Abstract
In the field of clinical neurology, automated detection of epileptic seizures based on electroencephalogram (EEG) signals has the potential to significantly accelerate the diagnosis of epilepsy. This rapid and accurate diagnosis enables doctors to provide timely and effective treatment for patients, significantly reducing the frequency of future epileptic seizures and the risk of related complications, which is crucial for safeguarding patients’ long-term health and quality of life. Presently, deep learning techniques, particularly Convolutional Neural Networks (CNNs) and Long Short-Term Memory networks (LSTMs), have demonstrated remarkable accuracy improvements across various domains. Consequently, researchers have utilized these methodologies in studies focused on recognizing epileptic signals through EEG analysis. However, current models based on CNN and LSTM still heavily rely on data preprocessing and feature extraction steps. Additionally, CNNs exhibit limitations in perceiving global dependencies, while LSTMs encounter challenges such as gradient vanishing in long sequences. This paper introduced an innovative EEG recognition model, that is the Spatio-temporal feature fusion epilepsy EEG recognition model with dual attention mechanism (STFFDA). STFFDA is comprised of a multi-channel framework that directly interprets epileptic states from raw EEG signals, thereby eliminating the need for extensive data preprocessing and feature extraction. Notably, our method demonstrates impressive accuracy results, achieving 95.18% and 77.65% on single-validation tests conducted on the datasets of CHB-MIT and Bonn University, respectively. Additionally, in the 10-fold cross-validation tests, their accuracy rates were 92.42% and 67.24%, respectively. In summary, it is demonstrated that the seizure detection method STFFD based on EEG signals has significant potential in accelerating diagnosis and improving patient prognosis, especially since it can achieve high accuracy rates without extensive data preprocessing or feature extraction.
Similar content being viewed by others
Introduction
Motivation
Epilepsy, a prevalent chronic neurological condition, results from abnormal discharge reactions in brain neurons1,2. Characteristically, during seizures, individuals with epilepsy experience a temporary loss of consciousness due to transient brain dysfunction, accompanied by either local or generalized convulsions, and possible fainting episodes3,4. Globally, approximately 65 million individuals are afflicted with epilepsy, with an estimated 2 million new cases diagnosed annually5. While drug therapy remains the primary means of managing epileptic seizures in most cases, recurrent seizures can inflict irreversible damage to the patient’s brain, profoundly impacting their physical and psychological well-being, and imposing a significant burden on their families. Therefore, there is an urgent need to detect epileptic seizures and provide scientific treatment guidance6 to mitigate their harmful impacts on patients and their families.
Although antiepileptic drugs and surgical interventions can alleviate symptoms to a certain extent, these treatment modalities remain ineffective in approximately 30% of epilepsy cases. Consequently, the establishment of a reliable epileptic seizure prediction system holds significant potential in mitigating seizure-related concerns. Early prediction of epileptic seizures ensures sufficient time for therapeutic intervention prior to their occurrence, thereby facilitating prompt treatment or prevention of seizures through the administration of appropriate medication.
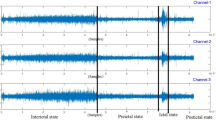
The EEG serves as a vital diagnostic tool in epilepsy, as it captures the voltage fluctuations of neurons within the brain, providing crucial information into brain disorders, including epilepsy. Based on EEG recordings, the brain activity of epilepsy patients is typically categorized into four distinct states: prior to seizure onset, during seizure episodes, the interictal state between seizures, and the state following seizure episodes.
Generally, neurologists diagnose epilepsy by meticulously analyzing abnormal shapes and amplitudes of EEG waves. However, relying solely on neurologists for epilepsy diagnosis through visual observation entails several drawbacks. Firstly, visual inspection is highly dependent on expert experience and professional knowledge, introducing potential subjectivity. Secondly, the frequent acquisition of brainwave signals and the subsequent accumulation of vast amounts of data necessitate prolonged visual observation7. Conversely, prolonged visual inspection can potentially impact the accuracy of clinical diagnosis and may give rise to dire consequences, including misdiagnosis and missed diagnosis. Moreover, the process of EEG acquisition is prone to strong noise generated by the surrounding electrical environment, thus increasing the difficulty and challenges associated with visual detection. Consequently, diagnosing epilepsy through visual analysis of EEG signals is not only time-consuming but also subjective, placing a significant burden on neuroscientific experts. Therefore, developing a reliable model capable of automatically detecting epileptic seizures from EEG signals holds immense practical significance for both research and clinical applications8.
Related work
In recent years, numerous methods have been developed to automatically predict epileptic seizures. The majority of these methods rely on the analysis of EEG, which is the electrical recording of brain activity and is widely regarded as the most reliable diagnostic and analytical tool for epilepsy due to its exceptional temporal resolution9. Some commonly used machine learning-based algorithms for classifying epileptic seizures utilizing EEG data include Gaussian Mixture Model (GMM), Artificial Neural Networks (ANN), Support Vector Machines (SVM), Random Forest (RF), Least Squares SVM (LS-SVM), Decision Trees (DT), K-Nearest Neighbors (KNN), and Naive Bayes, etc. Al Ghayab et al.10 proposed a novel algorithm that integrates frequency ___domain information with the information gain (InfoGain) technique to detect epileptic seizures from EEG data. Firstly, the EEG signals are decomposed utilizing the Fast Fourier Transform (FFT) and Discrete Wavelet Transform (DWT). Subsequently, the extracted features are ranked using InfoGain, and the most significant features are selected. Finally, the EEG signals are classified using a LS-SVM classifier. Tao et al.11 employed Local Mean Decomposition (LMD) to decompose the original EEG signal into a sequence of Product Functions (PFs). Each PF feature was then fed into five different classifiers: Back Propagation Neural Network (BPNN), KNN, Linear Discriminant Analysis (LDA), SVM, and SVM optimized by Genetic Algorithm (GA-SVM) for classification purposes16. On the other hand, Büyükçakır et al.12 developed a method based on Hilbert Vibration Decomposition (HVD) to predict EEG recordings of 10 patients from the CHB-MIT database. The 18-channel EEG signals were decomposed into 7 sub-components using the sliding window HVD method. These sub-components from all channels were subsequently utilized to calculate features that were then fed into the Multilayer Perceptron (MLP) classifier.
