Abstract
Despite of having rich reptilian fauna, the wild lizards from Pakistan remained unexplored for the presence of blood borne parasites. The present study was designed to report the molecular epidemiology and phylogenetic evaluation of various blood borne pathogens (Hepatozoon sp., Schellackia spp., Toxoplasma gondii, Plasmodium spp., Haemoproteus spp. and Leucocytozoon spp.) in blood samples of wild lizards (N = 84), trapped during March 2022 till June 2023 from district Karak and Dir in Khyber Pakhtunkhwa (Pakistan). Seven lizard species were identified during present study. Our results revealed that 11 out of 84 (13%) lizards were Haemogregarines infected. Infected lizards included Laudakia (L.) tuberculata (1/4, 25% prevalence), L. pakistanica (3/15, 20%) and L. agrorensis (7/53, 13%). DNA sequencing and BLAST analysis confirmed the presence of Hepatozoon sp. and Lankesterella sp. While the lizards were negative for the remaining screened pathogens. Phylogenetic analysis of both pathogens revealed genetic diversity among the Pakistani isolates and they clustered with isolates detected in reptiles, birds and rodents from different countries. For L. tuberculata, Haemogregarines prevalence significantly varied between the sample collection sites. In conclusion, this is the first report from Pakistan documenting a relatively higher Haemogregarines infection rate in wild Pakistani lizards. Further, comprehensive and large-scale studies must be conducted in unexplored geo-climatic regions of Pakistan to report the actual prevalence of Haemogregarines among the wild lizards as well as in other wildlife species. These findings will add to our knowledge regarding the genetic diversity and the interactions of these parasites with their hosts that will lead towards parasite control.
Similar content being viewed by others
Introduction
Pakistan is blessed to have rich reptilian fauna that has been represented by eight families including Chamaeleonidae, Varanidae, Scincidae, Lacertidae, Gekkonidae, Uromastycidae, Eublepharidae and Agamidae1. These reptiles have occupied a variety of habitats including wetlands, forests as well as deserts and they are widely considered as bio-indicators of the environmental quality2. Reptilian populations are globally declining due to number of factors including ultraviolet radiations, climate change and habitat destruction, chemical pollutants such as pesticides and fertilizers and pathogens3. Globally, ticks are among the most common ectoparasites but their prevalence is especially higher in tropical and sub-tropical parts of the World owing to the higher temperature and humidity in these areas that are optimal for tick growth and reproduction. Ticks are known to infest lizards and hence they are involved in the transmission of pathogens to lizards and can also carry infectious agents from lizards to the new hosts4.
Parasites have a negative impact on their hosts as they can disturb the physiology as well as reduce the reproductive success of the host5. Among the protozoan blood parasites, comprehensive studies had been carried out targeting the suborder Haemosporina (which includes genera Plasmodium, Haemoproteus and Leucocytozoon)6. Hepatozoon species belongs to family Hepatozoidae and they are the apicomplexan Protozoans that target the red and white blood cells of their diverse hosts including reptiles, amphibians and mammals5. Hepatozoon species have also been detected in a number of invertebrate species that mostly act as vectors and are involved in the transmission of these parasites from invertebrates to vertebrates and also from one vertebrate species to another7. Hepatozoon species are commonly transmitted via ingestion of an infected invertebrate or intermediate prey resulting in the liberation of developmental stages in the endothelial cells, hepatocytes and other visceral organs in a wide variety of vertebrate hosts8. Family Lankesterellidae are also apicomplexan protozoan that infect a variety of amphibians, reptiles and birds but they are relatively less explored as they are rarely found infecting domestic animals or humans9.
Despite the high prevalence and diversity of haemoprotozoan in lizards, limited information is available in literature regarding their interactions with hosts and the ecological roles they play10. Traditionally, haemosporidians and Hepatozoon species were used to be characterized based on their microscopic examination which cannot be conclusive as several species exhibits similar morphology11. The introduction of molecular tools has made it possible for researchers to effectively detect and differentiate haemosporidians and Hepatozoon species in lizards as well as in other reptiles6,7,9,12,13. A couple of studies has documenting the reptilian diversity in Pakistan along with the ecto-parasites infecting these reptiles4,14 but these animals have not been systematically investigated for the presence of blood borne pathogens leaving a significant knowledge gap. Hence, the present study was designed to report the prevalence of Hepatozoon sp., Toxoplasma gondii, Plasmodium spp., Haemoproteus spp., Leucocytozoon spp. and Schellackia spp. in seven wild lizard species that were trapped from various regions in district Karak and Dir of Khyber Pakhtunkhwa province in Pakistan.
