Abstract
Chronic kidney disease (CKD) is a major health concern, with renal interstitial fibrosis (RIF) as a key feature. Effective management of RIF is crucial for treating CKD. Yiqi Juanshen decoction (YQJSD), as traditional Chinese medicine, has shown promising results in CKD treatment. This study evaluates YQJSD’s effectiveness in ameliorating RIF and explores the underlying molecular mechanisms using the unilateral ureteral obstruction (UUO) model. YQJSD has been shown to effectively reduce serum creatinine and blood urea nitrogen levels, decrease extracellular matrix deposition, and down-regulate the expression of α-SMA, COL4α1, Fibronectin (FN). Mechanistically, YQJSD exerts its effects by modulating multiple pathways: it inhibits the NF-κB signaling pathway, inhibiting the expression of pro-inflammatory cytokines like NF-κB1, IL-1β, TNF-α, and CCR1. Simultaneously, YQJSD suppresses the epithelial-mesenchymal transition (EMT) by downregulating the expression of Snail1, Vimentin, Twist1, and FSP1, while increasing E-cadherin expression. Moreover, YQJSD can regulate the PI3K/AKT signaling pathway by decreasing the expression of LOXL2 and PIK3R1, along with p-AKT1/2/3. This modulation of the LOXL2/PI3K/AKT pathway contributes to the inhibition of both EMT and inflammation, highlighting a critical role in the therapeutic intervention against RIF. These findings suggest that YQJSD may serve as a promising therapeutic management of RIF in CKD patients.
Similar content being viewed by others
Introduction
Chronic kidney disease (CKD) has become a major global public health issue, significantly impacting human health1,2,3. CKD is defined as irreversible damage to kidney structure and/or a progressive decline in kidney function. Renal interstitial fibrosis (RIF), a key pathological process common in various forms of CKD, is marked by the infiltration of inflammatory cells, epithelial-mesenchymal transition (EMT), excessive accumulation of the extracellular matrix (ECM), and the loss of tubular structure. These processes collectively lead to renal failure and the alteration of normal kidney architecture4,5. Given the intricate nature of CKD pathogenesis and the limitations of existing therapeutic strategies, there is a pressing need for the development of effective pharmacological interventions aimed at the prevention and treatment of CKD.
Traditional Chinese Medicine (TCM) is increasingly employed in the management of CKD, with its effectiveness in slowing the progression of renal function decline and improving patient outcomes gaining recognition among both clinicians and patients. According to the fundamental principles of TCM, the primary pathogenic mechanisms underlying renal insufficiency include Qi deficiency, blood stasis, and phlegm turbidity. Numerous studies have investigated the mechanisms and therapeutic targets of TCM in the treatment of CKD, highlighting the multi-component and multi-target benefits of TCM formulations compared to single-component pharmaceuticals6,7,8. Notably, TCM’s efficacy in inhibiting inflammatory responses, regulating excessive ECM accumulation, and preventing renal fibrosis is considered a potentially effective strategy for the treatment of CKD9.
Yiqi Juanshen decoction (YQJSD) is an advanced formulation rooted in the principles of TCM, enhanced by extensive clinical practice and the principles of herbal synergy. This formulation consists of six medicinal herbs: Huang Qi (Astragalus membranaceus (Fisch.) Bunge, HQ), Bai Zhu (Atractylodes macrocephala Koidz, BZ), E Zhu (Curcuma phaeocaulis Valeton, EZ), Jiang Huang (Curcuma longa L., JH), Gua Lou Pi (Trichosanthes kirilowii Maxim, GLP), and Hai Zao (Sargassum horneri (Turner) C. Agardh, HZ) (see Table 1). Within this formulation, HQ and BZ are designated as the principal pharmacological agents, whereas EZ and JH serve as supplementary agents, and GLP and HZ function as supportive agents. This therapeutic combination aims to strengthen the spleen, replenish Qi, enhance blood circulation, and eliminate stasis, while also resolving phlegm and dispersing nodules, ultimately achieving a comprehensive regulation of renal function.
The Unilateral Ureteral Obstruction (UUO) mouse model is widely utilized for examining the underlying mechanisms of renal fibrosis and evaluating the effectiveness of anti-fibrotic therapies, due to its rapid induction of RIF that that closely resembles the pathological progression observed in human CKD10,11. Key indicators for assessing fibrosis progression include increased collagen deposition, elevated fibronectin expression, and tubular atrophy following obstruction12.
This study utilized the UUO mouse model to explore the therapeutic effects and underlying mechanisms of YQJSD in the treatment of RIF. The results indicated that YQJSD significantly reduces ECM deposition and alleviates RIF. Transcriptomic analysis suggested that YQJSD may attenuate inflammation and EMT by modulating the LOXL2/PI3K/AKT signaling pathway, thereby contributing to the treatment of RIF. Additionally, UHPLC-QE-MS technology was employed to identify the primary active components of YQJSD. Molecular docking techniques further elucidated the main active components involved in RIF treatment, providing a scientific basis for its therapeutic potential.
Results
YQJSD reduces UUO-induced kidney damage in a dose-dependent manner
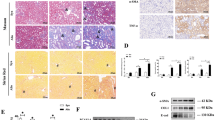
To assess the effects of YQJSD on kidney damage, treatment groups were organized as shown in Fig. 1A. A mouse model of UUO kidney injury was established, and kidney morphology and H&E staining were examined in Sham and UUO. The UUO group exhibited enlarged kidneys, thinned renal parenchyma, significant inflammatory cell infiltration, renal tubular epithelial cell detachment, and tubular atrophy, confirming successful model creation (Fig. 1A). Treatment of the UUO model with varying concentrations of YQJSD showed a dose-dependent improvement in kidney morphology, especially in the high-dose group, where kidney structure and weight ratio were almost normal (Fig. 1B). H&E staining confirmed that YQJSD significantly reduced inflammation, and tubular atrophy compared to the UUO group, indicating a dose-dependent effect. Telmisartan, a strong AT1 receptor antagonist that reduces renal fibrosis in the UUO model, was used as a positive control and improved ureteral obstruction-related changes13. YQJSD’s therapeutic effect was confirmed by significantly lower SCr and BUN levels in all treated groups compared to the UUO group, with the YQJSD-H group showing the greatest improvement, even exceeding telmisartan’s effects (Fig. 1C,D). To evaluate YQJSD’s hepatorenal toxicity, high doses were given to sham group mice, and serum levels of SCr, BUN, ALT, and AST were measured. These remained normal, indicating no hepatorenal toxicity (Fig. 1C–F). Thus, YQJSD effectively reduces renal injury from ureteral obstruction without evident toxicity.