Although machine learning-based seizure detection methods can achieve high accuracy, there are still numerous shortcomings. Firstly, manual selection of optimal features and classifiers is typically required. Secondly, feature extraction can be a time-consuming process, potentially reducing the efficiency of the model in practical applications. However, with the advent of deep learning, numerous seizure detection methods utilizing this technology have been proposed. Deep learning excels at accurately learning patterns through multi-layer neural network architectures, efficiently processing vast amounts of data. It has become an indispensable tool in the field of EEG signal processing, facilitating automatic feature extraction and classification, thereby enhancing seizure prediction capabilities and surpassing the performance achieved by traditional machine learning techniques. Truong et al.13 introduced a CNN model for feature extraction and classification, aiming to distinguish between pre-seizure and interictal segments. The preprocessing of 30-second EEG windows involved the use of the short-time Fourier transform (STFT) to extract frequency and time ___domain data. Khan et al.14 employed the DWT to transform EEG signals from a time-based representation to a time-frequency-based one. They subsequently integrated this with a CNN for feature extraction and differentiation between pre-seizure, seizure, and interictal states. This method was evaluated on 15 patients from the CHB-MIT dataset, achieving promising prediction results. Qiu et al.15 implemented an attention mechanism combined with a ResNet-LSTM network (DARLNet) for detecting epileptic seizures. The model uses ResNet and LSTM, which are inclined to capture temporal and spatial dependencies, and then employs a channel attention module to focus on relevant seizure information. Additionally, experiments were conducted using the Bonn EEG dataset to validate the results. Ji et al.16 proposed an end-to-end seizure prediction model by combining Graph Attention Networks (GAT) and Temporal Convolutional Networks (TCN). The EEG signals, after low-pass filtering, are input into the GAT module for spatial feature extraction, and then the TCN captures temporal features, allowing the end-to-end model to acquire spatio-temporal correlations from multi-channel EEG. The model was evaluated on the CHB-MIT database and achieved good results.
Contribution
Despite significant advancements in seizure detection, existing solutions still face limitations. CNN based approaches excel at extracting local features but often struggle to capture the global dependencies inherent in time series data. Conversely, neural network models designed to extract sequence features, such as RNNs and LSTMs, are adept at capturing long-term dependencies in time series data. However, their performance in capturing local spatial information remains suboptimal. To address these challenges, this paper introduces a novel deep learning model known as Space-time feature fusion with dual attention (STFFDA) for the classification of epileptic seizures in EEG signals. By integrating dual attention mechanisms and fusion techniques, the proposed model aims to enhance both local and global feature extraction, leading to improved seizure detection accuracy.
The main contributions of this paper are as follows: (1) The model directly uses EEG time series as input without requiring any time-frequency ___domain transformation for feature extraction. This simplifies the entire process and allows for timely seizure prediction when integrated into EEG measurement devices. (2) The use of Bidirectional Long Short-Term Memory (Bi-LSTM) and CNN enables feature extraction from both spatial and temporal dimensions. Specifically, the CNN module extracts spatial features from EEG signals, capturing channel correlations, while the Bi-LSTM module captures temporal dynamics and long-term dependencies. This fusion of spatial and temporal features enhances the model’s ability to understand EEG variations, improving classification accuracy. (3) The combination of one-dimensional Squeeze-and-Excitation Network (SENet) attention mechanism and dot-product attention mechanism significantly improves the model’s prediction accuracy. The former learns the relative importance of different channel features, while the latter learns the importance of spatio-temporal features. (4) The application of this model to two different datasets, namely the CHB-MIT and the University of Bonn, verifies its effectiveness across different seizure patterns and recording conditions, demonstrating its robust generalization performance.
Methodology
The structure of STFFDA model
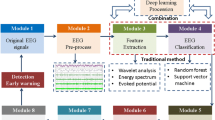
The epilepsy EEG recognition model, designated as STFFDA, comprises a CNN module, a one-dimensional Squeeze-and-Excitation (1D SE) module, a Bi-LSTM module, and an Attention module. Figure 1 delineates the architecture of the STFFDA model in detail.
STFFDA model structure. The model consists of four main modules: the CNN module is used to extract the spatial features of EEG signals, the Bi-LSTM module captures the temporal dynamics of the signals, the 1D SE module highlights important features through channel weighting, and the attention module integrates and weights both spatial and temporal features to enhance the significance of key features. Finally, the fully connected layer classifies the epileptic signals. With this architecture, the STFFDA model effectively addresses the dependency on preprocessing in traditional methods and significantly improves classification accuracy.
Figure 1 depicts the structure of the STFFDA model, which encompasses two primary modules. The CNN module is tasked with extracting spatial features, whereas the Bi-LSTM module focuses on extracting temporal features. The CNN module encodes the spatial characteristics of EEG signals by incorporating feature vectors from each EEG electrode channel. It comprises convolutional layers, each equipped with 32 kernels and 1 × 3 filters. These layers employ numerous convolutional kernels to sequentially filter the EEG signals of each channel, effectively capturing the sequential features within the EEG signals. The 1D SE module functions as a modular channel attention component, enhancing the channel features of the input feature map. Notably, the final output feature map size of the SE module remains consistent with the input feature map size, ensuring seamless integration within the model. The Bi-LSTM module is specifically designed to capture temporal dependencies within the EEG signals. This capability enables the model to learn intricate time-related correlations, thereby providing valuable insights into the dynamic nature of brain activity. The feature outputs from both modules are then flattened and concatenated. These combined features are processed through an attention mechanism, which assigns differential weights to the features. Ultimately, the fully connected layer produces the final output based on these weighted features.
