Abstract
Climate change, human activities, and habitat fragmentation—or even loss—pose ongoing threats to the survival of wildlife. Understanding the dietary habits of endangered species is a critical component of their conservation. In this study, we investigated the diet of water deer (Hydropotes inermis) by integrating traditional fecal microhistological analysis with a deep learning algorithm. Fecal samples were collected from water deer in northeastern China, with microscopic slides prepared for both the warm season (203 samples) and cold season (451 samples). A deep learning model was trained and tested using labeled images of 130 known plant species, achieving an accuracy of 99.83%. The results revealed that water deer consume 110 plant species from 86 genera and 40 families annually. The dietary patterns observed in this study align closely with those reported in other regions, reflecting species-specific foraging characteristics and further validating the reliability of deep learning algorithms in ecological research. Notably, significant seasonal variations were identified, highlighting the adaptability of water deer to changing environmental conditions. By examining the feeding ecology and seasonal dietary shifts of water deer in northeastern China, this study provides valuable insights for the development of targeted conservation strategies to support their populations in this region and beyond.
Similar content being viewed by others
Introduction
In the context of global climate change, increasing human disturbances, and ongoing habitat fragmentation, obtaining fundamental ecological data on endangered species is essential for wildlife conservation. These environmental pressures have significantly impacted the population distribution and abundance of numerous species, driving some to extinction1,2. Ungulates play a critical role in ecosystem functioning, maintaining ecological balance by shaping vegetation patterns, facilitating seed dispersal, and serving as prey for carnivores3,4. The dietary habits of ungulates are central to wildlife ecology, offering valuable insights into habitat selection, interspecific competition, and their potential resilience to climate change5,6,7,8. Therefore, understanding the diet of endangered species is vital for informing effective conservation strategies and habitat management.
The water deer (Hydropotes inermis), a small ungulate once widely distributed across China—including the Liaodong Peninsula, North China Plain, and areas along the Yangtze River9,10—is now primarily confined to southern and northeastern regions. Significant declines in their geographic range and population have been driven by habitat loss and fragmentation resulting from climate change and anthropogenic activities11. Historically, water deer were believed to be absent from northeastern China for the past 50 years, but recent research has confirmed their presence in this region12. Despite this rediscovery, critical gaps remain in understanding their ecology, particularly their dietary habits. Water deer are classified as concentrate selectors within the mixed feeder category, exhibiting a diverse diet dominated by tender leaves and shoots of woody plants, shrubs, and forbs13,14,15. This study aims to address these gaps by investigating their diet to elucidate how they adapt to the habitat and climatic conditions of northeastern China. Such knowledge is essential for developing scientifically informed conservation and management strategies to protect this endangered species.
Traditional methods for studying ungulate diets, such as fecal microhistological analysis and fecal DNA analysis16, remain widely used but are often time-consuming and costly. Fecal microhistological analysis, despite its labor-intensive nature, offers several advantages, including low cost, non-invasiveness, and methodological simplicity. These attributes have made it a popular choice for ungulate diet studies. However, its limitations are significant, particularly when researchers must compare numerous fecal microscopic slides with plant cell reference slides. The vast diversity of plant species and the similarity in cell structures often lead to identification challenges, even for experienced researchers. These limitations underscore the need for innovative approaches to improve accuracy and efficiency in dietary analysis. Deep learning, with its exceptional capabilities in self-supervised feature learning and multi-class classification, has shown great promise in addressing complex image recognition tasks. Its application to ecological research, particularly in the analysis of plant cell structures, has demonstrated both accuracy and efficiency17. By integrating deep learning with traditional fecal microhistological analysis, this study seeks to overcome the limitations of conventional methods and provide novel insights into the diet of water deer.
This study aims to: (1) evaluate the reliability of deep learning algorithms for microhistological diet analysis of wild herbivores, (2) identify the plant species and their proportions in the diet of water deer in northeastern China across seasons, and (3) explore seasonal dietary variations and their implications for the species’ adaptation to environmental changes. The findings are expected to provide a scientific foundation for conserving water deer populations by safeguarding their food resources in northeastern China. Furthermore, the integration of deep learning with traditional ecological methods holds potential for advancing research on other endangered species.
Materials and methods
Sampling area
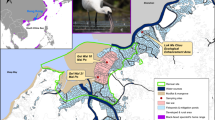
Fecal samples of water deer were collected from northeastern China, encompassing Changbai, Baishan, and Ji’an in Jilin Province, as well as Kuandian, Fengcheng, and Huanren in Liaoning Province (40°2′–42°13′N, 123°33′–128°18′E; Fig. 1). The sampling area in Jilin Province is characterized by a north temperate continental monsoon climate, with an average annual temperature of 4.6 °C and an average annual precipitation of 883.4 mm. In contrast, the sampling area in Liaoning Province experiences a warm temperate humid monsoon climate, with an average annual rainfall ranging from 800 to 1,200 mm and an average annual temperature of 6–9 °C. The study area is located along the north and northwest banks of the Yalu River, featuring a well-developed water system with numerous tributaries, including the Hun River, Ai River, and Cao River.
Map depicting sampling area and sampling sites generated with ArcGIS 10.8 (Environmental Systems Research Institute, Inc. in Redlands, California, USA. https://www.esri.com/en-us/arcgis/products).