Materials and methods
Study areas and subjects
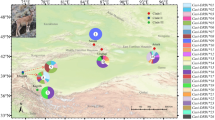
An active epidemiological survey was conducted to report the presence of blood-borne parasites in the blood samples of wild lizards that were collected from various sampling sites in district Karak and Dir in Khyber Pakhtunkhwa (KPK) province of Pakistan. The sampling sites included Chera Galai, Sheringal, Munda, Biyar, Usheri, Palosa Sar, Lardoag, Katair, Mayar, Samar Bagh and Shahi (Fig. 1). The study is reported in accordance with ARRIVE guidelines (https://arriveguidelines.org).
Map of Pakistan with Khyber Pakhtunkhwa (KPK) province highlighted in blue. In the map of KPK, the two sampling districts are shown green. While in the magnified map of district Dir Upper and and Kohat, the sampling sites are marked from where the wild lizard’s samples were collected during present study. Map was created by using the QGIS software (version 3.22.8-Białowieża https://qgis.org/). The entire shape file having the administrative, provincial and district bounders were downloaded from the website DIVA-GIS, under the tab free spatial Data.
Data and blood collection
A total of 84 wild lizards (38 male and 46 females) were trapped from March 2022 till June 2023 by using glue traps with baits following Ribeiro-Júnior et al.15. Standard taxonomic key was used to identify each reptile species1. The captured lizards included Laudakia tuberculata (Kashmir Rock Agama, N = 53), Laudakia pakistanica (Pakistani Agama, N = 15), Hemidactylus flaviviridis (Common House Gecko, N = 7), Laudakia agrorensis (Agror agama, N − 4), Calotes versicolor (Oriental Garden Lizard, N = 2), Laudakia nupta (Large-scaled Rock Agama, N = 2) and Varanus bengalensis (Bengal Monitor, N = 1) (Fig. 2).
An information sheet was filled out for each lizard on the sampling site to gather information about each animal (sampling site, species, sex, age, body weight, body length, presence or absence of ectoparasites) in order to calculate the risk factors associated with the prevalence of blood-borne parasites in trapped lizards. Ventral caudal or ventral coccygeal vein of each captured wild lizard was aseptically pricked with disposable syringe under Isoflurane anesthesia and animals were released following the data and blood collection. Blood (100–500 µL) was transferred into the screw-capped tubes containing 0.5 M EDTA as an anticoagulant and was used for further molecular analysis.
DNA extraction from blood
Genomic DNA was extracted from the blood of each lizard using the blood genomic DNA extraction kit (Solarbio, China) following the manufacturer’s instructions.
Molecular detection by PCR
The extracted DNA samples were analyzed for the presence of Hepatozoon spp. (by targeting 18 S rRNA gene)16, Toxoplasma gondii (target was ITS-1 gene, only internal primers of the nested PCR protocol were used)17, Plasmodium spp. (target was Cytochrome b gene)18, Haemoproteus spp. (by targeting Cytochrome b gene)18, Leucocytozoon spp. (by targeting COX1 gene) and Schellackia spp. (by targeting SSU rRNA gene)19 by using the protocols as mentioned in the respective citations. The detail of primers is provided in Table 1. DNA amplification was carried out in a DNA thermal cycler (Gene Amp® PCR system 2700 Applied Biosystems Inc., UK). Negative control (distilled water) and positive control DNA (from previous studies: Hepatozoon sp. accession numbers OR976039 and OR976040, Toxoplasma gondii accession number OR727859, Plasmodium spp. accession number OR725031. DNA of Haemoproteus spp., Leucocytozoon spp. and Schellackia spp. were kindly provided by Dr. Alireza Sazmand from Faculty of Veterinary Medicine, Bu-Ali Sina University, Hamedan, Iran) were included in each reaction.