Convolutional neural network
A CNN is a subtype of deep learning architecture that can effectively process one-dimensional, two-dimensional, or even three-dimensional data. CNNs utilize a combination of convolutional operations and nonlinear activation functions to extract meaningful features17. The fundamental components of a CNN typically include convolutional layers, pooling layers, and activation functions, among others.
The convolution operation is actually a linear operation used for feature extraction starting with the convolution kernel. Convolution kernel is a small weight matrix, which slides over the input data, performing matrix multiplication on the current input elements, and then aggregates the results into a single output pixel. Essentially, the calculation process of convolution is similar to fully connected layers as it involves linear combinations of neurons and non-linear transformations. But in convolution, the linear combination is performed on specific neurons at certain positions. Applying different convolution kernels to the input data can generate multiple feature maps. The pooling layer is responsible for downsizing the input image, reducing pixel information, and retaining essential details to decrease computational load, which mainly includes max pooling and average pooling. The activation layer, also known as the non-linear layer, uses non-linear activation functions like Tanh, Sigmoid, ReLU, respectively shown as Eq. (1)-Eq. (3). These functions determine how neurons should react to the input to produce outputs.
Bi-directional long short term memory network
The EEG signals reflect a vast amount of information regarding neuronal activity within the brain and exhibit high temporal sensitivity. This temporal characteristic enables EEG signals to capture the dynamic changes in brain activity, providing crucial clues for the diagnosis and treatment of brain disorders. However, the non-stationarity, nonlinearity, individual variability, and complexity of pathological conditions associated with EEG signals pose multiple challenges in practice for traditional EEG time feature extraction methods. The features extracted by deep neural networks (DNNs) and CNNs through hidden layer neurons are solely influenced by the current signal. Recurrent neural networks (RNNs) represent a specialized neural network architecture primarily used for processing sequential data, exhibiting features such as recurrent connections and historical information processing. These networks are extensively employed in natural language processing, speech recognition, time series analysis, and other domains. The hidden layer neurons in RNNs possess self-feedback loops, where their input is not solely derived from the speech signal at the current moment but also receives memory information from the hidden layer of the previous moment. This architecture mimics the human brain’s ability to predict speech information for the next moment based on historical speech data18. Nevertheless, during the training of long sequences, RNNs typically rely on backpropagation and gradient descent algorithms, which can lead to issues such as vanishing and exploding gradients.
LSTM was jointly proposed by Hochreiter and Schmidhuber in 199719, effectively addressing the issues of gradient vanishing and explosion, thereby making it particularly suitable for analyzing time series data. The schematic diagram of the LSTM unit structure is presented in Fig. 2. The LSTM classifier comprises four main components: the memory cell, the input gate, the forget gate, and the output gate. The memory cell is responsible for storing values across any given time interval, while the three gates regulate the flow of information entering and exiting the LSTM unit.
LSTM structure. The basic structure of a single LSTM unit includes three key components: the Forget Gate, the Input Gate, and the Output Gate. These gating mechanisms enable the LSTM to retain relevant information over long sequences, preventing gradient vanishing or explosion issues. In the STFFDA model, the bidirectional LSTM module leverages this mechanism to incorporate both forward and backward temporal information, enhancing the ability to model the dynamic changes in EEG signals.
The traditional LSTM20,21 is a unidirectional RNN that is limited to processing information in a strictly sequential manner, typically overlooking potential future contextual cues. Bi-LSTM is an improved version based on LSTM, initially proposed jointly by Graves and Schmidhuber in 200522. Bi-LSTM extends the traditional LSTM network, consisting of two layers of LSTM, one receiving inputs from the forward time sequence and the other from the backward time sequence, ensuring that the model simultaneously covers information transmission from past and future moments, thereby enhancing the model’s predictive ability. Bi-LSTM combines the advantages of Bi-RNN and LSTM, effectively increasing the amount of information available to the network, while considering both forward and backward dependencies, enabling the network to learn bidirectional dependencies of time sequences. This type of deep network can be used for speech recognition, text classification, and classification models. Furthermore, compared to unidirectional networks, the Bi-LSTM deep network can remember more information, providing better learning outcomes, especially when dealing with a large amount of data23.
In the proposed STFFDA model, the forward and backward parameters are independent of each other, but they share the feature vectors of the EEG sequence. At each time step, the forward and backward LSTM units respectively compute their hidden vectors, denoted as fht and bht, and then concatenate them to form the final hidden vector of the Bi-LSTM model. The output ht is given by the following Eq. (4):
illustrates the basic structure of the Bi-LSTM model, where {x1, x2,., xn} represents feature vectors, n is the number of time steps, {fh1, fh2,., fhn} and {bh1,bh2,.,bhn} represent forward hidden vectors and backward hidden vectors respectively, hn denotes the vector formed by concatenating fhn and bhn24.
Figure 3. Bi-LSTM structure. It consists of two LSTM layers: the forward layer processes the time sequence in the normal order, while the backward layer processes the sequence in reverse. By concatenating the outputs of both layers, the Bi-LSTM can learn the global temporal dependencies in EEG signals from the past to the future. This structure significantly enhances the extraction of temporal features in the STFFDA model.