The sampling area supports a diverse mammalian community, including species such as musk deer (Moschus moschiferus), roe deer (Capreolus pygargus), black bear (Ursus thibetanus), and lynx (Lynx lynx). The region is rich in vegetation resources and comprises varied habitat types, such as broad-leaved forests, mixed coniferous and broad-leaved forests, coniferous forests, shrubs, and grasslands. Dominant tree species include Gmelin larch (Larix gmelinii), Korean pine (Pinus koraiensis), and Mongolian oak (Quercus mongolica).
Fecal sampling procedure
Fresh fecal samples of water deer were collected from June to December 2021 within their suitable habitats in the study area, which included grassland/cropland, shrub/orchard, and Gmelin larch habitats12. Sampling was conducted along systematically planned transects to detect signs of water deer presence. Upon locating fresh feces, samples were collected using 50 ml sampling tubes, with elevation and environmental data recorded at each sampling point. To minimize the risk of false repetitions, a minimum distance of 500 m was maintained between consecutive fecal collection points when encountering grouped feces18. Although musk deer and roe deer also inhabit the study area, their preferred habitats differ from those of water deer, as they are typically found at higher elevations with steeper slopes. Water deer fecal pellets were identified following the methods described by Li et al.12,19. Compared to roe deer feces, water deer feces are characterized by smaller, more elongated pellets and denser fecal piles with shorter inter-pile distances. These distinct morphological and habitat features allowed for confident attribution of the collected fecal samples to water deer. Fresh feces excreted within three days were collected based on the approach outlined by Gao20. A total of 203 fresh fecal samples were collected during the warm season (July to August), and 451 samples were obtained during the cold season (November to December).
Microhistological analysis
The preparation of fecal samples for microhistological analysis followed a standardized protocol to ensure consistency and accuracy21. A few pellets were extracted from each sample to create composite samples for the warm and cold seasons, which were then crushed using a GIANXI coffee grinder. The crushed samples were sieved through 0.45 mm and 0.15 mm filters, with the material retained on the 0.15 mm filter stored for further analysis22. For each sample, 1 g of the sieved material was placed into a culture dish containing 40 ml of 10% sodium hypochlorite solution22. The mixture was thoroughly stirred to ensure homogeneity and processed for 3–5 h. After processing, the samples were poured onto a 0.075 mm filter and rinsed for 2 min22. The rinsed material was spread onto glass microscope slides using glycerol, and cover slips were secured with neutral gum. Three slides were prepared for each composite sample. The slides were examined under 100× magnification using a Leica DM1000 biomicroscope and Leica EZ software. Each slide was systematically photographed, capturing 10 to 20 fields of view in a left-to-right and top-to-bottom sequence to ensure comprehensive coverage.
Construction of plant cell image library
Plant cell images obtained from Ding23 and Zhou24 were incorporated into the image library for training the deep learning model. Ding’s study area overlapped with ours, while Zhou’s study area shared similar climatic and vegetation conditions, ensuring the appropriateness and reliability of these images for model training. The collected images were meticulously reviewed to exclude overlapping, unclear, or irrelevant samples, resulting in a final selection of 701 images representing 130 plant species. To optimize model training accuracy, each image was individually cropped to remove blank areas and converted to grayscale. Due to the limited number of images per plant species, data augmentation was performed using the cv2 library in OpenCV25 and the NumPy tool in PyCharm (2023.1.1)26. Specific augmentation techniques included resizing images by a factor of 1.5, horizontal and vertical flipping, 180° rotation, brightness adjustment, and the addition of salt-and-pepper noise27. These operations generated a total of 4,907 images for model training.
Model training, validation, and predicting
Model training was performed using the ResNet-50 (Residual Network with 50 layers) architecture28,29 in PyCharm (2023.1.1), with the training parameters specified in Table 1. ResNet-50, a deep convolutional neural network within the ResNet family, is distinguished by its residual learning blocks30. These blocks incorporate shortcut connections (skip connections) that enable the network to bypass one or more layers during training. This design addresses the vanishing gradient problem and facilitates the training of very deep networks. ResNet-50 has been widely applied in image recognition tasks, including image classification, object detection, and image segmentation31,32.
Before model training, the image library was partitioned into three subsets: a training set (60%, 3,019 images), a test set (20%, 903 images), and a validation set (20%, 985 images)33. The training set was used to train the ResNet-50 model over 20 epochs, with model accuracy evaluated using the validation set34. Subsequently, the trained deep learning model was employed to classify plant cell images from fecal samples. Prior to prediction, each image was individually cropped to remove blank areas and retain clear cellular structures. Additionally, images were converted to grayscale to improve prediction accuracy. Following processing, a total of 916 plant images were obtained from fecal composite samples during the warm season, and 625 images were obtained during the cold season.
Data analyses
The relative abundance of plant species a = Number of plant a identifications by deep learning model / Total plant images in warm or cold season. Differences in seasonal consumption proportions of specific plant types were assessed using the Mann-Whitney U test. To evaluate diet selectivity, α-diversity analyses were conducted by calculating the Shannon diversity index and inverse Simpson index using the vegan package (version 2.6-4)35. β-diversity analyses were performed to examine seasonal differences in diet composition, with compositional dissimilarities visualized through nonmetric multidimensional scaling (NMDS) plots based on Bray-Curtis distances. NMDS ordination was implemented via the metaMDS function in the vegan package35. Detected plants were categorized into four functional groups: woody species, ferns, graminoids, and forbs. All visualizations were generated using the ggplot2 package (version 3.3.5)36 and OriginPro 202337. Statistical analyses were performed in R (version 4.3.1)38, with significance thresholds set at P < 0.05.
