbiome and host proteins in the development of IR remains poorly characterized. In this study, we performed integrative analyses of clinical features, metagenomic sequencing and Olink proteomics to explore the complex functions of the gut microbiome in children with obesity and IR. Our findings support the idea that the gut microbiome associated with children with obesity may contribute to the development of IR by triggering metabolic inflammation.
Persistent gut microbiota imbalance has been identified as the primary endogenous factor affecting obesity and IR (16]. However, data in children are scarce. Firmicutes and Bacteroidetes are the dominant bacteria in the human intestine, and the F/B ratio is elevated in obese individuals. A higher F/B ratio in obese children than in healthy controls and a lower F/B ratio in obese children with IR than in obese children without IR were detected in our cohort, which is consistent with the findings in adults16,17. The relative abundances of s-Akkermansia muciniphilas and s-Bacteroides thetaiotaomicron were decreased in obese children, especially in obese children with IR. Compared with those with higher abundance, obese individuals who harboured a lower abundance of Akkermansia muciniphila presented a poor glucose profile18. Bacteroides thetaiotaomicron has been identified as a prebiotic that has the potential for carbohydrate utilization19. The relative abundance of s-Megamonas funiformis increased in adults with metabolic syndrome, which was consistent with our results20. After 1 month of intervention, a total of seven children improved their glucose metabolism. The composition of the gut microbiota also changed significantly, suggesting that microbes and host metabolism are closely related. The relative abundances of S. cloacae and S. johnsonii decreased, whereas those of S. butyricicoccus_sp. AM29-23AC was increased. As a Gram-negative bacterium, S. cloacae is considered a pathogenic bacterium related to obesity21. The short-chain fatty acid-producing bacteria Butyricicoccus has been annotated as a protective gut microbiota22. Since CAZymes can be encoded by the gut microbiota and digest host dietary carbohydrates, CAZymes act as a bridge between the gut microbiota and host glucose metabolism23. We found that CAZymes, including glycoside hydrolases, polysaccharide lyases and carbohydrate esterases, were altered after weight loss. These results illustrated that the gut microbiota is closely associated with obesity and IR in children.
Low-grade and chronic inflammation has been identified as one of the important factors that contributes to IR during obesity. The proinflammatory properties of TNFα play a detrimental role in the development of IR. We revealed a model by which Citrobacter freundii affects fasting insulin through TNFR1. Specifically, a relatively high level of serum TNFR1 has been found to partially explain the negative impact of Citrobacter freundii on host fasting insulin. Stimulation of umbilical cord blood mononuclear cells with the lipopolysaccharide of Citrobacter freundii increased the release of TNFα24. Although the causal relationship between inflammation and IR in children with obesity is still unclear, the gut microbiota has been reported to be one of the driving forces of host inflammation. Our results provide evidence that the gut microbiota may contribute to IR through metabolic inflammation.
A previous study revealed that DLK1 was positively correlated with body fat and HOMA-IR, which was consistent with our results25. Dlk1-deficient mice exhibit increased insulin sensitivity26. Palumbo et al.27 reported a negative correlation between DLK1 and HOMA-IR in girls with obesity. Sex-based differences may explain these inconsistencies. After IR improved, serum DLK1 levels significantly decreased, and a positive association between DLK1 and E. cloacae was observed. A previous study reported that Bifidobacterium promotes host metabolism via changes in Dlk1 expression. In addition, Lopez et al. reported that B. breve UCC2003 restored foetal glycaemia through FABP4, another significantly upregulated protein in our children28.
CCL16, also known as HCC4, was significantly decreased after weight loss. A protein array revealed higher levels of CCL16 in the subcutaneous adipose tissue of obese subjects than in that of lean subjects and was positively associated with HOMA-IR29. As a chemokine, few studies have focused on the pathomechanism of CCL16 in obesity and IR. ICAM2 is a member of the intercellular adhesion molecule family. The P9 protein produced by Akkermansia muciniphila can bind to ICAM2 and induce glucagon-like peptide-130. We found that CCL16 and ICAM2 were negatively associated with s-Actinomyces johnsonii. Further studies focused on the underlying potential causes need to be performed.
This study has several limitations. First, the HOMA-IR model was used in our study, whereas the gold standard for insulin resistance is the clamp test. Second, there was a lack of detailed dietary intake data to quantify preintervention dietary patterns or specific nutrient exposures. While the interventions included dietary counselling, individual compliance and baseline dietary habits were not systematically measured, which may have influenced the observed changes in the gut microbiome. Finally, the validation sample size was limited, and the subjects were regionalised. Given the difficulty in obtaining samples from children and the persistence of follow-up for weight loss in obese children, our study provides a multiomics perspective on weight loss in obese children with IR.