DNA sequencing and phylogenetic analysis
Amplified PCR products were sequenced by a commercial service provider (First Base, Malaysia). The obtained DNA sequences underwent initial trimming to eliminate primer-contaminated regions. Misread nucleotides at the start and end of the sequence were also removed using FinchTV (version 1.4.0). All sequences were submitted to NCBI’s GenBank and were assigned accession numbers: OR921620, OR921621, OR921622 and OR921623 (Hepatozoon sp.) and OR933561 and OR933562 (Lankesterella sp.). Similar sequences for both Hepatozoon sp. and Lankesterella sp. were retrieved using the Basic Local Alignment Search Tool (BLAST) from NCBI. Subsequently, all the sequences were aligned using the ClustalW multiple sequence alignment algorithm in BioEdit (version 7.2.5). The aligned sequences were imported into MEGA X (version 10.2.6). A model selection test was conducted for all sequences, using MEGA’s integrated model selection tool. The best-fit model was chosen based on Bayesian Information Criteria (BIC) and Akaike Information Criteria (AIC) values, with the model exhibiting the lowest BIC and AIC considered as the “best-fit” substitution model. Phylogenetic trees were constructed using the Maximum Likelihood algorithm in MEGA X with 1000 bootstraps. The final version of the inferred tree was generated using the iTOL server (https://itol.embl.de/, accessed on December12th 2023). 18 S rRNA partial sequences of Hepatozoon catesbianae (AF176837), Hepatozoon americanm (AF176836), Hepatozoon canis (AY461378) and Babesia anne (KT580785) were used as outgroups during. the phylogenetic analysis of Hepatozoon sp. Partial 18 S rRNA gene of Isospora tarentolae (KU180245), Isospora albogularis (KU180243), Isospora amphiboluri (KU180241), Caryospora ernsti (KU180247), Caryospora bigenetica (AF60975), Eimeria tenella (AF026388), Schellackia orientalis (KJ131414) and Goussia neglecta (FJ009242) served as an outgroup during phylogenetic analysis of Lankesterella sp.
Statistical analysis
Statistical package Minitab (Minitab, Pennsylvania, USA) was used for the statistical analysis of data. P ≤ 0.05 was considered significant. PCR based pathogen occurrence between various sampling sites and lizard species was compared through one way ANOVA. Association between the presence of each pathogen and studied epidemiological factors was assessed by contingency table analysis by using Fisher’s exact test (for 2 × 2 tables).
Results
Prevalence of haemogregarines in the blood samples of wild lizards
Analysis of the PCR results revealed that 11 out of 84 (13%) lizard blood samples screened during present study were Haemogregarines infected as they amplified a 600 bp amplicon from the 18 S rRNA gene of these pathogens (Supplementary Fig. 1). Among the infected lizards, seven were Laudakia tuberculata, three were Laudakia pakistanica and one was Laudakia agrorensis (Table 2).
Phylogenetic analysis of 18 S rRNA gene of haemogregarines
PCR products that amplified 18 S rRNA gene of Haemogregarines were confirmed by DNA sequencing. BLAST analysis of the amplified parasite sequences indicated that Pakistani lizards were infected with Hepatozoon sp. and Lankesterella sp. This was possible as we used the generalized primers that are capable of amplifying the 600 base pairs from the 18 S rRNA gene of a variety of organisms. BLAST results revealed that 4 lizard blood samples were infected with Hepatozoon sp. (98% similarity) whereas two were infected with Lankesterella sp. (97% similarity). Hence, separate phylogenetic analysis was carried out for both parasite species.
In the phylogenetic analysis, based on partial sequences of the 18 S rRNA gene of Hepatozoon sp., the four isolates (OR921620, OR921621, OR921622 and OR921623) detected in Pakistani lizards clustered together and showed similarities with Hepatozoon sp. isolates that were previously reported from Iran, Yamen, China,, Saudi Arabia, French Polynesia, Hungary, Chile, Brazil, Ghana, Algeria and Australia (Fig. 3).
Phylogenetic tree of Hepatozoon sp. based on the partial 18 S rRNA gene sequences. The four new sequences of Hepatozoon sp. (OR921620-23) generated in this study are marked with green squares. Scale bar represents 0.02 substitutions per nucleotide position. Bootstrap value is shown as number on the node.