YQJSD reduces UUO-induced kidney damage. (A) Treatment groups and kidney morphological changes were analyzed through appearance and H&E-stained sections (Scale bar = 100 μm, n = 6). Black arrows indicate inflammatory cell infiltration, red arrows show tubular epithelial cell detachment, and yellow arrows highlight tubule atrophy. (B) Kidney/body weight ratio (mg/g) for each mouse group (n = 6). (C–F) Serum biochemical indicators for each mouse group (n = 8) include SCr levels (C), BUN levels (D), AST in Sham and Sham + YQJSD-H groups (E), and ALT in Sham and Sham + YQJSD-H groups (F). Data are shown as mean ± SD. P-values were calculated using One-way ANOVA with Tukey’s test. ###p < 0.001 vs. Sham group. *p < 0.05, **p < 0.01 and ***p < 0.001 vs. UUO group.
YQJSD reduces renal fibrosis in a UUO model
We assessed YQJSD’s effects using Masson’s trichrome staining. Masson’s trichrome staining, which highlights collagen in blue, showed increased collagen and severe RIF in the UUO group. Both the YQJSD and telmisartan groups exhibited a marked reduction in blue areas, suggesting reduced renal interstitial damage and fibrosis, with the YQJSD-H group showing the greatest improvements (Fig. 2A,B). To assess YQJSD’s impact on ECM deposition, we examined COL4α1 and FN expression in kidney tissues using IHC and qPCR. IHC results revealed a significant increase in COL4α1 and FN levels in the UUO group compared to the sham control, while these levels significantly decreased in the YQJSD and telmisartan groups, especially in the YQJSD-H group (Fig. 2A,C,D). qPCR results confirmed a significant reduction in COL4α1 and FN levels in the YQJSD-H group (Fig. 2E,F), indicating that YQJSD treatment, especially at high doses, effectively reduced ECM accumulation.
α-SMA, a key fibrosis marker, were significantly lower in both the YQJSD and telmisartan groups, with the most substantial decrease in the YQJSD-H group (Fig. 2A,G). These results were consistent across qPCR, IHC, and Western blot analyses (Fig. 2H–J).
TGF-β1, crucial in renal fibrosis was assessed. Serum TGF-β1, were significantly higher in the UUO group but decreased with YQJSD treatment, especially in the YQJSD-H group (Fig. 2K), matching the sham control levels. Kidney tissue qPCR results supported this (Fig. 2L). Overall, YQJSD treatment effectively reduces RIF, with the YQJSD-H group showing the best therapeutic effects.
YQJSD alleviates UUO-induced renal fibrosis. (A) Masson staining and immunohistochemistry were used to evaluate renal collagen deposition (scale bar = 100 μm, n = 6) and protein levels of COL4α1, FN, and α-SMA (scale bar = 50 μm, n = 6). (B) Collagen volume fractions were determined via Masson staining (n = 6). (C–D) Protein levels of COL4α1 and FN were quantified (n = 6). (E–F) Relative mRNA levels of COL4α1 and FN in kidney tissues were assessed by qPCR (n = 3). (G) Protein levels of α-SMA were quantified (n = 6). (H) α-SMA protein levels in renal tissues via Western blot (n = 3). (I) Quantification of α-SMA protein signal from (H) (n = 3). (J) Relative α-SMA mRNA levels by qPCR (n = 3). (K) Serum TGF-β1 levels via ELISA (n = 8). (L) Relative TGF-β1 mRNA in renal tissues by qPCR (n = 3). Data are shown as mean ± SD. P-values were calculated using One-way ANOVA with Tukey’s test. ##p < 0.01 and ###p < 0.001 vs. Sham group. *p < 0.05, **p < 0.01 and ***p < 0.001 vs. UUO group.
ECM and immune response modulation by YQJSD in alleviating renal interstitial fibrosis
To clarify how YQJSD alleviates RIF, we performed RNA sequencing on kidney tissue from the sham, UUO, and YQJSD-H groups. Principal component analysis (PCA) showed distinct clustering of samples from each group, confirming consistent gene expression and reliable analysis (Fig. 3A). Comparing the sham and UUO groups revealed 4,437 upregulated and 3,130 downregulated genes in the UUO group (Supplementary Fig. 1A). A comparison between the UUO and YQJSD-H groups revealed 1,132 upregulated and 917 downregulated genes in the YQJSD-H group (Supplementary Fig. 1B). We suggest that genes showing opposite expression patterns between the groups may be key to YQJSD’s effects on renal fibrosis. This analysis identified 360 upregulated and 93 downregulated differentially expressed genes (DEGs) in the UUO group potentially responsive to YQJSD treatment (Fig. 3B). Clustering analysis showed significant group differences (Supplementary Fig. 1C). In the YQJSD-H group, there was a notable decrease in pro-fibrotic cytokines like Serpine1 and α-SMA, as well as ECM-related genes such as FN, COL4α1, and COL4α2 (Fig. 3C), aligning with Fig. 2 data.
Gene Ontology (GO) enrichment analysis of the 360 upregulated and 93 downregulated DEGs in the UUO group in response to YQJSD treatment revealed significant enrichment in ECM and immune response pathways, including collagen metabolic processes, regulation of leukocyte chemotaxis, regulation chemotaxis, B cell receptor signaling pathway, ECM organization, positive regulation of lymphocyte activation, and collagen fibril organization (Fig. 3D). KEGG pathway analysis14 of the 360 upregulated DEGs confirmed significant enrichment of with the ECM-receptor interaction and PI3K/AKT signaling pathway (Fig. 3E), aligning with YQJSD’s role in alleviating RIF, by suppressing the PI3K/AKT signaling pathway to reduce ECM accumulation in renal tissue.
YQJSD attenuates EMT and inflammation to alleviate RIF. (A) PCA among sham, UUO, and YQJSD groups (n = 4). (B) Venn diagram of overlapping DEGs. (C) Heatmap of pro-fibrotic cytokines and ECM-related genes (n = 4). (D) GO enrichment of 360 upregulated and 93 downregulated DEGs related to (B). (E) KEGG (www.kegg.jp/kegg/kegg1.html) pathway enrichment of 360 upregulated DEGs related to (B). (F) Enriched gene sets in UUO vs. Sham by GSEA. (G) EMT and inflammation-related gene sets were downregulated in the YQJSD vs. UUO by GSEA.