Results
Accuracy of trained model
The accuracy of the training set progressively increased, reaching 99.83% after 20 training epochs, while the validation set accuracy surpassed 84% (Fig. 2). Additionally, the model was evaluated using a test set of plant cell images, achieving an accuracy of 88.8%. These results demonstrate that the deep learning model developed in this study is highly effective for identifying plant cell images in water deer fecal samples. During the warm season, the model successfully identified 96 distinct plant species, compared to 93 species recognized in the cold season (Fig. 3).
Comparison between the images in the training set and the images photographed from the fecal microscope slides. a, b, c: The plant cell images collected from Ding23 and Zhou24, which was set as training set of the deep learning model; d, e, f: the images photographed from the fecal microscope slides in current study, which were recognized by the deep learn model; a, d: Clematis terniflora; b, e: Populus davidiana; c, f: Polygonum sieboldii.
Plant species composition
The results indicated that water deer consumed a total of 110 plant species, encompassing 86 genera from 42 families throughout the year (S1). During the warm season, their diet consisted of 96 species from 78 genera and 39 families, whereas in the cold season, they consumed 93 species from 75 genera and 38 families (S1). The most frequently consumed families, along with their respective relative abundances over the year, were Fabaceae (10.38%), Asteraceae (8.76%), Poaceae (7.72%), Polygonaceae (6.88%), Salicaceae (6.16%), Rosaceae (5.45%), and Malvaceae (5.26%) (Fig. 4a, S1). In the warm season, Kummerowia striata (2.95%) was the most frequently consumed plant within the Fabaceae family, while in the cold season, Amphicarpaea edgeworthii (3.2%, S1) was the most consumed. Within the Poaceae family, Poa sibirica (4.26%) was the predominant plant during the warm season, whereas Alopecurus aequalis (1.28%) was more prevalent during the cold season (S1).
Similarly, Fabaceae and Asteraceae exhibited high feeding ratios in both the warm and cold seasons, accounting for 10.07% and 8.95% of Fabaceae, and 9.92% and 8.48% of Asteraceae, respectively (Fig. 4a, S1). However, the feeding ratios for Poaceae (10.15%, P = 0.004) and Salicaceae (9.5%, P = 0.002, S2) were significantly higher during the warm season, while they decreased to 4.16% and 1.28%, respectively, during the cold season (Fig. 4a, S1). In the cold season, water deer consumed 8.3% of Rosaceae (P = 0.004) and 8.48% of Cannabaceae (P = 0.002, S2), whereas during the warm season, only 3.17% and 2.29% of these two families were consumed, respectively (Fig. 4a, S1).
At the genus level, Populus (5.45%) was the most dominant throughout the year, followed by Tilia (5.26%), Polygonum (5.26%), Humulus (4.8%), Euonymus (4.54%), Acer (4.09%), and Picea (4.02%, Fig. 4b, S1). During the warm season, the top four genera most consumed by water deer were Populus (8.52%, P = 0.002), Tilia (6.44%, P = 0.036), Polygonum (6.44%, P = 0.036), and Juncus (5.68%, P = 0.002). In contrast, during the cold season, the top four genera were Humulus (8.48%, P = 0.002), Euonymus (6.24%, P = 0.240), Picea (5.76%, P = 0.054), and Acer (4%, P = 0.733) (Fig. 4b, S1, S2).
Plant form composition
The overall composition of plants, classified by growth morphology, was dominated by woody species (47.63%) and forbs (41.99%), with smaller proportions of graminoids (7.72%) and ferns (2.66%). This compositional pattern remained consistent across seasons. During the warm season, the proportions were 47.38% woody species, 39.3% forbs, 10.15% graminoids, and 3.17% ferns. In the cold season, the proportions shifted slightly to 48% woody species, 45.92% forbs, 4.16% graminoids, and 1.92% ferns (Fig. 5).
Plant species diversity
Diversity analysis illustrated the significant differences in plant diversity between the warm and cold seasons, as measured by the Shannon diversity index (Fig. 6a) and the Simpson index (Fig. 6b). For the Shannon diversity index, the values were significantly higher during the warm season compared to the cold season (P = 0.00028). Similarly, the Simpson index also showed significantly higher values during the warm season (P = 0.0012). The NMDS plot illustrated the differences in plant composition between the cold and warm seasons (Fig. 7). Samples from the warm season cluster on the right side of NMDS1, while cold season samples are more dispersed on the left, indicating a gradient driven by seasonal changes. The stress value of 0.089 suggested a good fit of the model to the data, supporting the reliability of the observed patterns.