A total of two Lankesterella sp. isolates were identified during present investigation and analysis of their partial 18 S rRNA gene sequences indicated that these isolates clustered in two different clades in our phylogenetic framework. Isolate 1 (OR933562) closely resembled the Lankesterella species reported from lizards and rodents in Pakistan, Yemen, Spain, Argentina, Chile, USA, Mexico and Oman. Whereas isolate 2 (OR933561) clustered in a separate clade with Lankesterella sp. isolates that were detected in bird species from Norway, Czech Republic, Lithuania and rodent in Pakistan (Fig. 4).
Phylogenetic tree of Lankesterella sp. based on the partial 18 S rRNA gene sequences. The two new sequences of Lankesterella sp. (OR933561-62) generated in this study are marked with green squares. Scale bar represents 0.05 substitutions per nucleotide position. Bootstrap value is shown as number on the node.
Molecular detection of haemosporidians and Schellackia spp. in the lizard blood samples
None of the trapped wild lizard during present study was found infected with Plasmodium spp., Haemoproteus spp., Leucocytozoon spp. or Schellackia spp.
Risk factor analysis
When the prevalence of Haemogregarines was compared among the captured lizards, one way ANOVA result revealed that the prevalence of Haemogregarines was not limited to particular lizards species (P = 0.8) (Table 2). As Laudakia tuberculata and Laudakia pakistanica were the only species for which more than one lizard were infected with Hepatozoon sp., hence the risk factor analysis was carried out for them individually as well. It was observed that the prevalence of Haemogregarines in Laudakia tuberculata varied significantly with the sampling sites (P = 0.001). All the infected Laudakia tuberculata were captured from Biyar. While none of the lizards captured from Lardoag, Katair, and Shahi were found Haemogregarines infected (Table 3).
Risk factor analysis indicated that age, sex, and presence of ectoparasites on Laudakia tuberculata as well as for Laudakia pakistanica had no association with Hepatozoon sp. infection (Supplementary Table 1).
Discussion
Lizards can serve as the reservoir for large number of disease causing agents including protozoa, helminths, pentastomida and arthropods. Lizards can be involved, directly or indirectly in the transmission of these pathogens to the definitive as well as new hosts20. Despite of having reptilian diversity, neither traditional (like blood smear screening) nor the modern tools (e.g., ELIZA and PCR) has been used to screen blood borne parasites among the Pakistani lizard fauna till date.
During the present investigation, 13% of screened wild lizards were found infected with Haemogregarines. The infected lizards included Laudakia tuberculata, Laudakia pakistanica and Laudakia agrorensis (Table 1). A number of wild lizard species are known to be infected with Hepatozoon parasites and these parasites have the potential to be transmitted onwards by a wide range of possible vectors (e.g. mosquitoes, ticks, mites and leeches)8,21. It is also an established fact that Hepatozoon species can reach new hosts through vector ingestion or when the host consumes an infected animal22. In a study conducted in Morocco, Er-Rguibi et al.23 had captured wild lizards including Timon tangitanus, Atlantolacerta andreanskyi, Podarcis vaucheri, Scelarcis perspicillata, Acanthodactylus dumerilli, Tarentola mauritanica and Quedenfeldtia trachyblepharus and screened them for the presence of blood borne parasites. They found that 1.26% lizards were infected with Hepatozoon sp. During a molecular epidemiological survey conducted in various regions of Spain and Portugal, Maia et al.24. trapped 133 lizards including Algyroides marchi, Podarcis bocagei, Podarcis hispanica and Podarcis lilfordi and found that 58% of them were infected with Hepatozoon sp. In another study, Maia et al.8 had reported 5% infection rate of Hepatozoon sp. among reptilian blood samples that were collected from North Africa. Infected reptiles belonged to genera Chalcides, Eumeces, Lacertidae, Atlantolacerta, Timon, Podarcis, Scelarcis, Quedenfeldtia and Tarentola. A study was conducted at Stephens Island in New Zealand by Godfrey et al.25 during which they found that 36/208 lizards (16.8%) were infected with Hepatozoon tuatara. A low Hepatozoon sp. prevalence was reported by Harris et al.21 in two endemic reptilian species that were trapped from the Seychelles Island in east Africa: one of them was a lizard species (Mabuya wrightii) and the other was a local snake (Lycognathophis sp.). Similarly, the presence of Lankesterella sp. has been reported in various lizard/reptilian species from various parts of the world. From Nicaragua, Chang et al.10 screened the blood samples from six black spiny-tailed iguanas (Ctenosaura similis) that were exhibiting clinical signs and reported the presence of Lakesterella spp. and Hepatozoon gamezi in the blood of iguana. From San Benito Oeste Island in Mexico, Quillfeldt et al.5 found that 85% of screened side-blotched lizards, Uta stansburiana, were infected with Lankesterella sp. Megía-Palma et al.12 had also found Lankesterella sp. infecting lizards that were trapped from North and South America. Maia et al.26 had also reported 13 Haemogregarines isolates from amphibians and reptiles that were captured from Oman. Phylogenetic analysis revealed that 6/13 of the generated isolates were never reported before from the study area and they belonged to Lankesterella sp.