YQJSD intervention mitigates renal fibrosis by modulating EMT and inflammation pathways
Research shows that blocking EMT and inflammation benefits renal fibrosis treatment15,16,17. GSEA results indicated that EMT and inflammation-related genes were highly expressed in the UUO group but significantly reduced in the YQJSD group (Fig. 3F–G), suggesting YQJSD effectively inhibits fibrosis-promoting processes.
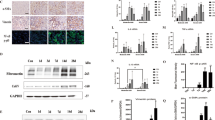
Key markers like Snail1, Twist1, FSP1, and Vimentin are used to evaluate EMT18. Significant transcriptional changes in these markers were observed between the UUO model and YQJSD-treated groups (Supplementary Fig. 1D and Fig. 4A). Immunofluorescence showed increased in Snail1 and α-SMA in the UUO group, which normalized after YQJSD treatment (Fig. 4B). IHC analysis indicated higher Vimentin in the UUO group compared to controls, with reduced levels post-YQJSD treatment (Fig. 4C,D). This finding was substantiated through Western blot analysis (Fig. 4E,F) and qPCR (Fig. 4A). Previous research has indicated that elevated Vimentin expression correlates with a reduction in the epithelial marker E-cadherin19. IHC analysis showed reduced E-cadherin expression in the UUO group, consistent with EMT-associated loss of epithelial characteristics (Fig. 4C,D). Treatment with YQJSD restored E-cadherin expression (Fig. 4C,D), demonstrating its effectiveness in inhibiting EMT and reducing renal fibrosis.
Previous studies highlight the importance of inflammation in kidney injuries20. Supplementary Fig. 1D shows that YQJSD treatment reduces UUO-induced inflammatory gene expression. This study examined YQJSD’s impact on inflammation by measuring interleukin-1β (IL-1β) and tumor necrosis factor-α (TNF-α). Serum ELISA and qPCR results demonstrated that increasing doses of YQJSD led to a dose-dependent reduction in IL-1β and TNF-α levels with increasing YQJSD dosage (Fig. 5A–D), indicating its effectiveness in inhibiting UUO-induced inflammatory cytokines at the molecular level. Additionally, GSEA showed significant downregulation of the NF-κB gene set in the YQJSD group (Data not shown). IHC analysis, Western blotting, and qPCR confirmed that NF-κB1 levels in renal tissues were elevated in the UUO group but suppressed in the YQJSD group (Fig. 5E–I). GO analysis showed that YQJSD treatment highly enriched genes related to leukocyte chemotaxis regulation. We focused on CCR1, crucial for leukocyte recruitment and fibrosis in the UUO mouse model21. Serum protein levels and mRNA expression of CCR1 significantly increased in the UUO group, but YQJSD treatment led to a dose-dependent decrease in CCR1 expression (Fig. 5J–K). The results indicate the YQJSD inhibits the NF-κB signaling pathway.
YQJSD modulates EMT-related genes to alleviate RIF. (A) qPCR measured mRNA levels of Snail1, Vimentin, Twist1, and FSP1 in renal tissues (n = 3 or 4). (B) Immunofluorescence showed Snail1 (green) and α-SMA (red) in each group (Scale bar = 50 μm, n = 6). (C) Immunohistochemistry assessed Vimentin and E-cadherin protein levels (scale bar = 50 μm, n = 6). (D) Quantified protein levels of Vimentin and E-cadherin staining (n = 6). (E) Western blotting determined Vimentin protein levels in renal tissues (n = 3). (F) Quantified Vimentin protein signal from (E) (n = 3). Data are shown as mean ± SD. P-values were calculated using One-way ANOVA with Tukey’s test. #p < 0.05 and ###p < 0.001 vs. Sham group. *p < 0.05, **p < 0.01 and ***p < 0.001 vs. UUO group.
YQJSD modulates inflammation-related genes to alleviate RIF. (A–B) Serum IL-1β and TNF-α levels in mice (n = 8). (C–D) Renal tissues mRNA levels of IL-1β and TNF-α via qPCR (n = 4). (E) Renal mRNA levels of NFKB1 via qPCR (n = 3). (F) NF-κB1 protein levels via IHC (scale bar = 50 μm, n = 6). (G) NF-κB1 protein quantification (n = 6). (H) Renal NF-κB1 levels via Western blot (n = 3). (I) NF-κB1 protein signal quantification from Fig. 5H (n = 3). (J) Serum CCR1 levels in mice (n = 8). (K) Renal mRNA levels of CCR1 via qPCR (n = 3). Data are shown as mean ± SD. P-values were calculated using One-way ANOVA with Tukey’s test. ##p < 0.01 and ###p < 0.001 vs. Sham group. *p < 0.05, **p < 0.01 and ***p < 0.001 vs. UUO group.
YQJSD reduces RIF by targeting the LOXL2/PI3K/AKT signaling pathway
The PI3K/AKT signaling pathway is crucial in renal fibrosis through fibroblast proliferation, apoptosis inhibition, ECM accumulation, and EMT facilitation22,23. Our study found that KEGG analysis of DEGs showed significant enrichment of the PI3K/AKT signaling pathway (Fig. 3E), which GSEA confirmed was upregulated in the UUO model and downregulated after YQJSD treatment (Fig. 6A,B). This suggests YQJSD’s regulatory effect on the pathway, which is linked to the EMT process and NF-κB-mediated inflammation. We assessed PIK3R1 and AKT1 expression changes in each group. The qPCR results demonstrated a nearly 10-fold upregulation of these genes in the UUO group (Fig. 6C). IHC and Western blotting (Fig. 6D–G) showed that YQJSD significantly lowered PIK3R1, phosphorylated AKT1/2/3 (p-AKT1/2/3) levels, suggesting it reduces renal fibrosis by affecting the PI3K/AKT pathways. Lysyl Oxidase-Like 2 (LOXL2), crucial for ECM collagen fiber formation, can activate the PI3K/AKT pathway and promote fibrosis24,25. Transcriptomic analysis revealed increased LOXL2 in the UUO group, which decreased with YQJSD treatment (Data not shown). Further experiments showed that serum LOXL2 levels declined with higher YQJSD doses (Fig. 6H), as confirmed by qPCR and Western blotting results in renal tissue (Fig. 6I–K). This suggests YQJSD reduces renal fibrosis by inhibiting the LOXL2-mediated PI3K/AKT pathway, offering new clinical treatment strategies.