Discussion
Diet comparison of water deer between warm and cold season
Water deer exhibited a consistent preference for Fabaceae and Asteraceae plants across both warm and cold seasons (Fig. 4a, S1), likely due to the year-round availability of these plant families. However, notable seasonal variations were observed in the consumption of other plant families. During the warm season, the proportion of Poaceae and Salicaceae in the diet was significantly higher, whereas their consumption decreased during the cold season (Fig. 4a, S1, S2). This pattern may reflect the vigorous growth, high palatability, and nutritional value of Poaceae and Salicaceae plants during the warm season, as well as their widespread distribution. In contrast, water deer consumed a higher proportion of Rosaceae and Cannabaceae during the cold season, with significantly lower proportions observed in the warm season (Fig. 4a, S1, S2). This shift may be driven by the availability of fallen Rosaceae leaves and dried Humulus scandens (Cannabaceae) during the cold season. Conversely, the greater diversity of food resources available in the warm season likely reduced the reliance on these plant families.
The relative abundance of plant genera consumed by water deer differed significantly between warm and cold seasons. During the warm season, the top four genera in their diet were Populus, Tilia, Polygonum, and Juncus (Fig. 4b, S1, S2). In contrast, during the cold season, the dominant genera shifted to Humulus, Euonymus, Picea, and Acer (Fig. 4b, S1, S2). These patterns suggest that the palatability and availability of these plants may be more favorable during the cold season, driving their increased consumption by water deer. Notably, water deer exhibited similar genus-level dietary preferences to other ungulates in northeastern China, such as red deer (Cervus elaphus) and sika deer (Cervus nippon), which also primarily consumed Euonymus, Acer, Abies, Pinus, and Corylus39. This aligns with the broader behavior of ungulates in northern and cold regions, which adjust their food choices seasonally to meet nutritional demands39. Winter, in particular, represents a critical period for ungulate nutrition in these regions, as cold climates and limited food resources pose significant challenges40,41. Consequently, the ability to adapt dietary preferences to seasonal changes is essential for the survival of ungulates under such harsh environmental conditions.
The diversity indices of plant species consumed by water deer were significantly higher during the warm season compared to the cold season (Fig. 6). This pattern can be attributed to the vigorous growth of plants in the warm season, which increases the availability and variety of forage. Additionally, the birthing period of water deer, typically occurring in May-June, coincides with elevated energy demands for lactation and offspring rearing, further driving the need for a diverse and abundant diet. Analysis of the specific plants consumed revealed a relatively balanced distribution of plant species during the warm season (Fig. 8). In contrast, the cold season exhibited pronounced variability in the relative abundance of consumed plants, ranging from 8.48% (Humulus scandens, P = 0.002) to as low as 0.16% (Populus cathayana, P = 0.002; Fig. 8, S2). This disparity reflects the reduced diversity and availability of plant species during the cold season, compounded by the limited nutritional value of deciduous leaves from woody plants such as Populus cathayana, which are high in fiber but low in essential nutrients.
Notably, significant differences were observed in the consumption of specific plant species between seasons. Plants more frequently consumed during the warm season, such as Picea koraiensis (P = 0.054), Euonymus macropterus (P = 0.002), and Filipendula palmata (P = 0.014), showed reduced proportions in the cold season, with differences reaching up to 6.19% (Fig. 8, S2). Conversely, plants preferred during the cold season, including Juncus effusus (P = 0.002), Populus davidiana (P = 0.002), Poa sibirica (P = 0.008), and Populus cathayana (P = 0.002), were less prominent in the warm season, with maximum differences of 5.2% (Fig. 8, S2). These shifts highlight the adaptive foraging strategies of water deer, which adjust their dietary preferences in response to seasonal changes in plant availability and nutritional quality. In summary, the reduced variety and quantity of plant species during the cold season necessitate increased reliance on a limited number of available forage options. Water deer demonstrate remarkable dietary flexibility, modulating their consumption patterns to meet energy and nutritional requirements in response to fluctuating environmental conditions.
Diet comparison of water deer between current study and other regions
From a plant family perspective, our findings demonstrated a high degree of dietary similarity between water deer in northeastern China and those in other regions. In this study, the dominant plant families consumed by water deer align closely with results from a fecal DNA analysis of water deer in Korea, which reported a diet dominated by Salicaceae (27.5%), Fabaceae (17.5%), and Poaceae (14.2%)42. Similarly, another fecal DNA study in Korea identified Rosaceae (22.0%), Moraceae (19.0%), Betulaceae (10.0%), and Fabaceae (8.7%) as the primary plant families in their diet43. Further supporting this pattern, a microhistological study of water deer in the Zhoushan Islands, Zhejiang Province, China, documented their consumption of 137 plant species from 115 genera and 61 families, with Fabaceae, Fagaceae, Ericaceae, and Rosaceae representing a significant proportion across all four seasons13. Additionally, multiple dietary studies of water deer in China have consistently highlighted the prominence of Fabaceae, Asteraceae, Rosaceae, and Poaceae throughout the year23,44. Collectively, these findings indicate that water deer in the present study, as well as those in Korea and southern China, exhibit a strong preference for Fabaceae and Rosaceae plants. This consistency in dietary patterns across geographically distinct populations suggests a potential influence of inherent genetic factors on food selection. Furthermore, the observed dietary similarities may also reflect convergent evolutionary adaptations to similar ecological niches, as water deer in these regions face comparable environmental pressures, such as seasonal resource availability and predation risks. The preference for Fabaceae and Rosaceae plants, which are often rich in nutrients and widely distributed, may represent an optimal foraging strategy shaped by natural selection.