Phylogenetic analysis was performed separately for Hepatozoon spp. and Lankesterella sp. as they belong to different taxonomic group of Apicomplexa. Genetic diversity of Hepatozoon spp. has never been reported from Pakistani reptiles in general and in wild lizards specifically. So, our data is a significant contribution in this unexplored research ___domain. Interestingly, our phylogenetic analysis revealed that several rodent, tick and reptilian species were infected with Hepatozoon sp6,28,29. Our literature review and data search in GenBank indicated that most Hepatozoon sequences from reptiles have not been identified at the species level and same happened during the present investigation. The sequences amplified in this study resembled the 18 S rRNA sequences of Hepatozoon sp. isolated from Iran (Accession number MN723845, unpublished data), reptiles in Yamen (Accession number MW07644928), king rat snakes in China (Accession numbers KF939621, KF939626 and KF9396276), lizards and rodents in Saudi Arabia (Accession number MN497412 and OR717488, unpublished data), rodents in French Polynesia (Accession number MT919388, unpublished data), rodents in Hungary (Accession number JX644997, unpublished data), marsupial and ticks in Chile (Accession number FJ71981829, Accession number MH174345, unpublished data) rodents and crocodiles in Brazil (Accession number OM033661 and OM033663, unpublished data; Accession number KJ413113 and MF435048, unpublished data), Ghana (Accession number EF157822, unpublished data), lizards in Algeria (Accession number HQ7347878), and in reptiles and their ticks in Australia (Accession number EU43023430) (Fig. 3). Our study also highlights that distinguishing Hepatozoon species based on host species alone is challenging due to overlap in parasite sequences.
The present investigation is the first report from Pakistan regarding the presence of Lankesterella sp. in local wild lizards. The isolates detected in this study showed genetic diversity as they resided in different clad indicating endemic diversity within this parasite (Fig. 4). The two isolates amplified from Pakistani lizards resembled the 18 S rRNA gene sequences of Lankesterella sp. deposited in GenBank from rodents in Pakistan (Accession number OR818017 and OR818019, unpublished data), reptiles in Yemen (Accession number MW07644228), lizards in Spain (Accession number KJ1314179), Lizards in Argentina (Accession number MF16755412), lizards in Chile (Accession number MF16755512), , lizards in USA (Accession number MF16755212). , side blotched lizard in Mexico (Accession number MH4592925), and reptiles from Oman (Accession number KX453652, KX453653, KX453658 and KX45366026), bird species from Norway (Accession number MG808273 and MG808274, unpublished data), Czech Republic (Accession number Accession number ON319023 and ON31902431, Accession number ON31903232), Passeriformes birds in Lithuania (Accession number MW72767913). and rodent in Pakistan (Accession number OR81801833) (Fig. 4). This data suggested that although the isolates generated from Pakistani lizards during present investigation were distinct from each other but phylogenetically they are related to known Lankesterella species found in various lizards and bird species and our local isolates might represent new taxonomic entities suggesting differences in host–parasite compatibility between these lineages and probably represent undescribed endemic species. Our results also indicate that host ecology and host relatedness may influence Lankesterella species distributions and, more generally, highlight the importance of screening wild hosts from remote and unexplored regions in Pakistan to provide new insights into parasite diversity.