Squeeze-and-Excitation(SE)-Block
Attention mechanism is a technique used for data processing, which is one of the fundamental abilities of human perception and decision-making. In the field of cognitive science, individuals selectively focus on some data while ignoring other visible information, thus effectively utilizing limited resources for visual information processing. By assigning weights to different parts of the data, the attention mechanism autonomously selects and focuses on the key information in the input data. Attention mechanisms have a wide range of applications in enhancing the performance of deep neural networks25.
The 2D data SE attention mechanism network proposed by Hu et al.26 won first place in the ILSVRC 2017 classification task. The full name of the SE attention mechanism is Squeeze-and-Excitation (SE) attention mechanism. Its main idea is to compress and excite the input features to improve the model’s performance. In Squeeze step, the input feature maps are compressed into a single vector through global average pooling operation, and then mapped to a smaller vector through a fully connected layer. In Excitation step, each element in this vector is compressed to between 0 and 1 using a sigmoid function, then multiplied with its original input feature map to obtain the weighted feature map.The SE attention mechanism is shown in Fig. 4.
The specific process is as follows, taking 2D SE as an example. In the first step, the feature map U is globally average pooled through Squeeze(Fsq(·)), generating a 1 × 1×C vector, where each channel is represented by a single value. In the second step, two fully connected layers are employed to generate the desired weight information using the learned weight W, indicating the feature relevance. The vector z obtained from the previous step is processed through two fully connected layers, W1 and W2, to obtain the desired channel weight values s. After passing through these layers, different values in s represent different channel weight information, assigning different weights to each channel. zc can be represented as Eq. (5).
The third step involves assigning weights from the second step’s weight vector s to the feature map U. s can be represented as Eq. (6).
By multiplying the generated feature vector s (1 × 1×C) with the feature map U (H×W×C) channel-wise, i.e., each of the H×W values in each channel of the feature map U is multiplied by the corresponding weight in s, the desired feature map \(\mathop X\limits^{\sim }\) can be obtained, which has the same dimensions as the feature map U. The SE module does not alter the size of the feature map. Through this method, the model can dynamically learn the importance of each channel, thereby enhancing its performance. The value of \(\mathop X\limits^{\sim }\) is given by Eq. (7).
SE attention mechanism. The module first compresses the input feature map using global average pooling (Squeeze) to generate a compact channel descriptor vector. Then, it applies two fully connected layers with a Sigmoid activation function to weight the features (Excitation), highlighting important feature channels. Finally, the channel weights are multiplied with the original feature map to obtain the weighted feature map. In the STFFDA model, this module enhances the recognition of key features.
Dot-product attention mechanism
When segmenting and extracting features from input signals, it is possible to extract some redundant information due to the suddenness of epileptic seizures and inaccurate positioning of seizure markers. Therefore, not all feature vectors at each time step contribute equally, making it difficult to identify which part is more important for seizure detection. To address this issue, this paper introduces a dot-product attention mechanism at the end of the model, which enhances the effectiveness of useful information by adaptively assigning different weights and suppressing the effects of irrelevant and redundant information.
In the final stage of the STFFDA model, spatial and temporal features extracted by the CNN and Bi-LSTM modules are processed into hidden vectors qt. To evaluate the importance of each feature at different time steps, the model employs a multilayer perceptron (MLP) to perform a nonlinear mapping of the hidden vectors, generating new hidden representations ut. Subsequently, these representations are normalized using a Softmax function to produce a probability vector ∂t, which represents the weights of the features at each time step. These weights directly reflect the contribution of the features to the classification task.The model then uses the attention weights ∂t to perform a weighted summation of the hidden vectors qt, resulting in the final output vector s. The calculation formulas are shown in Eqs. (8)–(10).
Among them, Ww is the attention weight matrix, and bw is the bias matrix.
This process ensures that the model focuses on the most critical features for epilepsy seizure detection while ignoring irrelevant ones. During training, the parameters of the attention mechanism, including weight matrices and biases, are optimized to enable the model to dynamically learn the importance of features at each time step, adapting to the complexity of different EEG signals.
Materials and experimental setup
Dataset
The EEG signals used in this work are from the CHB-MIT database27,28. The database was obtained from 22 patients and recorded at Boston Children’s Hospital. Recordings were made in pediatric patients with epilepsy (5 males, ages 3–22 years; 17 females, ages 1.5–19 years) over several days of monitoring after discontinuation of antiepileptic drug therapy to describe their seizures and determine their suitability for surgery. EEG signals were recorded using the international 10–20 electrode placement system from 23 channels (FP1-F7, F7-T7, T7-P7, P7-O1, FP1-F3, F3-C3, C3-P3, P3-O1, FP2-F4, F4-C4, C4-P4, P4-O2, FP2-F8, F8-T8, T8-P8, P8-O2, FZ-CZ, CZ-PZ) for seizure detection without any filtering or preprocessing, with a sampling rate of 256 and a resolution of 16 bits. Each channel’s recording duration is 1 h, so each channel contains 256 samples/s × 3600 s = 921,600 samples. In the raw, unprocessed form, the input shape of the data is (42139, 23, 921600), Among them, the number of samples refers to the total number of samples in the dataset, which varies depending on the patients’ data selected for the experiment.