The number of plant species consumed by water deer in the current study was relatively lower than that reported in southern China (137 species)13, but the number of genera was higher than that observed in Korea (63 genera)43. This discrepancy may be attributed to climatic and vegetation differences across regions. Both northeastern China and Korea fall within the temperate monsoon climate zone, resulting in similar vegetation compositions. In contrast, the distribution area of water deer in southern China experiences a subtropical monsoon climate, characterized by higher plant diversity and distinct vegetation conditions. Consequently, water deer in southern China consume a significantly greater number of plant species compared to those in northeastern China. This regional variation in dietary breadth may reflect adaptive responses to local ecological conditions, such as resource availability and competition. For instance, the higher plant diversity in southern China likely provides water deer with a broader range of foraging options, while the temperate regions may favor specialization on fewer, more abundant plant species.
Variations in the number of plant species consumed by water deer across regions may also be influenced by factors such as study duration, methodologies, and geographic locations. For instance, a comparison between traditional microhistological analysis and DNA-based methods reveals that the latter tends to overemphasize chloroplast-rich tissues, potentially underestimating the contribution of seeds or roots to the diet. Additionally, DNA analysis exhibits limitations in detecting non-angiosperm materials, such as mosses and ferns, and is entirely ineffective for identifying fungi45. Notably, fecal microhistological analysis has been widely employed in studies of deer feeding ecology, including those referenced above. For example, Guo et al. identified the highest number of plant species consumed by deer13, a finding that may reflect either the methodological reliability of microhistological analysis or the exceptional plant diversity in their study area. This underscores the precision and effectiveness of fecal microhistological analysis in identifying plant species, further supporting its utility in dietary studies.
The dietary composition of water deer consistently featured a high prevalence of woody plants and forbs across all studied regions. These findings align closely with those of Ding et al.23, who reported a diet dominated by forbs (52.94%) and woody plants (35.29%). In contrast, Guo et al.13 observed somewhat divergent patterns, with woody plants representing a substantial proportion of the diet year-round (up to 61.15%), while forbs were prominent only in spring (48.39%) and summer (49.73%), declining significantly in autumn (~ 30%) and winter (< 20%). These differences may reflect the contrasting climatic conditions. The prevalence of evergreen woody plants in their study area likely contributed to the elevated consumption of woody plants during autumn and winter. This suggests that water deer may exhibit dietary plasticity, adapting their foraging strategies to maximize energy intake under varying environmental conditions. Further variations were observed in the study by Lee et al.43 on Korean water deer, where woody plants dominated the diet (68.6%), while forbs and graminoids accounted for only 7.0% and 0.7%, respectively. These disparities may be attributed to subspecies differences, as the Korean water deer (H. i. argyropus)46,47,48 is distinct from the Chinese water deer (H. i. inermis)12,49, which was the focus of other studies. The observed dietary differences between subspecies may reflect evolutionary divergence driven by local adaptation to distinct ecological niches, highlighting the role of environmental factors in shaping foraging behavior.
The application of deep learning in ecological studies
Deep learning has emerged as a highly efficient and accurate tool for ecological research. Among deep learning models, ResNet-50 has proven particularly valuable, playing a pivotal role in species identification and population monitoring through the analysis of camera trap images50. Its applications extend to biodiversity assessment, where it enables the precise identification and classification of both plant and animal species within complex ecological datasets51. Furthermore, ResNet-50 contributes to evaluating the impacts of climate change on ecosystems by monitoring shifts in vegetation patterns and wildlife distributions52. In the current study, the ResNet-50 model achieved an impressive accuracy of 99.83%, providing robust support for our findings and highlighting the transformative potential of deep learning in ecological studies. The utility of deep learning is further exemplified by its application in identifying morphologically similar species within the Podarcis hispanicus complex of wall lizards, where it achieved high accuracy rates53. These results demonstrate that deep learning models are powerful tools for field identification and monitoring of cryptic species complexes, offering a reliable alternative to traditional methods reliant on expert knowledge or genetic analysis.
Deep learning is increasingly being integrated into wildlife ecological research, offering significant reductions in researchers’ workloads54,55. For instance, deep convolutional neural networks have been employed to identify, count, and describe the behaviors of 48 species within the Snapshot Serengeti dataset, which comprises 3.2 million images. This approach saved over 17,000 h of manual labeling effort while achieving an identification accuracy exceeding 93.8% 51. In the current study, we innovatively applied deep learning algorithms to plant cell recognition, achieving high accuracy and substantially reducing the time required for plant image classification. Traditional methods for comparing 1,541 images across 130 plant species could take several months and often yielded inconsistent results. In contrast, our deep learning-based approach completed data organization, labeling, model training, and prediction within a few days. These findings underscore the potential of deep learning algorithms to significantly enhance the efficiency and accuracy of wildlife ecological research.
The collection of high-quality datasets is a critical step in training deep learning models. In this study, the acquisition of plant cell images played a pivotal role in the initial stages, significantly contributing to model development. We utilized plant cell images from prior studies, sourced either from the study region or from areas with similar plant compositions and climatic conditions. This approach eliminated the need for dedicated collection of plant samples and preparation of cell slides, substantially reducing both the workload and time required for the study. For other research leveraging deep learning in image recognition, such as species identification from infrared camera images, the initial identification and labeling of large image datasets remain labor-intensive and time-consuming. Therefore, the establishment of comprehensive resource databases—such as plant cell image databases, animal image databases, and animal sound databases—could streamline preliminary research efforts. However, this strategy is not without limitations. For instance, the plant cell images used in this study may not encompass all plant species consumed by water deer in the study area, potentially restricting the model’s ability to recognize certain food sources. To address this, future studies should focus on expanding the model library by accumulating cell images from a wider range of plant species. This will enable a more comprehensive representation of the dietary composition of water deer and improve the accuracy of the results.