YQJSD regulates LOXL2/PI3K/AKT pathway. (A) PI3K/AKT Enrichment in the UUO group via GSEA. (B) PI3K/AKT gene sets are downregulated in the YQJSD-H group via GSEA. (C) Renal mRNA level of PIK3R1 and AKT1 via qPCR (n = 3). (D) PIK3R1, AKT1/2/3, and p-AKT1/2/3 protein levels via IHC analysis (scale bar = 50 μm, n = 6). (E) Protein levels of PIK3R1, AKT1/2/3, and p-AKT1/2/3 are quantified (n = 6). (F) Renal PIK3R1, AKT1/2/3, and p-AKT1/2/3 protein levels via Western blotting (n = 3). (G) PIK3R1, AKT1/2/3, and p-AKT1/2/3 protein signals are quantified from (F) (n = 3). (H) Serum levels of LOXL2 via ELISA (n = 8). (I) Renal mRNA levels of LOXL2 via qPCR (n = 4). (J) Renal LOXL2 protein levels via Western blotting (n = 3). (K) LOXL2 protein signals are quantified from (J) (n = 3). Data are shown as mean ± SD. P-values were calculated using One-way ANOVA with Tukey’s test. #p < 0.05, ##p < 0.01 and ###p < 0.001 vs. Sham group. *p < 0.05 and ***p < 0.001 vs. UUO group.
Verification of YQJSD components’ docking with AKT1 and PIK3R1
UHPLC-QE-MS was used for chemical characterization, analyzing chromatograms in both ionization modes. We identified 370 active components with a composite score above 0.6 (Supplementary file), mainly terpenoids, flavonoids, phenylpropanoids, and alkaloids. Key components like Kaempferide, Hydroxygenkwanin, Demethoxycurcumin, Apigenin, and 1,7-bis(4-hydroxyphenyl) heptane-3-one, were docked to confirm interactions with AKT1 and PIK3R1. Docking results showed five compounds strongly binding to AKT1 (Fig. 7A–E), with Hydroxygenkwanin, Kaempferide, and Apigenin also binding to PIK3R1 (Fig. 7F–H). All affinity Scores were below − 6, and binding energies were less than − 30 kcal/mol, indicating high affinity of these ligands for target proteins (Table 2). The interaction pattern map revealed stable binding of key targets to active components, suggesting YQJSD’s effective interaction with core targets and supporting its therapeutic potential in RIF treatment.
Discussion
RIF, a key pathological feature in chronic kidney disease, leads to damage and dysfunction in kidney structures due to ECM accumulation, EMT, and ongoing inflammation26,27. Addressing RIF is crucial, making the exploration of effective treatments urgent. This study examines the traditional Chinese medicine formula YQJSD, which includes Astragalus membranaceus (Fisch.) Bunge, Atractylodes macrocephala Koidz, Curcuma phaeocaulis Valeton, Curcuma longa L., Trichosanthes kirilowii Maxim, and Sargassum horneri (Turner) C. Agardh. And we found the main active components in YQJSD detected using UHPLC-QE-MS have antifibrotic properties. For instance, Hydroxygenkwanin promotes E-cadherin expression in MG-63 and U2OS cells while simultaneously inhibiting the N-cadherin and Vimentin expression, thereby reducing EMT occurrence. This suggesting its potential for antifibrotic effects28. Additionally, Demethoxycurcumin is considered as effective ingredient of Curcuma longa L. for treating hepatic fibrosis29, whereas kaempferide, as the main pharmaceutical ingredient of Yougui pills, has been shown to alleviate renal fibrosis via the TGF-β/Smad signaling pathway, demonstrating significant antifibrotic effecacy30. Notably, Apigenin not only alleviates renal fibrosis and improves hyperuricemic nephropathy through the Wnt/β-catenin pathway31 but also improves kidney damage caused by diabetes by inhibiting oxidative stress and fibrosis processes32.
The study shows that the YQJSD effectively reduces RIF, especially in high doses, by inhibiting ECM deposition caused by UUO treatment, including COL4α1 and FN. This was confirmed through Masson’s staining, IHC, and qPCR, indicating YQJSD’s anti-fibrotic effects. Moreover, high-dose YQJSD did not harm liver and kidney function in the Sham mice, highlighting its safety and potential as a new RIF treatment strategy.
TGF-β1 plays a crucial role in RIF progression by promoting fibroblast growth, inducing ECM production, and inhibiting its degradation33,34. This study found that UUO group elevated serum TGF-β1 levels, which were significantly reduced by high-dose YQJSD treatment, aligning with decreased TGF-β1 mRNA in kidney tissues. TGF-β1 also affects chemokine expression like CCR1, impacting inflammation35,36,37,38. Research shows CCR1’s role in kidney diseases, with antagonist BX471 reducing fibrosis in UUO mice39, highlighting CCR1 as a potential therapeutic target21. YQJSD reduces serum CCR1, aligning with transcriptional changes, and effectively inhibiting RIF by regulating TGF-β1 pathways.
In RIF, EMT promotes mesenchymal phenotype and myofibroblast function in renal tubular epithelial cells, accelerating ECM accumulation40. YQJSD reduced the expression of EMT and inflammation-related genes (Snail1, Twist1, FSP-1, IL-1β, and TNF-α) and regulated Vimentin and E-cadherin, suggesting it alleviates fibrosis by inhibiting EMT and inflammation. The fibrosis marker α-SMA was negatively correlated with YQJSD, especially in high doses, highlighting its anti-fibrotic effect.
The PI3K/AKT pathway is crucial in fibrotic, affecting cell survival, apoptosis, migration, and EMT41,42,43. Inhibiting the PI3K/AKT/NF-κB pathway reduces inflammation and renal fibrosis, and Leonurine has been shown to inhibit this pathway, alleviating osteoarthritis44,45. Notably, NF-κB is key in Snail activation and its inhibition reduces EMT17. The study found that YQJSD inhibits PI3K/AKT activity and LOXL2 expression, reducing EMT and inflammation in UUO mice, mitigating RIF, and improving fibrosis outcomes.