Conclusion
In conclusion, an integrative analysis using metagenomic sequencing and Olink proteomics revealed the abnormal composition of the gut microbiota and serum proteins in obese children with IR. The gut microbiome associated with children with obesity may contribute to the development of IR by triggering metabolic inflammation. Our data indicate that the link between the gut microbiota and host proteins may represent a novel target for therapeutic strategies for childhood obesity and IR.
Data availability
Sequence data that support the findings of this study have been deposited in the National Center for Biotechnology Information with the primary accession code PRJNA1090975.
References
Jebeile, H., Kelly, A. S., O’Malley, G. & Baur, L. A. Obesity in children and adolescents: epidemiology, causes, assessment, and management. Lancet Diabetes Endocrinol. 10, 351–365. https://doi.org/10.1016/S2213-8587(22)00047-X (2022).
Aagaard, K. M. et al. Understanding risk and causal mechanisms for developing obesity in infants and young children: A National institutes of health workshop. Obes. Rev. 25, e13690. https://doi.org/10.1111/obr.13690 (2024).
Zhang, A. M. Y., Wellberg, E. A., Kopp, J. L. & Johnson, J. D. Hyperinsulinemia in obesity, inflammation, and Cancer. Diabetes Metab. J. 45, 285–311. https://doi.org/10.4093/dmj.2020.0250 (2021).
Qin, J. et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490, 55–60. https://doi.org/10.1038/nature11450 (2012).
Ayala-García, J. C. et al. Mediation analysis of waist circumference in the association of gut microbiota with insulin resistance in children. Child. (Basel). 10 https://doi.org/10.3390/children10081382 (2023).
Yuan, X. et al. Gut microbiota of Chinese obese children and adolescents with and without insulin resistance. Front. Endocrinol. (Lausanne). 12, 636272. https://doi.org/10.3389/fendo.2021.636272 (2021).
Aleksandrova, K., Egea Rodrigues, C., Floegel, A. & Ahrens, W. Omics biomarkers in obesity: novel etiological insights and targets for precision prevention. Curr. Obes. Rep. 9, 219–230. https://doi.org/10.1007/s13679-020-00393-y (2020).
Pietzner, M. et al. Synergistic insights into human health from aptamer- and antibody-based proteomic profiling. Nat. Commun. 12, 6822. https://doi.org/10.1038/s41467-021-27164-0 (2021).
Andrade, M. I. et al. Identification of cutoff points for homeostatic model assessment for insulin resistance index in adolescents: systematic review. Rev. Paul Pediatr. 34, 234–242. https://doi.org/10.1016/j.rpped.2015.08.006 (2016).
Ji, C. Y. Working group on obesity in, C. Report on childhood obesity in China (1)--body mass index reference for screening overweight and obesity in Chinese school-age children. Biomed. Environ. Sci. 18, 390–400 (2005).
Liu, C., Cui, Y., Li, X. & Yao, M. Microeco: an R package for data mining in microbial community ecology. FEMS Microbiol. Ecol. 97, FIAA255. https://doi.org/10.1093/femsec/fiaa255 (2021).
Rohart, F., Gautier, B., Singh, A., KA, L. C. & mixOmics An R package for ‘omics feature selection and multiple data integration. PLoS Comput. Biol. 13, e1005752. https://doi.org/10.1371/journal.pcbi.1005752 (2017).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. ClusterProfiler: an R package for comparing biological themes among gene clusters. Omics 16, 284–287. https://doi.org/10.1089/omi.2011.0118 (2012).
Kursa, M. B. & Rudnicki, W. R. Feature selection with the Boruta package. J. Stat. Softw. 36, 1–13. https://doi.org/10.18637/jss.v036.i11 (2010).
Tofighi, D. & MacKinnon, D. P. RMediation: an R package for mediation analysis confidence intervals. Behav. Res. Methods. 43, 692–700. https://doi.org/10.3758/s13428-011-0076-x (2011).
Larsen, N. et al. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One. 5, e9085. https://doi.org/10.1371/journal.pone.0009085 (2010).
Haro, C. et al. Consumption of two healthy dietary patterns restored microbiota dysbiosis in obese patients with metabolic dysfunction. Mol. Nutr. Food Res. 61 https://doi.org/10.1002/mnfr.201700300 (2017).