Conclusions
This study demonstrates the efficacy of integrating fecal microhistological analysis with deep learning algorithms to elucidate the dietary ecology of water deer in northeastern China. Our deep learning model, trained on plant cell images from prior studies, achieved an identification accuracy of 99.83%, enabling the detection of 110 plant species (86 genera, 42 families) consumed by water deer across seasons. Notably, the observed dietary composition exhibited strong congruence with water deer populations in other regions13,23,42,43,44,56, validating both the methodology and the ecological consistency of the species’ foraging behavior. Significant seasonal shifts in plant consumption patterns were identified, with woody species and forbs dominating the diet in both warm (July–August) and cold (November–December) seasons. These adaptive dietary adjustments underscore the importance of temporal resource dynamics in shaping the species’ foraging strategies. Such insights are critical for informing habitat management plans aimed at supporting water deer populations, particularly in the context of climate-driven vegetation changes. The integration of deep learning with traditional microhistological techniques has substantially accelerated dietary analysis while maintaining high precision. This approach not only advances research on water deer ecology but also establishes a scalable framework for studying dietary patterns in other endangered herbivores. As deep learning algorithms continue to evolve, their application in ecological studies promises to enhance our capacity to monitor biodiversity and design targeted conservation strategies.
Data availability
The raw data supporting the conclusions of this article are stored in College of Wildlife and Protected Areas, Northeast Forestry University and it will be made available by the corresponding author without undue reservation.
References
Mills, K. E., Pershing, A. J., Sheehan, T. F. & Mountain, D. Climate and ecosystem linkages explain widespread declines in North American Atlantic salmon populations. Glob. Change Biol. 19, 3046–3061. https://doi.org/10.1111/gcb.12298 (2013).
Simon-Nutbrown, C. et al. Species distribution modeling predicts significant declines in coralline algae populations under projected climate change with implications for conservation policy. Front. Mar. Sci. 7 https://doi.org/10.3389/fmars.2020.575825 (2020).
Chitwood, M. C., Baruzzi, C. & Lashley, M. A. Ecology of fear in ungulates: opportunities for improving conservation. Ecol. Evol. 12 https://doi.org/10.1002/ece3.8657 (2022).
Velamazan, M. et al. Contrasting effects of wild and domestic ungulates on fine-scale responses of vegetation to climate and herbivory. Landscape Ecol. 38, 3463–3478. https://doi.org/10.1007/s10980-023-01676-0 (2023).
Jung, T. S., Stotyn, S. A. & Czetwertynski, S. M. Dietary overlap and potential competition in a dynamic ungulate community in Northwestern Canada. J. Wildl. Manage. 79, 1277–1285. https://doi.org/10.1002/jwmg.946 (2015).
Odadi, W. O., Karachi, M. K., Abdulrazak, S. A. & Young, T. P. African wild ungulates compete with or facilitate cattle depending on season. Science 333, 1753–1755. https://doi.org/10.1126/science.1208468 (2011).
Espunyes, J. et al. Different effects of alpine Woody plant expansion on domestic and wild ungulates. Glob. Change Biol. 25, 1808–1819. https://doi.org/10.1111/gcb.14587 (2019).
Mysterud, A. Ungulate migration, plant phenology, and large carnivores: the times they are a-changin’. Ecology 94, 1257–1261. https://doi.org/10.1890/12-0505.1 (2013).
Ohtaishi, N. & Sheng, H. Deer of China: Biology and Management: Proceedings of the International Symposium on Deer of China, Held in Shanghai, China,Elsevier,. (1993).
Hu, J., Fang, S. G. & Wan, Q. H. Genetic diversity of Chinese water deer (Hydropotes inermis inermis): implications for conservation. Biochem. Genet. 44, 161–172. https://doi.org/10.1007/s10528-006-9020-7 (2006).
Xu, H., Zheng, X. & Lu, H. Impact of human activities and habitat changes on distribution of Chinese water deer along the Coast Arsa in Northern Jiangsu. Acta Theriol. Sinica. 18, 161–167. https://doi.org/10.16829/j.slxb.1998.03.001 (2006).
Li, Z. et al. Distribution update of water deer (Hydropotes inermis) and prediction of their potential distribution in Northeast China. Sci. Rep. 13, 5610. https://doi.org/10.1038/s41598-023-32314-z (2023).
Guo, G. & Zhang, E. Diet of the Chinese water deer (Hydropotes inermis)in Zhoushan Archipelago, China. Acta Theriol. Sinica. 25, 122–130. https://doi.org/10.3969/j.issn.1000-1050.2005.02.003 (2005).
Hofmann, R. in Biology of deer production. Proceedings of an International Conference held at Dunedin, New Zealand, 13–18 February 1983. 393–407.