Materials and methods
YQJSD preparation
YQJSD comprises six medicinal herbs: Astragalus membranaceus (Fisch.) Bunge, Atractylodes macrocephala Koidz, Curcuma phaeocaulis Valeton, Curcuma longa L., Trichosanthes kirilowii Maxim, and Sargassum horneri (Turner) C. Agardh. This decoction was produced from Sichuan New Green Pharmaceutical Co., LTD., granular preparation, and obtained from the Chongqing Traditional Chinese Medicine Hospital in Chongqing, China. Detailed information about the herbal constituents and their respective proportions is presented in Table 1. To prepare varying concentrations, YQJSD was dissolved in ultrapure water.
Reagents and antibodies
Telmisartan was sourced from Boehringer Ingelheim (C84089, Germany). Analytical Kits for Glutamic Oxaloacetic Transaminase/Aspartate Aminotransferase (GOT/AST, G0424W), Glutamic Pyruvic Transaminase/Alanine Aminotransferase (GPT/ALT, G0423W), serum Creatinine (SCr) (G1204W), and blood urea nitrogen (BUN) (G1201W) were acquired from Grace Biotechnology (Suzhou, China). ELISA kits to detect TGF-β1 (QZ-10402), LOXL2 (QZ-13785), CCR1 (QZ-12234), IL-1β (QZ-10247), and TNF-α (QZ-10225) were purchased from Jiubang Biological Technology (Quanzhou, China). For Western blot analysis, PIK3R1 and COL4α1 antibodies were obtained from Proteintech (60225-1-lg, Wuhan, China) and Novus Biologicals (NB120-6586SS, USA), respectively. Antibodies including α-SMA (ab124964), AKT1/2/3 (ab179463), NF-κB1 (ab32360), Vimentin (ab92547), Fibronectin (FN) (ab45688), and GAPDH (ab181602) were sourced from Abcam (Waltham, MA, USA). Additionally, the p-AKT1/2/3 (Ser473) antibody was acquired from Affinity Biosciences (AF0016, Jiangsu, China). E-cadherin (GB12083) and Snail1 (GB11260) antibodies were procured from Servicebio Technology (Wuhan, China). LOXL2(382092) antibody, secondary antibodies anti-rabbit (511203) and anti-mouse (511103) were obtained from ZENBIO Biotechnology (Chengdu, China).
UHPLC-QE-MS
YQJSD granules equivalent to 85 g of herbal ingredients were thoroughly dissolved into 38.4 ml of ultrapure water. The sample was vortexed for 30 s to make the stock solution with concentration of 2.21 g/mL and centrifuged at 4 ℃, 12,000 rpm for 15 min. A 300 µl upper layer was transferred to an EP tube, and 1000 µl of extraction liquid was added. The mixed liquid was swirled for 30 s, ultrasound in an ice water bath for 5 min, and then the sample was centrifuged for 15 min at 12000 rpm at 4 ℃ after standing at -40 ℃ for 1 h. The supernatant was carefully taken and filtered through a 0.22 μm microporous filter membrane, and 200 µL of each sample was mixed into QC samples, which were then tested on the machine. LC-MS/MS was conducted on a UHPLC system (Thermo Fisher Scientific, USA). The acquisition of MS and MS/MS data was performed utilizing an Orbitrap Exploris 120 mass spectrometer and Xcalibur software (Thermo Fisher Scientific, USA), employing the IDA acquisition mode.
Statement on ethics and compliance
All animal experiments were approved by the Ethics Committee of Chongqing Medical University (no. 2022075, approval date: 27 Jan 2022). All procedures were conducted in strict accordance with the ARRIVE guidelines (https://arriveguidelines.org) and protocols of the office of Research Integrity. Humane endpoints were established, and euthanasia was performed using CO2 inhalation followed by cervical dislocation, in accordance with the AVMA Guidelines for the Euthanasia of Animals.
Animal experiments
Male C57BL/6J mice, aged 6–8 weeks and weighing 20–25 g, were obtained from the Experimental Animal Center of Chongqing Medical University. The mice were housed in a controlled environment at 22 °C with a 12-hour light/dark cycle. A mouse model of RIF was established using a method reported in the literature15. The specific experimental procedures are as follows: The mice were randomly divided into two groups: a sham surgery group (n = 20) and the UUO group (n = 50). For the UUO group, surgery involved making an incision along the left side of the abdominal midline to expose the left kidney and ureter, ligating and cutting the ureter bilaterally with 4 − 0 suture at a third of the distance from the renal end, following by layer-by-layer continuous suturing of the skin. In the sham surgery group, the same surgical steps were performed without ligating or cutting the ureter. The sham surgery group was randomly divided into two subgroups: sham control group (n = 10) and sham + YQJSD-H group (n = 10). The UUO group was randomly divided into five subgroups: UUO model group (n = 10), telmisartan group (n = 10, telmisartan, 3 mg/kg/day), YQJSD-L group (n = 10), YQJSD-M group (n = 10) and YQJSD-H group (n = 10). The next day after surgery, the mice were medicated for two weeks according to their groups, administered by gavage once a day, starting at 9 am. The dosage of YQJSD was determined based on the body surface area ratio of a 70 kg adult and a 20 g mouse, approximately 1:0.0026. The dosage of YQJSD for the mice was divided into three groups: 5.525 g/kg/day (low dose, YQJSD-L), 11.05 g/kg/day (medium dose, YQJSD-M), 22.1 g/kg/day (high dose, YQJSD-H). The sham surgery group and UUO group were given physiological saline by gavage, while the other groups were treated with corresponding drugs by gavage. The gavage volume was proportional to the body weight of the mice. After two weeks treatment, the mice were euthanized, and kidney tissues and serum were collected for subsequent analysis.
RNA sequencing
Kidney tissues were harvested from the sham group, UUO group, and YQJSD-H group for RNA sequencing (RNA-seq). RNA-seq was performed by Meiji Biomedical Technology Co., LTD (Shanghai, China) using the Illumina platform. Differentially expressed genes (DEGs), identified with a false discovery rate (FDR) less than 0.05, were clustered and analyzed using the Gene Ontology (GO) database (http://www.geneontology.org/) and the Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg/). Subsequently, Gene Set Enrichment Analysis (GSEA) was conducted, accessible at http://software.broadinstitute.org/gsea/index.jsp. All sequencing data are available through the NCBI Sequence Read Archive under the accession number PRJNA1201334.