As this is the first report regarding the risk factors associated with the prevalence Haemogregarines in Pakistani lizards, so no comparable data is available in literature. During the present investigation, we have observed a significant variation in the prevalence of Haemogregarines infecting Laudakia tuberculata with the sampling sites. Contrary to our observations, Wozniak and Telford34 had reported that the prevalence of Hepatozoon parasites in lizards varied between the species and similar to our observations they also found that parasite prevalence varied between the various geographical locations from where the samples were collected. Similar to our observations, Moreira22 had reported that Hepatozoon spp. prevalence in lizards varied with the sampling sites in Morocco. Maia et al.24 had also reported that Hepatozoon spp. prevalence varied between the host species and geographical locations but age, sex and nature of the habitat had no association with Hepatozoon spp. prevalence. Godfrey et al.25 had reported that Hepatozoon tuatarae (Apicomplexa) infection pattern varied predominantly with the host size and tick infestation during their study that was conducted in Stephens Islands, New Zealand.
Conclusion
In conclusion, to the best of our knowledge, this is the very first report regarding the presence of Hepatozoon sp. and Lankesterella sp. in wild Pakistani lizards. However, other targeted parasitic species including Plasmodium spp., Haemoproteus spp., Leucocytozoon spp. or Schellackia spp. were not detected in trapped lizards. Laudakia tuberculata, Laudakia pakistanica, and Laudakia agrorensis were among the infected species. Further, comprehensive large-scale studies are warranted in various unexplored geo-climatic regions of Pakistan to adjudge the prevalence of Hepatozoon sp. and Lankesterella sp. in the wild lizards as well as other reptiles, amphibians and small mammals.
Data availability
The datasets generated and/or analyzed during the current study are available in the GenBank repository, with Accession numbers OR921620, OR921621, OR921622 and OR921623 (Hepatozoon sp.) and OR7933561 and OR933562 (Lankesterella sp.). https://www.ncbi.nlm.nih.gov/nuccore/OR921620. https://www.ncbi.nlm.nih.gov/nuccore/OR921621. https://www.ncbi.nlm.nih.gov/nuccore/OR921622. https://www.ncbi.nlm.nih.gov/nuccore/OR921623 . https://www.ncbi.nlm.nih.gov/nuccore/OR933561. https://www.ncbi.nlm.nih.gov/nuccore/OR933562.
References
Khan, M. S. Key and checklist to the lizards of Pakistan (Reptilia: squamata: Sauria. Herpetozoa 15 (3/4), 99–119 (2002).
Sajjad, A. et al. Urban herpetofauna and public attitude towards their conservation in Rawalpindi and Islamabad, Pakistan. Int. J. Conserv. Sci. 12 (4), 1503–1514 (2021).
Masroor, R. A. Contribution to the herpetology of Northern Pakistan: the amphibians and reptiles of Margalla hills NationalPark and surrounding region. Soc. Stud. Amphib Rep. N. Y.. 4 (6), 2670–2672 (2012).
Khan, M. et al. New records of Amblyomma gervaisi from Pakistan, with detection of a reptile-associated Borrelia Sp. Tick. Tick. Born Dis. 13 (6), 102047. https://doi.org/10.1016/j.ttbdis.2022.102047 (2022).
Quillfeldt, P. et al. Molecular Survey of Coccidian infections of the side-blotched Lizard Uta stansburiana on San Benito Oeste Island, Mexico. Étude moléculaire des infections à coccidies du Lézard Uta stansburiana Sur L’île de San Benito Oeste, mexique. Parasit 25, 43. https://doi.org/10.1051/parasite/2018043 (2018).
Han, H. et al. First report of Hepatozoon (Apicomplexa: Adeleorina) from King rat snakes (Elaphe carinata) in Shanghai, with description of a new species. Acta Parasitol. 60(2), 266–274. https://doi.org/10.1515/ap-2015-0038
O’Dwyer, L. H. et al. Description of three new species of Hepatozoon (Apicomplexa, Hepatozoidae) from rattle snakes (Crotalus durissus terrificus) based on molecular, morphometric and morphologic characters. Exp. Parasitol. 135, 200–207. https://doi.org/10.1016/j.exppara.2013.06.019 (2013).
Maia, J. P., Harris, D. J. & Perera, A. Molecular survey of Hepatozoon species in lizards from North Africa. J. Parasitol. 97 (3), 513–517. https://doi.org/10.1645/GE-2666.1 (2011).