Hofmann, R., Ludwig, K. O. C. K. R. A., Axmacher, H. & J. & Seasonal changes in rumen papillary development and body condition in free ranging Chinese water deer (Hydropotes inermis). J. Zool. 216, 103–117 (1988).
Höss, M., Kohn, M., Pääbo, S., Knauer, F. & Schröder, W. Excrement analysis by PCR. Nature 359, 199–199. https://doi.org/10.1038/359199a0 (1992).
Filella, J. B., Bonilla, Q., Quispe, C. C., Dalerum, F. & E. & Artificial intelligence as a potential tool for micro-histological analysis of herbivore diets. Eur. J. Wildl. Res. 69, 11. https://doi.org/10.1007/s10344-022-01640-4 (2023).
Zuberogoitia, I. et al. Ungulate vehicle collisions in a Peri-Urban environment: consequences of transportation infrastructures planned assuming the absence of ungulates. Plos One. 9 https://doi.org/10.1371/journal.pone.0107713 (2014).
Li, Z., Liu, Z., Mi, S., Wu, J. & Teng, L. Habitat selection of the Chinese water deer at Baishan musk deer natural reserve in spring and summer. Acta Ecol. Sin. 41, 1625–1633. https://doi.org/10.5846/stxb201911122385 (2021).
Gao, H. Studies on Population Genetics and Factors Driving Differentiation of Alashan Red Deer (Northeast Forestry University, 2021).
Forsyth, D. M. & Davis, N. E. Diets of Non-Native deer in Australia estimated by macroscopic versus Microhistological rumen analysis. J. Wildl. Manage. 75, 1488–1497. https://doi.org/10.1002/jwmg.179 (2011).
Luo, Y. Compared Study on the Diet, Habitat Selection of Sympatric Blue sheep(Pseudois nayaur) and Red deer(Cervus elaphus alxaicus) in Helan Mountains, China Doctor thesis, Northeast Forestry University, (2011).
Ding, Y. Study on Diets Analysis of Water Deer (Hydropotes inermis) in the Baishan Musk Deer National Nature Reserve in Jilin Province (Northeast Forestry University, 2022).
Zhou, L. Winter Nutritional Ecological Pattern of Wild Red Deer in the Forest-Grassland Ecotone in Inner Mongolia (Northeast Forestry University, 2022).
Bradski, G. The OpenCV library. Dr Dobb’s Journal: Softw. Tools Prof. Program. 25, 120–123. https://elibrary.ru/item.asp?id=4934581
Islam, Q. N. Mastering PyCharm (Packt Publishing Ltd, 2015).
Shorten, C. & Khoshgoftaar, T. M. A survey on image data augmentation for deep learning. J. Big Data. 6 https://doi.org/10.1186/s40537-019-0197-0 (2019).
Zeng, W. & Li, M. Crop leaf disease recognition based on Self-Attention convolutional neural network. Comput. Electron. Agric. 172, 105341. https://doi.org/10.1016/j.compag.2020.105341 (2020).
He, K., Zhang, X., Ren, S. & Sun, J. in Proceedings of the IEEE conference on computer vision and pattern recognition. 770–778.
Shafiq, M. & Gu, Z. Deep residual learning for image recognition: A survey. Appl. Sciences-Basel. 12 https://doi.org/10.3390/app12188972 (2022).
Ismael, A. M. & Şengür, A. Deep learning approaches for COVID-19 detection based on chest X-ray images. Expert Syst. Appl. 164, 114054. https://doi.org/10.1016/j.eswa.2020.114054 (2021).
Xu, S. et al. Pig face recognition based on trapezoid normalized pixel difference feature and trimmed mean attention mechanism. IEEE Trans. Instrum. Meas. 72, 1–13. https://doi.org/10.1109/TIM.2022.3232093 (2022).
Xu, Y. & Goodacre, R. On splitting training and validation set: A comparative study of Cross-Validation, bootstrap and systematic sampling for estimating the generalization performance of supervised learning. J. Anal. Test. 2, 249–262. https://doi.org/10.1007/s41664-018-0068-2 (2018).
Byra, M. et al. Transfer learning with deep convolutional neural network for liver steatosis assessment in ultrasound images. Int. J. Comput. Assist. Radiol. Surg. 13, 1895–1903 (2018).
Oksanen, J. Vegan: an introduction to ordination. (2015). https://doi.org/10.1109/WSC.1999.823070
Wickham, H. ggplot2. Wiley Interdisciplinary Reviews: Comput. Stat. 3, 180–185. https://doi.org/10.1002/wics.147 (2011).
Moberly, J. G., Bernards, M. T. & Waynant, K. V. Key features and updates for origin 2018. J. Cheminform. 10, 1–2. https://doi.org/10.1186/s13321-018-0259-x (2018).
Ihaka, R. & Gentleman, R. R: a Language for data analysis and graphics. J. Comput. Graphical Stat. 5, 299–314. https://doi.org/10.1080/10618600.1996.10474713 (1996).
Zhong, L. Winter Foraging and Nutritional Strategy, Feeding Habitat Suitability Assessment of Sympatric Red Deer (Cervus elaphus) and Sika Deer (Cervus nippon) in the Muling Forest Region, Northeast China (Northeast Forestry University, 2020).
Seto, T., Matsuda, N., Okahisa, Y. & Kaji, K. Effects of population density and snow depth on the winter diet composition of Sika deer. J. Wildl. Manage. 79, 243–253. https://doi.org/10.1002/jwmg.830 (2015).