Measurement of serum biochemical indicators
Blood samples collected from all groups were centrifuged at 3500 rpm for 5 min to prepare serum. Subsequently, serum levels of ALT, AST, SCr, and BUN were quantitatively determined using GPT/ALT, GOT/AST, SCr, and BUN analytical kits, respectively, according to the manufacturer’s instructions.
Histological and immunohistochemistry (IHC) analysis
Kidney tissue samples were fixed in 4% paraformaldehyde, followed by paraffin embedding and sectioning. The sections were stained with hematoxylin and eosin (H&E) staining and Masson’s trichrome to observe overall structural changes in the renal tissues using an Olympus BX53 microscope (Olympus, Tokyo, Japan).
For IHC analysis, the sections underwent antigen retrieval followed by incubation in 3% hydrogen peroxide for 25 min to block endogenous peroxidases. After washing with PBS at room temperature, the sections were blocked with 3% bovine serum albumin for 30 min. They were then incubated with primary and corresponding secondary antibodies. Observation of the sections was also performed using the Olympus BX53 microscope.
For the analysis of Masson’s trichrome staining and IHC staining, five positive areas were randomly selected from each slide, and the positive stained regions were quantitatively analyzed using Image-Pro Plus 6.0 (Media Cybernetics, Bethesda, MD, USA).
Double immunofluorescence staining
The co-localization of Snail1 and a-SMA was examined by double immunofluorescence staining. The slices were dewaxed, soaked in EDTA antigen repair buffer to repair antigens, circled, and hermetically sealed with a mixture of 3% hydrogen peroxide and 3% BSA. Following that, the slices were left overnight with anti-Snail1, added Alexa Fluor 488-conjugated goat anti-rabbit IgG. Subsequently, Cy3-TSA was applied for a 10 min incubation. The slices were then rinsed, subjected to antigen restoration once again, and subsequently incubated with primary antibodies, followed by the addition of goat anti-rabbit antibodies and staining with DAPI. Finally, fluorescence microscopy (eclipse CI; Nikon, Tokyo, Japan) was used to observe the slices.
ELISA
Quantitative determination of serum levels of TGF-β1, TNF-α, IL-1β, CCR1, and LOXL2 was performed using ELISA kits. The procedures were carried out according to the manufacturer’s instructions. Absorbance was measured at 450 nm, and the concentrations of the proteins were calculated based on the standard curves.
RNA extraction and real-time quantitative PCR (qPCR) analysis
RNA from kidney tissues was extracted using a modified Trizol method. RNA concentration and purity were measured with NanoDrop ND-2000. cDNA was synthesized using the PrimeScript RT Reagent Kit with gDNA Eraser (RR047Q, Takara). Gene expression levels were quantified by qPCR using TB Green Premix Ex Taq Kit (RR420Q, Takara) on a CFX96 detection system (Bio-Rad). To ensure data reliability, each sample was set up with three technical replicates, and gene expression normalization was performed using GAPDH. Relative gene expression levels were calculated using the 2-ΔΔCT method. The specific primer sequences used are listed in Table 3.
Western blot analysis
Western blotting was employed to analyze the relative protein levels in renal tissue. Proteins were extracted using RIPA lysis buffer and subsequently separated by electrophoresis, then transferred onto PVDF membranes. Membranes were incubated with primary antibodies including α-SMA (1:10,000), ΝF-κΒ1 (1:10,000), PIK3R1 (1:5000), AKT1/2/3 (1:10,000), phosphorylated-AKT1/2/3 (p-AKT1/2/3) (1:1000), LOXL2 (1:1000), Vimentin (1:1000), and GAPDH (1:10,000). Following incubation, membranes were washed with TBST and incubated with secondary antibodies at 20–25 °C for 1 h. Proteins were detected using a hypersensitive luminescence solution and imaged with an imaging system (Odyssey FC; Licor Biosciences, Lincoln, USA). Band gray was calculated and quantified relative to GAPDH using ImageJ software (National Institutes of Health, Bethesda, MD, USA).
Molecular docking analysis
Molecular docking was conducted to elucidate interactions between key components of YQJSD and principal genes involved in RIF. Select components from YQJSD, including Kaempferide, Hydroxygenkwanin, Demethoxycurcumin, Apigenin, and 1,7-bis(4-hydroxyphenyl) heptan-3-one, were docked with key targets AKT1 and PIK3R1 to validate the accuracy of the predicted indications. Crystal structures of the targets were obtained from the RCSB Protein Data Bank (https://www.rcsb.org/). Using Schrödinger’s Maestro 13.5, the crystal structures were processed for docking: they were dehydrated, hydrogen atoms were added, and existing ligands were removed using the Protein Preparation Wizard. The LigPrep module was employed to process 2D structure files of the compounds, generating all possible 3D stereoisomers of the ligands. Binding sites on the protein surface were identified using the SiteMap module. Molecular docking was then executed with the prepared ligands and defined targets using high-precision XP docking. The docking poses were evaluated by XP GScore; a score less than − 6 suggests higher stability of the compound-protein complex. Binding free energies were calculated using the MM-GBSA method, with values lower than − 30 kcal/mol indicating particularly strong and stable binding.
Statistical analysis
Statistical analysis was conducted using GraphPad Prism 9.0.0 software (GraphPad Prism Inc., La Jolla, CA, USA). To determine significant differences between the groups, a one-way ANOVA followed by Tukey’s post-hoc test was employed. p < 0.05 was considered statistically significant.
Conclusions
This study found that YQJSD can improve kidney function and reduce RIF in UUO mice. Using transcriptomics and network pharmacology, it was suggested that YQJSD works by inhibiting the LOXL2/PI3K/AKT pathway, which suppresses EMT and inflammation. Thus, YQJSD could be a promising treatment for CKD by targeting RIF (Fig. 8).
Data availability
All data supporting the finding of this paper are present in the article and/or supplementary information files. The further queries are available from the corresponding author on request.
References
Chen, T. K., Knicely, D. H. & Grams, M. E. Chronic kidney disease diagnosis and management: a review. JAMA 322, 1294–1304. https://doi.org/10.1001/jama.2019.14745 (2019).