Megía-Palma, R., Martínez, J. & Merino, S. Molecular characterization of haemococcidia genus Schellackia (Apicomplexa) reveals the polyphyletic origin of the family Lankesterellidae. Zool. Scr. 43 (3), 304–312 (2014).
Chang, Y. C. et al. Reevaluation of hemoparasites in the black Spiny-Tailed Iguana (Ctenosaura similis) with the first pathological and molecular characterizations of Lankesterella desseri N. Sp. and redescription of Hepatozoon Gamezi. Microorg 11 (10), 2374. https://doi.org/10.3390/microorganisms11102374 (2023).
Harl, J. et al. Avian haemosporidian parasites of accipitriform raptors. Malar. J. 21. https://doi.org/10.1186/s12936-021-04019-z (2022).
Megía-Palma, R. et al. Phylogenetic analyses reveal that Schellackia parasites (Apicomplexa) detected in American lizards are closely related to the genus Lankesterella: is the range of Schellackia restricted to the old world?? Parasit. Vect. 10 (1), 470. https://doi.org/10.1186/s13071-017-2405-0 (2017).
Chagas, C. R. F. et al. Lankesterella (Apicomplexa, Lankesterellidae) blood parasites of passeriform birds: prevalence, molecular and morphological characterization, with notes on sporozoite persistence in vivo and development in vitro. Anim 18, 1451. https://doi.org/10.3390/ani11051451 (2021).
Khalil, S., Rana, A. H. & Hussain, T. Morphometric analysis of spiny tailed Lizard (saara hardwickii) from lesser Cholistan desert, Bahawalpur, Punjab, Pakistan. J. Anim. Plant. Sci. 32 (1), 301–310 (2022).
Ribeiro-Júnior, M. A., Gardner, T. A. & Ávila-Pires, T. C. S. The effectiveness of glue traps to sample lizards in a tropical rainforest. South. Amr J. Herpetol. 1 (2), 131–137 (2006).
Ujvari, B., Madsen, T. & Olsson, M. High prevalence of Hepatozoon spp. (Apicomplexa, Hepatozoidae) infection in water pythons (Liasis fuscus) from tropical Australia. J. Parasitol. 90, 670–672 (2004).
Zintl, A. et al. In vitro culture combined with quantitative TaqMan PCR for the assessment of Toxoplasma gondii tissue cyst viability. Vet. Parasitol. 164, 167–172 (2009).
Ciloglu, A., Ellis, V. A., Bernotienė, R., Valkiūnas, G. & Bensch, S. A new one-step multiplex PCR assay for simultaneous detection and identification of avian haemosporidian parasites. Parasitol. Res. 118 (1), 191–201 (2018).
Zechmeisterová, K., Javanbakht, H., Kvičerová, J. & Široký, P. Against growing synonymy: identification pitfalls of Hepatozoon and Schellackia demonstrated on North Iranian reptiles. Eur. J. Protistol. 79, 125780 (2021).
Swei, A., Ostfeld, R. S., Lane, R. S. & Briggs, C. J. Impact of the experimental removal of lizards on Lyme disease risk. Proc. R. Soc. B: Biol. Sci. 278 (1720), 2970–2978 (2011).
Harris, D. J., Maia, J. P. & Perera, A. Molecular survey of Apicomplexa in Podarcis wall lizards detects hepatozoon, sarcocystis, and Eimeria species. J. Parasitol. 98 (3), 592–597 (2012).
Moreira, M. I. V. B. D. Fitness effects of Hepatozoon blood parasites in selected lizard species. M. Sc thesis. Uni. Porto. https://core.ac.uk/download/pdf/302931917.pdf (2013).
Er-Rguibi, O. et al. Molecular survey and microscopic examination of haemoparasites infecting lizards from Morocco. Acta Parasitol. 68 (3), 593–603. https://doi.org/10.1007/s11686-023-00688-9 (2023).
Maia, J. P., Perera, A. & Harris, D. J. Molecular survey and microscopic examination of Hepatozoon Miller, 1908 (Apicomplexa: Adeleorina) in lacertid lizards from the Western mediterranean. Folia Parasitol. 59 (4), 241 (2012).