Taillon, J., Sauve, D. G. & Cote, S. D. The effects of decreasing winter diet quality on foraging behavior and life-history traits of white-tailed deer fawns. J. Wildl. Manage. 70, 1445–1454. https://doi.org/10.2193/0022-541x(2006)70 (2006). [1445:Teodwd]2.0.Co;2.
Kim, J., Joo, S. & Park, S. Diet composition of Korean water deer (Hydropotes inermis argyropus) from the Han river estuary wetland in Korea using fecal DNA. Mammalia 85, 487–493. https://doi.org/10.1515/mammalia-2021-0010 (2021).
Lee, S. K., Woo, C., Lee, E. J. & Yamamoto, N. Using high-throughput sequencing to investigate the dietary composition of the Korean water deer (Hydropotes inermis argyropus): a Spatiotemporal comparison. Sci. Rep. 12, 22271. https://doi.org/10.1038/s41598-022-26862-z (2022).
Huang, Y. Food-habits of Chinese water deer (Hydropotes inermis) in Poyang Lake area Master thesis, Jiangxi Normal University, (2016).
Soininen, E. M. et al. Analysing diet of small herbivores: the efficiency of DNA barcoding coupled with high-throughput pyrosequencing for Deciphering the composition of complex plant mixtures. Front. Zool. 6, 1–9. https://doi.org/10.1186/1742-9994-6-16 (2009).
Allen, G. M. The Mammals of China and Mongolia (American Museum of Natural History, 1938).
Jo, Y. S., Baccus, J. T. & Koprowski, J. L. Mammals of Korea: a review of their taxonomy, distribution and conservation status. Zootaxa 4522, 1–216. https://doi.org/10.11646/zootaxa.4522.1.1 (2018).
Geist, V. Deer of the World: their Evolution, Behaviour, and Ecology (Stackpole Books, 1998).
Kim, H. R., Kim, E. K., Jeon, M. G. & Park, Y. C. Intraspecific phylogeny of the Korean water deer, Hydropotes inermis argyropus (Artiodactyla, Cervidae). Anim. Syst. Evol. Divers. 31, 118–121. https://doi.org/10.5635/ASED.2015.31.2.118 (2015).
Norouzzadeh, M. S. et al. Automatically identifying, counting, and describing wild animals in camera-trap images with deep learning. Proc. Natl. Acad. Sci. U.S.A. 115, E5716–E5725. https://doi.org/10.1073/pnas.1719367115 (2018).
Beery, S., Van Horn, G. & Perona, P. in Proceedings of the European conference on computer vision (ECCV). 456–473.
Anderson, K. & Gaston, K. J. Lightweight unmanned aerial vehicles will revolutionize Spatial ecology. Front. Ecol. Environ. 11, 138–146. https://doi.org/10.1890/120150 (2013).
Pinho, C., Kaliontzopoulou, A., Ferreira, C. A. & Gama, J. Identification of morphologically cryptic species with computer vision models: wall lizards (Squamata: lacertidae: Podarcis) as a case study. Zool. J. Linn. Soc. 198, 184–201. https://doi.org/10.1093/zoolinnean/zlac087 (2023).
Wang, H. et al. Automatically detecting the wild giant panda using deep learning with context and species distribution model. Ecol. Inf. 72 https://doi.org/10.1016/j.ecoinf.2022.101868 (2022).
Carl, C., Schoenfeld, F., Profft, I., Klamm, A. & Landgraf, D. Automated detection of European wild mammal species in camera trap images with an existing and pre-trained computer vision model. Eur. J. Wildl. Res. 66 https://doi.org/10.1007/s10344-020-01404-y (2020).
Kim, B. J., Lee, N. S. & Lee, S. D. Feeding diets of the Korean water deer (Hydropotes inermis argyropus) based on a 202 bp RbcL sequence analysis. Conserv. Genet. 12, 851–856. https://doi.org/10.1007/s10592-011-0192-2 (2011).
Acknowledgements
We thank the assistance supplied by the Hongshi Forestry Bureau, Helong Forestry Bureau, Changbai Forest Management Bureau, Linjiang Forestry Bureau, Baishan Forestry Bureau, Ji’an Forestry Bureau, Benxi Forestry Development Service Center, Kuandian Manchu Autonomous County Forestry and Grassland Bureau, and Fengcheng forestry and grassland Bureau.
Funding
This research was funded by “National Natural Science Foundation of China” (Grant nos. 32071649, 32070519).
Author information
Authors and Affiliations
Contributions
LZZ, LZS, YZC and TLW conceived the ideas and designed the study and its methods, LZZ, ZMC, LJM and LZS performed data analyses, prepared figures and tables, and wrote the manuscript, LZZ, LZS, RHK, LZY, SJ and HXZ conducted the fieldwork and data collection, CC, JJ, LZS, and TLW interpreted the results and edited the manuscript to its final submitted draft. All authors contributed to the article and approved the submitted version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, Z., Yao, Z., Zhang, M. et al. The dietary patterns of water deer recently rediscovered in Northeast China exhibit remarkable similarities to those observed in other regions. Sci Rep 15, 9351 (2025). https://doi.org/10.1038/s41598-025-92473-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-92473-z