Another EEG database used in this study is available from the University of Bonn29. It consists of five datasets A, B, C, D and E. The dataset from the University of Bonn comprises five sets labeled A to E. Each sequence contains 100 single-channel EEG signal recordings, totaling 23.6s. Noise caused by various muscle movements in the EEG recordings has been eliminated. Group A(with eyes open) and Group B(with eyes close) recorded scalp EEGs from 5 healthy subjects. Group C, D and E were intracranial EEG recordings from 5 patients with definite epilepsy. Group C includes EEG records from the hippocampus of the lateral hemisphere of the brain in the interictal period of the patients. Group D includes EEG recordings of seizure-free areas during the interictal period in patients with epilepsy. Group E includes the EEG recording collected from epileptic patients during the epileptic seizure. EEG recordings are a blend of scalp and intracranial recordings. The EEG data consists of 11,500 samples, sampled at a frequency of 173.61 Hz, and comprises 178 channels. The Bonn University EEG database consists of 5 categories x 100 files x 4097 data points (23.6 s).
Table 1 Summary of CHB-MIT epilepsy EEG dataset. *The initial seizure is excluded due to its occurrence within the first hour, limiting the availability of sufficient pre-seizure time. **The two seizures are merged when the second seizure occurs within the interval following the first one30.
Data preprocessing
In the CHB-MIT database, the start and end times of seizures in EEG signal recordings are marked based on expert judgment, and the frequency and duration of seizures vary from person to person. However, to ensure the reliability of the experiments, the researchers addressed the issue of data imbalance by training patient-specific models using an equal number of interictal and preictal samples31. The specific information for the CHB-MIT database is as follows: the data for CHB01 and CHB21 were collected from the same patient, with a 1.5-year gap between collections. This study selected a larger group of subjects using 23 channels to collect EEG signals, categorizing these signals into three types. The first type is normal EEG, the second type is EEG during seizures, and the third type is EEG signals recorded before seizures. These different types of EEG play a crucial role in the prevention and treatment of seizures. In this study, EEG signals of each category were extracted twice for each patient, with each extraction lasting 5 s. Data that did not meet the 5-second time requirement for ictal and interictal periods were excluded from the statistics.
The Bonn University EEG signal dataset is saved based on sampling points, where one sampling point represents one sample. Similarly, the CHB-MIT database is also divided based on sampling points. All these datasets were collected together, resulting in over 40,000 sampling data points. It is worth noting that no filtering or preprocessing was applied to these recordings prior to analysis32. The Bonn University EEG database consists of 5 categories x 100 files x 4097 data points (23.6 s). EEG signals are divided based on sampling points, with one sampling point representing one sample.
In this study, the time window for the pre-seizure phase is not specifically defined, but rather processed based on the 1 Hz sampling frequency of the dataset, where each data point represents the EEG signal at a given time. The duration of the pre-seizure phase is selected based on the integrity of the dataset and the time sequence, so there are no clear boundaries. Instead, it is processed according to the actual time distribution of the data and the relative timing of the seizures. This approach enhances the flexibility of the data and avoids biases that may arise from manually setting a time window. Normal data is obtained by selecting the EEG signals from the patient during the interictal period, which refers to the time intervals when the patient does not experience a seizure. These data are collected during the periods between seizures. Therefore, normal data is not based on a fixed time window, but rather on the actual interictal data.
Experimental setup
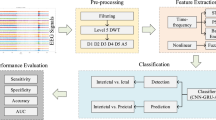
To fully validate the performance of the model, a single validation test and a 10-fold cross-validation test were performed on each dataset. The single validation test divides each dataset into a 90% training set and a 10% test set. However, due to the random division of the dataset, the results of the single validation test may not be sufficient to indicate the performance of the model. Therefore, this paper also adopted 10-fold cross-validation, which first randomly divides the dataset into 10 equally sized and disjoint subsets. In each round, 9 subsets are randomly selected as the training set, and the remaining 1 subset is used as the test set. After each round, 9 subsets are randomly selected again as the training set for training. Through multiple rounds of calculation, the results of 10 test sets are obtained, and the average value of the evaluation index is used to assess the performance of the model.The specific parameters of the model are shown in Table 2.
The duration of epileptic seizures is relatively short, which may lead to an imbalance in the dataset. To address this issue in the experiment, we used the Weighted Cross-Entropy Loss. By assigning higher weights to the minority class (epileptic seizures), the model can pay more attention to these less frequent seizure samples, avoiding bias towards the majority class (e.g., normal state), thus improving the model’s performance in seizure detection.
Evaluation indexes
This study primarily employs deep learning models for decoding EEG signals, thus specific diagnostic statistical metrics were used to assess the performance of the model. The confusion matrix is mainly composed of true positive(TP), false negative(FN), false negative(FP) and true positive(TN).
Based on the confusion matrix, the accuracy, precision, recall, F1-score, and Matthews correlation coefficient (MCC) of the test results are described, with evaluation metrics formulas shown in (11)-(15).
In this study, the evaluation metrics are averaged using the macro-average method. Macro-average is a commonly used performance evaluation method in multi-class classification tasks. It calculates the evaluation metrics (such as precision, recall, F1 score, etc.) for each class and then averages them to obtain an overall score. Unlike micro-average, macro-average independently computes the metrics for each class and averages them, without being affected by sample size differences. This makes it a more fair way to assess the model’s performance in cases of class imbalance, as each class has equal weight regardless of its sample size.
Experimental results and analysis
Single test result of STFFDA model
This study aims to examine the performance of the proposed STFFDA model in identifying epileptic signals in EEG data, and to demonstrate its effectiveness through comparisons with several existing deep learning and traditional machine learning algorithms. It comprehensively assesses the performance of STFFDA model. Specifically, DNN, CNN, convolutional neural network-recurrent neural network (CNN-RNN) and inception v1 using a one-dimensional convolution kernel (1D Inception v1) were selected as representatives of deep learning algorithms for comparative experiments. In addition to deep learning methods, this study also considers several traditional machine learning algorithms, including the Bayesian classifier (Bayes), DT, KNN, and SVM.