Collaboration, G. B. D. C. K. D. Global, regional, and national burden of chronic kidney disease, 1990–2017: a systematic analysis for the global burden of Disease Study 2017. Lancet 395, 709–733. https://doi.org/10.1016/S0140-6736(20)30045-3 (2020).
Kalantar-Zadeh, K., Jafar, T. H., Nitsch, D., Neuen, B. L. & Perkovic, V. Chronic kidney disease. Lancet 398, 786–802. https://doi.org/10.1016/S0140-6736(21)00519-5 (2021).
Panizo, S. et al. Fibrosis in chronic kidney disease: Pathogenesis and consequences. Int. J. Mol. Sci. 22. https://doi.org/10.3390/ijms22010408 (2021).
Liu, Y. Cellular and molecular mechanisms of renal fibrosis. Nat. Rev. Nephrol. 7, 684–696. https://doi.org/10.1038/nrneph.2011.149 (2011).
Yuan, H., Wu, X., Wang, X. & Yuan, C. Chinese herbal decoction astragalus and angelica exerts its therapeutic effect on renal interstitial fibrosis through the inhibition of MAPK, PI3K-Akt and TNF signaling pathways. Genes Dis. 9, 510–521. https://doi.org/10.1016/j.gendis.2020.06.001 (2022).
Li, R. et al. Fufang Shenhua tablet inhibits renal fibrosis by inhibiting PI3K/AKT. Phytomedicine 116, 154873. https://doi.org/10.1016/j.phymed.2023.154873 (2023).
Zheng, L. et al. Distinct responses of gut microbiota to Jian-Pi-Yi-Shen decoction are associated with improved clinical outcomes in 5/6 nephrectomized rats. Front. Pharmacol. 11, 604. https://doi.org/10.3389/fphar.2020.00604 (2020).
Liu, X. Y., Zhang, X. B., Zhao, Y. F., Qu, K. & Yu, X. Y. Research progress of Chinese herbal medicine intervention in renal interstitial fibrosis. Front. Pharmacol. 13, 900491. https://doi.org/10.3389/fphar.2022.900491 (2022).
Aranda-Rivera, A. K., Cruz-Gregorio, A., Aparicio-Trejo, O. E., Ortega-Lozano, A. J. & Pedraza-Chaverri, J. Redox signaling pathways in unilateral ureteral obstruction (UUO)-induced renal fibrosis. Free Radic. Biol. Med. 172, 65–81. https://doi.org/10.1016/j.freeradbiomed.2021.05.034 (2021).
Geng, X. Q. et al. Ganoderic acid hinders renal fibrosis via suppressing the TGF-beta/Smad and MAPK signaling pathways. Acta Pharmacol. Sin. 41, 670–677. https://doi.org/10.1038/s41401-019-0324-7 (2020).
Chevalier, R. L., Forbes, M. S. & Thornhill, B. A. Ureteral obstruction as a model of renal interstitial fibrosis and obstructive nephropathy. Kidney Int. 75, 1145–1152. https://doi.org/10.1038/ki.2009.86 (2009).
Sugiyama, H. et al. Telmisartan inhibits both oxidative stress and renal fibrosis after unilateral ureteral obstruction in acatalasemic mice. Nephrol. Dial. Transpl. 20, 2670–2680. https://doi.org/10.1093/ndt/gfi045 (2005).
Kanehisa, M., Furumichi, M., Tanabe, M., Sato, Y. & Morishima, K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 45, D353–D361. https://doi.org/10.1093/nar/gkw1092 (2017).
Bai, Y. et al. Ruxolitinib alleviates renal interstitial fibrosis in UUO mice. Int. J. Biol. Sci. 16, 194–203. https://doi.org/10.7150/ijbs.39024 (2020).
Li, R. et al. Salidroside ameliorates renal interstitial fibrosis by inhibiting the TLR4/NF-kappaB and MAPK signaling pathways. Int. J. Mol. Sci. 20. https://doi.org/10.3390/ijms20051103 (2019).
Grande, M. T. et al. Snail1-induced partial epithelial-to-mesenchymal transition drives renal fibrosis in mice and can be targeted to reverse established disease. Nat. Med. 21, 989–997. https://doi.org/10.1038/nm.3901 (2015).
Stone, R. C. et al. Epithelial-mesenchymal transition in tissue repair and fibrosis. Cell. Tissue Res. 365, 495–506. https://doi.org/10.1007/s00441-016-2464-0 (2016).
Mendez, M. G., Kojima, S. & Goldman, R. D. Vimentin induces changes in cell shape, motility, and adhesion during the epithelial to mesenchymal transition. FASEB J. 24, 1838–1851. https://doi.org/10.1096/fj.09-151639 (2010).
Jiang, G. T., Chen, X., Li, D., An, H. X. & Jiao, J. D. Ulinastatin attenuates renal interstitial inflammation and inhibits fibrosis progression in rats under unilateral ureteral obstruction. Mol. Med. Rep. 10, 1501–1508. https://doi.org/10.3892/mmr.2014.2323 (2014).
Eis, V. et al. Chemokine receptor CCR1 but not CCR5 mediates leukocyte recruitment and subsequent renal fibrosis after unilateral ureteral obstruction. J. Am. Soc. Nephrol. 15, 337–347. https://doi.org/10.1097/01.asn.0000111246.87175.32 (2004).
Xu, W., Yang, Z. & Lu, N. A new role for the PI3K/Akt signaling pathway in the epithelial-mesenchymal transition. Cell. Adhes. Migr. 9, 317–324. https://doi.org/10.1080/19336918.2015.1016686 (2015).
Wang, H. et al. The role of PI3K/Akt signaling pathway in chronic kidney disease. Int. Urol. Nephrol. https://doi.org/10.1007/s11255-024-03989-8 (2024).
Grau-Bove, X., Ruiz-Trillo, I. & Rodriguez-Pascual, F. Origin and evolution of lysyl oxidases. Sci. Rep. 5, 10568. https://doi.org/10.1038/srep10568 (2015).
Erasmus, M. et al. Linking LOXL2 to cardiac interstitial fibrosis. Int. J. Mol. Sci. 21. https://doi.org/10.3390/ijms21165913 (2020).
Webster, A. C., Nagler, E. V., Morton, R. L. & Masson, P. Chronic kidney disease. Lancet 389, 1238–1252. https://doi.org/10.1016/S0140-6736(16)32064-5 (2017).