García-Gamboa, R. et al. Associations between bacterial and fungal communities in the human gut microbiota and their implications for nutritional status and body weight. Sci. Rep. 14, 5703. https://doi.org/10.1038/s41598-024-54782-7 (2024).
Bedu-Ferrari, C. et al. In-depth characterization of a selection of gut commensal bacteria reveals their functional capacities to metabolize dietary carbohydrates with prebiotic potential. mSystems, e0140123. https://doi.org/10.1128/msystems.01401-23 (2024).
Sheng, S. et al. Gut Microbiome is associated with metabolic syndrome accompanied by elevated gamma-glutamyl transpeptidase in men. Front. Cell. Infect. Microbiol. 12, 946757. https://doi.org/10.3389/fcimb.2022.946757 (2022).
Gu, M. et al. Lactobacillus pentosus MJM60383 inhibits lipid accumulation in Caenorhabditis elegans induced by Enterobacter cloacae and glucose. Int. J. Mol. Sci. 24, 280. https://doi.org/10.3390/ijms24010280 (2022).
Luo, J. et al. Integrative metabolomics highlights gut microbiota metabolites as novel NAFLD-related candidate biomarkers in children. Microbiol. Spectr. e0523022 https://doi.org/10.1128/spectrum.05230-22 (2024).
Zhou, Z., Sun, B., Yu, D. & Zhu, C. Gut microbiota: an important player in type 2 diabetes mellitus. Front. Cell. Infect. Microbiol. 12, 834485. https://doi.org/10.3389/fcimb.2022.834485 (2022).
Reuschel, E., Toelge, M., Entleutner, K., Deml, L. & Seelbach-Goebel, B. Cytokine profiles of umbilical cord blood mononuclear cells upon in vitro stimulation with lipopolysaccharides of different vaginal gram-negative bacteria. PLoS One. 14, e0222465. https://doi.org/10.1371/journal.pone.0222465 (2019).
Jensen, C. H. et al. The imprinted gene Delta like non-canonical Notch ligand 1 (Dlk1) associates with obesity and triggers insulin resistance through Inhibition of skeletal muscle glucose uptake. EBioMedicine 46, 368–380. https://doi.org/10.1016/j.ebiom.2019.07.070 (2019).
Abdallah, B. M., Ditzel, N., Laborda, J., Karsenty, G. & Kassem, M. DLK1 regulates Whole-Body glucose metabolism: A negative feedback regulation of the Osteocalcin-Insulin loop. Diabetes 64, 3069–3080. https://doi.org/10.2337/db14-1642 (2015).
Palumbo, S. et al. Circulating levels of DLK1 and glucose homeostasis in girls with obesity: A pilot study. Front. Endocrinol. (Lausanne). 13, 1033179. https://doi.org/10.3389/fendo.2022.1033179 (2022).
Lopez-Tello, J. et al. Maternal gut microbiota Bifidobacterium promotes placental morphogenesis, nutrient transport and fetal growth in mice. Cell. Mol. Life Sci. 79, 386. https://doi.org/10.1007/s00018-022-04379-y (2022).
Skopková, M. et al. Protein array reveals differentially expressed proteins in subcutaneous adipose tissue in obesity. Obes. (Silver Spring). 15, 2396–2406. https://doi.org/10.1038/oby.2007.285 (2007).
Si, J., Kang, H., You, H. J. & Ko, G. Revisiting the role of Akkermansia muciniphila as a therapeutic bacterium. Gut Microbes. 14, 2078619. https://doi.org/10.1080/19490976.2022.2078619 (2022).
Funding
This study was supported by the National Natural Science Foundation of China (82373594 and 81903340).
Author information
Authors and Affiliations
Contributions
Conceptualization, CY.Y. and YF.X.; methodology, LJ.L.; software, YJ.Q.; data curation, M.L., LJ.L., BY.L., RL.G.; writing—original draft preparation, LJ.L.; writing—review and editing, CY.Y., YS.L; s-pervision, YF.X.; All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
This study was approved by the Second Affiliated Hospital of Xi’an Jiaotong University (No. 2022245).
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.


Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Liu, L., Li, M., Qin, Y. et al. Childhood obesity and insulin resistance is correlated with gut microbiome serum protein: an integrated metagenomic and proteomic analysis. Sci Rep 15, 21436 (2025). https://doi.org/10.1038/s41598-025-07357-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-07357-z