In the experiment, we used Decision Tree, k-NN, and Bayesian models for comparison. The main parameters of the Decision Tree model include criterion, which determines the splitting criterion; it defaults to ‘gini’, using Gini impurity to measure the quality of splits. Splitter determines the strategy for feature selection and defaults to ‘best’, meaning the best feature is chosen for splitting. Max_depth limits the maximum depth of the tree, and defaults to None, meaning there is no limit on the depth. Min_samples_split specifies the minimum number of samples required to split an internal node, with a default value of 2. Min_samples_leaf indicates the minimum number of samples required in a leaf node, with a default value of 1. Max_features defines the maximum number of features to consider when splitting a node, and defaults to None, meaning all features are considered. For the k-NN model, the main parameters include n_neighbors, which determines the number of neighbors used for classification, with a default value of 5. Weights controls the weight of the neighbors, with the default set to ‘uniform’, meaning all neighbors are weighted equally. Algorithm determines the algorithm used to compute neighbors, with the default set to ‘auto’. Metric defines the distance metric, and it defaults to ‘minkowski’, using the Minkowski distance. p is the parameter for the Minkowski distance, with a default value of 2, which corresponds to the Euclidean distance.The main parameters of the SVM include the C parameter, which controls the regularization strength, with a default value of 1.0. A larger value of C results in a smaller margin but fewer classification errors, potentially leading to overfitting, while a smaller C allows more errors, which may lead to underfitting. The kernel parameter determines the type of kernel function used, with the default being ‘rbf’ (Radial Basis Function). Other parameters include shrinking, which enables a heuristic shrinking strategy to accelerate the optimization process, with the default value set to True. The probability parameter is set to False by default, but when set to True, it allows for probability estimation and enables the ‘predict_proba()’ method for probability predictions.
Although traditional machine learning algorithms have been widely used in various fields, these methods generally require manual feature extraction from raw data, which is both time-consuming and may affect the accuracy of the final results due to the limitations of ___domain knowledge. In contrast, deep learning methods have unique advantages in processing high-dimensional and large-scale data by automatically learning hierarchical feature representations of the data, enabling them to directly extract more complex and abstract information from raw data.
The STFFDA model was extensively validated and tested on two epilepsy datasets: the CHB-MIT and Bonn University datasets, as evident from Tables 3 and 4. Specifically, the CHB-MIT dataset encompasses three distinct labels: normal EEG signals, pre-seizure EEG signals, and seizure EEG signals. Conversely, the Bonn University dataset comprises five diverse types of EEG signals. The CHB-MIT dataset primarily focuses on predicting seizure signals, whereas the Bonn University dataset mainly aims at the diagnosis and classification of EEG signals.
As seen in Table 3, the STFFDA model proposed in this paper outperforms the other models, achieving an accuracy of 95.18%, precision of 95.16%, recall of 95.16%, F1 score of 95.16%, and MCC of 92.77%. Closely trailing the STFFDA is the KNN model, with an accuracy of 92.85%, precision of 92.94%, recall of 92.87%, F1 score of 92.84%, and MCC of 89.34%. Bayes, on the other hand, exhibits the poorest performance, with an accuracy of 46.67%, precision of 47.44%, recall of 46.55%, F1 score of 46.50%, and MCC of 21.47%.
In the five-classification task on the Bonn University dataset, the performance of the STFFDA model is also the best, with an accuracy of 77.65%, precision of 77.52%, recall of 77.67%, an F1 score of 77.47%, and MCC of 72.13%. The results of the STFFDA model and other models in both the three and five-classification tasks indicate that the proposed STFFDA model is superior to other models. This is because the parallel structure of the STFFDA model can simultaneously extract the spatio-temporal features of the input signal, thereby improving the accuracy of the model.
Ten-fold cross validation result of STFFDA model
To ensure the reliability of model testing and reduce random errors, this study also employed a 10-fold cross-validation method to assess the performance of each model. Specifically, the performance of the STFFDA model is presented in Figs. 5 and 6.
Tables 5 and 6 summarize the average results of the 10-fold cross-validation. As can be seen from Table 5, on the CHB-MIT dataset, for the three-class classification task, the STFFDA model proposed in this paper demonstrates remarkable performance. Specifically, it achieved an accuracy of 92.42%, precision of 92.47%, recall of 92.42%, an F1 score of 92.42%, and a Matthews Correlation Coefficient (MCC) of 88.66%. Under the same 10-fold cross-validation conditions, the KNN model ranked second, with an accuracy of 91.44%, precision of 91.75%, recall of 91.45%, an F1 score of 91.46%, and an MCC of 87.29%. As shown in Table 6, on the Bonn dataset, for the five-class classification task, the STFFDA model achieved an accuracy of 67.24%, precision of 67.24%, recall of 67.11%, an F1 score of 67.13%, and an MCC of 58.95%. However, on the five-classification task, 1D Inception-v1 ranks second only to the STFFDA model, with an accuracy of 64.78%, precision of 65.25%, recall of 64.82%, F1 score of 64.41%, and MCC of 56.29%. Therefore, combining the results from Tables 3, 4, 5 and 6, it can be concluded that compared to other comparative models, the STFFDA model exhibits excellent performance in epileptic seizure recognition classification tasks using EEG signals, both in single-validation and 10-fold cross-validation, thus verifying the stability and reliability of the STFFDA model.