Ha, S., Chung, K. W., Lee, J., Chung, H. Y. & Moon, H. R. Renal tubular PAR2 promotes interstitial fibrosis by increasing inflammatory responses and EMT process. Arch. Pharm. Res. 45, 159–173. https://doi.org/10.1007/s12272-022-01375-5 (2022).
Dong, X., Wang, Y., Zhuang, H. & An, G. Hydroxygenkwanin suppresses proliferation, invasion and migration of osteosarcoma cells via the miR–320a/SOX9 axis. Mol. Med. Rep. 26. https://doi.org/10.3892/mmr.2022.12815 (2022).
Han, Q., Zhu, J. & Zhang, P. Mechanisms of main components in Curcuma longa L. on hepatic fibrosis based on network pharmacology and molecular docking: a review. Med. (Baltim). 102, e34353. https://doi.org/10.1097/MD.0000000000034353 (2023).
Wang, L. et al. You-gui pill ameliorates renal tubulointerstitial fibrosis via inhibition of TGF-beta/Smad signaling pathway. J. Ethnopharmacol. 169, 229–238. https://doi.org/10.1016/j.jep.2015.04.037 (2015).
Li, Y. et al. Apigenin ameliorates hyperuricemic nephropathy by inhibiting URAT1 and GLUT9 and relieving renal fibrosis via the Wnt/beta-catenin pathway. Phytomedicine 87, 153585. https://doi.org/10.1016/j.phymed.2021.153585 (2021).
Malik, S. et al. Apigenin ameliorates streptozotocin-induced diabetic nephropathy in rats via MAPK-NF-kappaB-TNF-alpha and TGF-beta1-MAPK-fibronectin pathways. Am. J. Physiol. Ren. Physiol. 313, F414–F422. https://doi.org/10.1152/ajprenal.00393.2016 (2017).
Qi, W., Chen, X., Poronnik, P. & Pollock, C. A. Transforming growth factor-beta/connective tissue growth factor axis in the kidney. Int. J. Biochem. Cell. Biol. 40, 9–13. https://doi.org/10.1016/j.biocel.2007.01.006 (2008).
Meng, X. M., Nikolic-Paterson, D. J. & Lan, H. Y. TGF-beta: the master regulator of fibrosis. Nat. Rev. Nephrol. 12, 325–338. https://doi.org/10.1038/nrneph.2016.48 (2016).
Furuichi, K. et al. Distinct expression of CCR1 and CCR5 in glomerular and interstitial lesions of human glomerular diseases. Am. J. Nephrol. 20, 291–299. https://doi.org/10.1159/000013603 (2000).
Segerer, S., Nelson, P. J. & Schlondorff, D. Chemokines, chemokine receptors, and renal disease: from basic science to pathophysiologic and therapeutic studies. J. Am. Soc. Nephrol. 11, 152–176. https://doi.org/10.1681/ASN.V111152 (2000).
Anders, H. J., Vielhauer, V. & Schlondorff, D. Chemokines and chemokine receptors are involved in the resolution or progression of renal disease. Kidney Int. 63, 401–415. https://doi.org/10.1046/j.1523-1755.2003.00750.x (2003).
Han, Y., Wang, J., Zhou, Z. & Ransohoff, R. M. TGFbeta1 selectively up-regulates CCR1 expression in primary murine astrocytes. Glia 30, 1–10 (2000).
Anders, H. J. et al. A chemokine receptor CCR-1 antagonist reduces renal fibrosis after unilateral ureter ligation. J. Clin. Invest. 109, 251–259. https://doi.org/10.1172/JCI14040 (2002).
LeBleu, V. S. et al. Origin and function of myofibroblasts in kidney fibrosis. Nat. Med. 19, 1047–1053. https://doi.org/10.1038/nm.3218 (2013).
Hsu, H. S. et al. Involvement of ER stress, PI3K/AKT activation, and lung fibroblast proliferation in bleomycin-induced pulmonary fibrosis. Sci. Rep. 7, 14272. https://doi.org/10.1038/s41598-017-14612-5 (2017).
Du, Y. M. et al. Effect of bradykinin on rats with thromboangiitis obliterans through PI3K/Akt signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 23, 10169–10176. https://doi.org/10.26355/eurrev_201911_19587 (2019).
Zhu, J. F. et al. Annexin A1-suppressed autophagy promotes nasopharyngeal carcinoma cell invasion and metastasis by PI3K/AKT signaling activation. Cell. Death Dis. 9, 1154. https://doi.org/10.1038/s41419-018-1204-7 (2018).
Lei, L. et al. Wogonin alleviates kidney tubular epithelial injury in diabetic nephropathy by inhibiting PI3K/Akt/NF-kappaB signaling pathways. Drug Des. Devel Ther. 15, 3131–3150. https://doi.org/10.2147/DDDT.S310882 (2021).
Hu, Z. C. et al. Inhibition of PI3K/Akt/NF-kappaB signaling with leonurine for ameliorating the progression of osteoarthritis: in vitro and in vivo studies. J. Cell. Physiol. 234, 6940–6950. https://doi.org/10.1002/jcp.27437 (2019).
Acknowledgements
The authors thank the foundations for the financial support.
Funding
This work was supported by grants from the National Natural Science Foundation of China (Grant Nos.: 82100131, 81573860), Natural Science Foundation Project of Chongqing (Grant No.: CSTB2023NSCQ-MSX0136).
Author information
Authors and Affiliations
Contributions
Kaiyue Tan: methodology, validation, formal analysis, visualization, writing-original draft preparation. Jingwei Deng: software, validation. Yi Liu: methodology. Yudi Zhang: methodology, supervision, data curation. Yu Xiong: software, validation. Su Yuan: resources, data curation. Jun Liu: data curation. Zhiwei Chen: formal analysis, writing-review and editing. Yuanyuan Liu: Project administration, writing-review and editing, funding acquisition. Wenfu Cao: project administration, supervision, writing-review and editing, funding acquisition. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Tan, K., Deng, J., Liu, Y. et al. Yiqi Juanshen decoction alleviates renal interstitial fibrosis by targeting the LOXL2/PI3K/AKT pathway to suppress EMT and inflammation. Sci Rep 15, 4248 (2025). https://doi.org/10.1038/s41598-025-86622-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-86622-7