Godfrey, S. S., Nelson, N. J. & Bull, C. M. Ecology and dynamics of the blood parasite, Hepatozoon Tuatarae (Apicomplexa), in Tuatara (Sphenodon punctatus) on Stephens Island. New. Z. J. Wildlif Dis. 47 (1), 126–139 (2011).
Maia, J. P., Harris, D. J., Carranza, S. & Goméz-Díaz, E. Assessing the diversity, host-specificity and infection patterns of apicomplexan parasites in reptiles from Oman. Arabia Parasitol. 143 (13), 1730–1747. https://doi.org/10.1017/S0031182016001372 (2016).
de Ferreira, A. R., Perles, D., Machado, L., Prado, R. Z., André, M. R. & C.P.A. & Molecular detection of apicomplexan hemoparasites in anurans from Brazil. Parasitol. Res. 119, 3469–3479. https://doi.org/10.1007/s00436-020-06835-9 (2020).
Tomé, B., Maia, J., Perera, A., Carranza, S. & Vasconcelos, R. Parasites in a hotspot: diversity and specificity patterns of apicomplexans infecting reptiles from the Socotra Archipelago. Parasitol 148 (1), 42–52. https://doi.org/10.1017/S0031182020002000 (2021).
Merino, S. et al. Martinez de La Puente, J. Molecular characterization of an ancient Hepatozoon species parasitizing the ‘living fossil’ marsupial ‘monito Del Monte Dromiciops gliroides from Chile. Biol. J. Linn. Soc. Lond. 98 (3), 568–576 (2009).
Vilcins, I. M., Ujvari, B., Old, J. M. & Deane, E. Molecular and morphological description of a Hepatozoon species in reptiles and their ticks in the Northern territory, Australia. J. Parasitol. 95 (2), 434–442. https://doi.org/10.1645/GE-1725.1 (2009).
Martínez, J. et al. Hemoparasites and immunological parameters in snow bunting (Plectrophenax nivalis) nestlings. Polar Biol. 41 (9), 1855–1866 (2018).
Venkatachalam, A. K. S. B., Čepička, I., Hrazdilová, K. & Svobodová, M. Host specificity of passerine Lankesterella (Apicomplexa: Coccidia). Eu J. Protistol. 90, 126007. https://doi.org/10.1016/j.ejop.2023.126007 (2023).
Ijaz, M. et al. First report of Hepatozoon and Lankesterella spp. Infections in wild rodents from Pakistan, and their potential impact on blood parameters and oxidative stress markers in vital organs. Vet. Res. Commun. 49 (1), 45. https://doi.org/10.1007/s11259-024-10611-w (2024).
Wozniak, E. J. & Telford, S. R. Jr. The fate of Hepatozoon species naturally infecting Florida black racers and watersnakes in potential mosquito and soft tick vectors, and histological evidence of pathogenicity in unnatural host species. Int. J. Parasitol. 21 (5), 511–516 (1991).
Acknowledgements
The authors extend their appreciation to the Researchers Supporting Project number (RSP2025R197), King Saud University, Riyadh, Saudi Arabia.
Author information
Authors and Affiliations
Contributions
F.I. and M.L. had designed and supervised this study. A.U.K. and S.K. collected the lizard samples and epidemiological data. R.S., M.K., H.A., M.S., M.D., S.I., T.M.D. and H.F. performed the wet lab experiments. H.M. and A.K. performed the phylogenetic analysis. All authors contributed to the writing of manuscript and approved the final version for submission.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
Ethical Research Committee of the Institute of Zoology at Bahauddin Zakariya University Multan (Pakistan) approved all the experimental procedures and protocols applied in this study via letter number Zool./ Ethics/22-7. Notably, all methods adhered to the ethical guidelines outlined in the “Guide for the Care and Use of Laboratory Animals,” as stipulated by the National Academy of Sciences.
ARRIVE guidelines
The study is reported in accordance with ARRIVE guidelines (https://arriveguidelines.org).
Statement of informed consent
There are no human subjects in this article and informed consent is not applicable.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Shabbir, R., Latif, M., Khan, A.U. et al. Leading report on the molecular prevalence of emerging pathogens Hepatozoon sp. and Lankesterella sp. in the blood samples of seven wild lizard species. Sci Rep 15, 9014 (2025). https://doi.org/10.1038/s41598-025-91185-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-91185-8