Ten-fold cross validation result of STFFDA model
To validate the proposed STFFDA model, this paper conducted ablation experiments using ten-fold cross-validation on two datasets: the Bonn University dataset and the CHB-MIT dataset, with the results shown in Tables 7 and 8. The spatial feature module achieved an Accuracy of 91.50%, Precision of 91.51%, Recall of 91.51%, F1-score of 91.50%, and MCC of 87.26% on the Bonn University epilepsy dataset. The temporal feature module had an Accuracy of 89.01%, Precision of 89.02%, Recall of 89.02%, F1-score of 89.01%, and MCC of 83.53% on the Bonn University epilepsy dataset. Similarly, the spatial feature module on the CHB-MIT epilepsy dataset achieved an Accuracy of 65.46%, Precision of 65.60%, Recall of 65.52%, F1-score of 65.38%, and MCC of 56.90%. The accuracy of the temporal feature module on the CHB-MIT epilepsy dataset is 64.15%, precision is 64.35%, recall is 64.18%, F1-score is 64.14%, and MCC is 55.22%. It can be seen that the classification performance of the spatial feature module and the temporal feature module shows the same trend across both datasets, with the spatial feature module performing slightly better than the temporal feature module in both datasets. However, the STFFDA model proposed in this paper is still the best among all the models compared in the ablation experiments, further demonstrating the effectiveness of the proposed method.
When comparing the performance of the model on the CHB-MIT dataset and the Bonn University dataset, we found that the STFFDA model achieved higher accuracy on the CHB-MIT dataset. This difference may be attributed to several factors. The CHB-MIT dataset contains more samples and includes data from different patients with various types of epileptic seizures, providing more diverse training data, which helps the model extract more robust features. In contrast, the Bonn University dataset is relatively smaller, with fewer categories, and the task is more complex, adding additional challenges for the model. The EEG signal quality in the CHB-MIT dataset is higher, reducing noise and artifacts, whereas the Bonn University dataset may be more affected by noise, leading to a decline in model performance. Additionally, the CHB-MIT dataset mainly involves a three-class classification task, which is relatively simpler, while the Bonn University dataset involves a five-class classification task, further increasing the complexity of the model.
Conclusion
This paper proposes a spatio-temporal feature fusion model for epileptic EEG recognition based on a dual attention mechanism—STFFDA. The STFFDA model aims to enhance the accuracy and reliability of detecting epileptic seizures from EEG signals. Through extensive experiments and analyses, we demonstrate the superiority of our model compared to other existing methods. First, the fusion of spatio-temporal features allows the CNN module in the STFFDA model to capture spatial features of the EEG signals and the weights of the channels, while the Bi-LSTM module captures the temporal dynamics of the EEG signals. This comprehensive feature representation significantly enhances the model’s ability to distinguish between epileptic seizures and normal EEG patterns, with the CNN and Bi-LSTM modules adapting to different EEG channel data. Compared to traditional detection methods based on time or frequency features, spatiotemporal feature extraction can more comprehensively describe the seizure patterns in EEG signals. Especially when dealing with complex or noisy signals, spatiotemporal feature extraction can effectively reduce noise interference and improve the model’s sensitivity to real epileptic seizures. Secondly, the introduction of the dual attention mechanism further improves the performance of the STFFDA model. The SE attention module focuses on identifying the EEG channels with the most informative content, while the attention mechanism module after feature fusion emphasizes recognizing important spatio-temporal features in the EEG data, assigning greater weights to these significant features. The combination of these two attention mechanism modules enables better extraction of EEG signal features. Finally, the experimental results on the CHB-MIT and Bonn datasets demonstrate the effectiveness of the STFFDA model. Our model achieves state-of-the-art performance in accuracy, precision, recall, F1 score, and MCC, outperforming other comparative models in both single validation and 10-fold cross-validation experiments. The method was evaluated on two types of epilepsy datasets from CHB-MIT and Bonn University, achieving classification accuracies of 92.42% and 67.24%, respectively. Finally, ablation experiments further validated the superiority of the spatio-temporal feature fusion method and confirmed the effectiveness of the dual attention mechanism.
In summary, the STFFDA model proposed in this paper offers a promising solution for identifying epilepsy seizures from EEG signals. The integration of spatio-temporal features with a dual attention mechanism enhances the model’s classification accuracy and reliability. Future work could consider using datasets with larger sample sizes to further validate the robustness and stability of the method. Additionally, this study primarily deals with a single type of epileptic seizure, and the seizure types in the dataset are relatively simple. In reality, there are various types of epileptic seizures, including focal and generalized seizures. Future research could expand to the detection of multiple types of epileptic seizures. To handle different types of seizures, further optimization of the spatiotemporal feature extraction approach may be needed, or more refined classifiers may need to be designed to distinguish between different seizure types.
Data availability
Data Availability Statement: This study is an experimental analysis of a publicly available data set. The data can be found in this web page: https://physionet.org/content/chbmit/1.0.0/ andhttps://www.ukbonn.de/epileptologie/arbeitsgruppen/ag-lehnertz-neurophysik/downloads/.
References
Gao, X. et al. Automatic detection of epileptic seizure based on approximate entropy, recurrence quantification analysis and convolutional neural networks. Artif. Intell. Med. 102, 101711 (2020).
Gabeff, V. et al. Interpreting deep learning models for epileptic seizure detection on EEG signals. Artif. Intell. Med. 117, 102084 (2021).
Fisher, R. S. et al. Epileptic seizures and epilepsy: definitions proposed by the International League against Epilepsy (ILAE) and the International Bureau for Epilepsy (IBE). Epilepsia 46 (4), 470–472 (2005).
MacAllister, W. S. & Schaffer, S. G. Neuropsychological deficits in childhood epilepsy syndromes. Neuropsychol. Rev. 17, 427–444 (2007).
He, H., Liu, X. & Hao, Y. A progressive deep wavelet cascade classification model for epilepsy detection. Artif. Intell. Med. 118, 102117 (2021).
Shayegh, F. et al. A brief survey of computational models of normal and epileptic eeg signals: a guideline to model-based seizure prediction. J. Med. Signals Sens. 1 (1), 62–72 (2011).
Acharya, U. R. et al. Application of entropies for automated diagnosis of epilepsy using EEG signals: a review. Knowl. Based Syst. 88, 85–96 (2015).
Janjarasjitt, S. Epileptic seizure classifications of single-channel scalp EEG data using wavelet-based features and SVM. Med. Biol. Eng. Comput. 55 (10), 1743–1761 (2017).
Assali, I. et al. CNN-based classification of epileptic states for seizure prediction using combined temporal and spectral features. Biomed. Signal Process. Control 82, 104519 (2023).
Al Ghayab, H. R. et al. Epileptic seizures detection in EEGs blending frequency ___domain with information gain technique. Soft. Comput. 23, 227–239 (2019).
Zhang, T. & Chen, W. LMD based features for the automatic seizure detection of EEG signals using SVM. IEEE Trans. Neural Syst. Rehabil. Eng. 25 (8), 1100–1108 (2016).
Büyükçakır, B., Elmaz, F. & Mutlu, A. Y. Hilbert vibration decomposition-based epileptic seizure prediction with neural network. Comput. Biol. Med. 119, 103665 (2020).
Truong, N. D. et al. A generalised seizure prediction with convolutional neural networks for intracranial and scalp electroencephalogram data analysis. Preprint at arXiv:1707.01976 (2017).
Khan, H. et al. Focal onset seizure prediction using convolutional networks. IEEE Trans. Biomed. Eng. 65 (9), 2109–2118 (2017).
Qiu, X., Yan, F. & Liu, H. A difference attention ResNet-LSTM network for epileptic seizure detection using EEG signal. Biomed. Signal Process. Control 83, 104652 (2023).
Ji, D. et al. Epileptic seizure prediction using Spatiotemporal Feature Fusion on EEG. Int. J. Neural Syst. (2024).
Indolia, S., Goswami, A. K., Mishra, S. P. & Asopa, P. Conceptual understanding of convolutional neural network – a deep learning approach. Int. Conf. Comput. Intell. Data Sci. 132, 679–688 (2018).
Li, X. et al. A mild cognitive impairment diagnostic model based on IAAFT and BiLSTM. Biomed. Signal Process. Control 80, 104349 (2023).
Hochreiter Sepp, and Jürgen Schmidhuber. Long short-term memory. Neural Comput. 9(8), 1735–1780 (1997).
Kong, W. et al. Short-term residential load forecasting based on LSTM recurrent neural network. IEEE Trans. Smart grid 10(1), 841–851 (2017).
Elessawy, R. H., Eldawlatly, S. & Hazem, M. Abbas. A long short-term memory autoencoder approach for EEG motor imagery classification. 2020 international conference on computation, automation and knowledge management (ICCAKM). (IEEE, 2020).
Graves, A. & Jürgen Schmidhuber. and. Framewise phoneme classification with bidirectional LSTM and other neural network architectures. Neural Netw. 18(5-6), 602–610 (2005).
Göker, H. Automatic detection of migraine disease from EEG signals using bidirectional long-short term memory deep learning model. Signal. Image Video Process. 17 (4), 1255–1263 (2023).
Tang, Y. et al. Epileptic seizure detection based on path signature and Bi-LSTM Network with attention mechanism. IEEE Trans. Neural Syst. Rehabil. Eng. (2024).
Li, X. et al. A Novel Hybrid YOLO Approach for Precise Paper defect detection with a dual-layer template and an attention mechanism. IEEE Sens. J. (2024).
Hu, J., Shen, L. & Sun, G. Squeeze-and-excitation networks. In: Proc. IEEE conference on computer vision and pattern recognition. 7132–7141. (2018).
Shoeb, A. H. & Guttag, J. V. Application of machine learning to epileptic seizure detection. In: Proc. 27th international conference on machine learning (ICML-10). 975–982. (2010).
Shoeb, A. et al. Patient-specific seizure onset detection. Epilepsy Behav. 5 (4), 483–498 (2004).
Andrzejak, R. G. et al. Indications of nonlinear deterministic and finite-dimensional structures in time series of brain electrical activity: dependence on recording region and brain state. Phys. Rev. E 64, 061907 (2001).
Zhang, J. et al. A scheme combining feature fusion and hybrid deep learning models for epileptic seizure detection and prediction. Sci. Rep. 14 (1), 16916 (2024).
Quadri, Z. F., Akhoon, M. S. & Loan, S. A. Epileptic Seizure Prediction Using Stacked CNN-BiLSTM: A Novel Approach. (IEEE Transactions on Artificial Intelligence, 2024).
Abdulwahhab, A. H. et al. Detection of epileptic seizure using EEG signals analysis based on deep learning techniques. Chaos Solitons Fractals 181, 114700 (2024).
Acknowledgements
This work is partially supported by the General Projects of Shaanxi Science and Technology Plan(No.2023-JC-YB-504), the National Natural Science Foundation of China (No. 62172338), the Xijing University special talent research fund (No.XJ17T03) and the Natural Science Foundation of Chongqing CSTC(No. CSTB2022NSCQ-MSX1581).
Author information
Authors and Affiliations
Contributions
Z.H. ,Y.M. ,Y.Y.and L.H. contributed to conceptualization, investigation, analysis and interpretation, and writing—original draft preparation. Q.D.,J.S, H.S, J.S. and S.Z, contributed to editing, reviewing, and validation.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Huang, Z., Yang, Y., Ma, Y. et al. EEG detection and recognition model for epilepsy based on dual attention mechanism. Sci Rep 15, 9404 (2025). https://doi.org/10.1038/s41598-025-90315-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-90315-6