Abstract
Sepsis is closely linked to immunity. Our research aimed to identify key genes associated with sepsis immunity utilizing single-cell RNA sequencing (scRNA-seq) data. This study obtained the GSE167363 and GSE54514 datasets from the Gene Expression Omnibus (GEO). The GSE167363 dataset was subjected to cluster analysis, cell proportion analysis, cell interaction analysis, and gene set enrichment analysis (GSEA). The differentially expressed genes (DEGs) of CD8+ T cells were intersected with the DEGs in the GSE54514 dataset, and key genes related to immunity in sepsis patients were identified through Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis. Finally, we validated the gene expression levels in a mouse model of sepsis caused by cecum ligation and puncture (CLP). Findings indicated that intercellular communication of Cytotoxic CD8+ T cells was reduced in the sepsis survivors compared to non-survivors. The expression of 3 down-regulated key DEGs (ITGB2, SELL and ICAM3) was negatively correlated with the abundance of CD8+ T cells. Moreover, Cytotoxic CD8+ T cells with low expression of ITGB2, SELL and ICAM3 were more adverse to the survival of sepsis as compared to those with high expression of the above genes. These genes may predict increased survival in sepsis by regulating intercellular communication in cytotoxic CD8+ T cells, suggesting that they are potential therapeutic targets for improving sepsis prognosis.
Similar content being viewed by others
Introduction
Sepsis is a critical condition characterized by organ dysfunction resulting from an imbalanced host inflammatory response to infection1, which can involve multiple organs, leading to organ damage or failure. The prevalence of sepsis, a critical illness, is a substantial threat to human health. According to epidemiological reports, sepsis has been recognized as a global health burden since 2017 due to its high morbidity and mortality. To date, approximately 20% of annual deaths globally are associated with sepsis, which greatly affects quality of life2.
Since the pathogenesis of sepsis is still unclear, an increasing number of studies have begun to explore the key genes involved in sepsis pathogenesis by using second-generation sequencing technology, providing potential possibilities for the treatment of sepsis and improving the prognosis of sepsis patients3. For example, transcriptome sequencing has been used to identify pivotal genes in adult patients with sepsis4, and single-cell RNA sequencing (scRNA-seq) has been used to characterize the status of various immune cells during the development of sepsis5. Undoubtedly, the rapid development of bioinformatics has greatly promoted the process of exploring the pathogenesis of sepsis.
In recent years, studies have shown that the immune response is crucial for the development of sepsis6. With in-depth research on immunity, immunotherapy has also been proven to be an effective method for treating sepsis7. Therefore, exploring the role of immune cells in sepsis has become a hot research topic. Studies have shown that autophagy can induce neutrophils to form neutrophil extracellular traps during sepsis, and increased neutrophil autophagy can improve the survival rate of patients with sepsis8,9. Moreover, studies have confirmed that Sestrin2 has the potential to improve the prognosis of sepsis patients by inhibiting the pyroapoptosis of dendritic cells10.
Qiu X et al. extracted peripheral blood mononuclear cells (PBMCs) from healthy controls (HCs) and survivors and nonsurvivors of sepsis at 0 h and 6 h for single-cell RNA sequencing (scRNA-seq) (GSE167363) to explore the dynamic changes in human single-cell transcription characteristics during sepsis11. As a result, PBMCs have been proven to play key roles in the immune response to infection and have been widely used in the scRNA-seq of sepsis12. Parnell et al. extracted whole blood from HCs and sepsis survivors and nonsurvivors for RNA-seq (GSE54514) and identified key regulatory genes in the whole blood of patients with sepsis to monitor potential immune dysfunction13. Our study used 6 samples from the GSE167363 dataset, including 2 HCs, 2 survivors, and 2 nonsurvivors of sepsis. Bioinformatics analysis was performed on 163 samples from 18 HCs, 26 sepsis survivors, and 9 sepsis nonsurvivors in the GSE54514 dataset. Key genes related to immunity in sepsis patients were identified through cell proportion analysis, cell interaction analysis, gene set enrichment analysis (GSEA), immune score analysis, etc. Finally, a mouse model of cecal ligation and puncture (CLP) sepsis was constructed to detect the expression levels of these key genes through qRT-PCR.
Materials and methods
Acquisition of scRNA-seq data
The human scRNA-seq dataset GSE167363 was downloaded from the GEO database. Six scRNA-seq datasets obtained from human PBMCs were selected, including 2 healthy controls (GSM5102900、GSM5102901), 2 survivors (GSM5102902、GSM5102903) and 2 nonsurvivors of sepsis (GSM5102904、GSM5102905). The microarray dataset GSE54514 was downloaded, and 163 whole-blood RNA expression datasets were selected from 18 HCs and 26 sepsis survivors and 9 sepsis nonsurvivors.
Analyzing the scRNA-seq data
We used the “Seurat” package (v3.2.2) to analyze the GSE167363 dataset. First, the quality control of the cells was carried out based on the following criteria: (1) we excluded genes detected in < 5 cells; (2) we eliminated cells that had fewer than 200 genes; and (3) we removed cells with mitochondrial gene expression ≥ 10%. Subsequently, the gene expression of the remaining cells was standardized using a linear regression model. The samples were subjected to batch effect correction using the IntegrateData method of the “Seurat” package (v3.2.2), and 6 samples were integrated. A total of 23,881 cells and 20,078 genes were included in the processed data. Principal component analysis (PCA) was employed to identify the dimensions that were statistically significant, with a P value less than 0.05. After that, the t-distributed stochastic neighbor embedding (t-SNE) algorithm was utilized to reduce the dimensionality of 30 initial principal components (PCs) while performing cluster classification analysis. Finally, the cell clusters were classified and annotated according to the marker genes of the cells14.
Cell proportion analysis
We compared the proportions of different cells among HCs, sepsis survivors and nonsurvivors.
Analysis of intercellular interactions
Using the R package “CellChat”, GSE167363 among HCs and among sepsis survivors and non-survivors data were analysed for cell-to-cell communication, selecting all the communication pathway information in the database, including: Secreted Signaling, ECM-Receptor, Cell-Cell Contact, Heterodimers, Others, KEGG15,16,17, and Literature. The differences between survivors and nonsurvivors of sepsis were compared.
GSEA
The Fgsea package was used for GSEA of cytotoxic CD8+ T cells in HCs, sepsis survivors and nonsurvivors, and C2 in the Msigdb (https://www.gsea-msigdb.org/gsea/index.jsp) was selected as the database.
Analysis of DEGs in cytotoxic CD8+ T cells
Using the Seurat FindMarkers (CD8A) function, the DEGs of cytotoxic CD8+ T cells were compared between the sepsis survivors and nonsurvivors, with the parameters logfc.threshold = 0.25 and min.pct = 0.2.
Analysis of DEGs in the GSE54514 dataset
Limma (v3.16.1)18was used to analyze the DEGs between sepsis survivors and nonsurvivors in the GSE54514 dataset, with |logFC| > 0.5 and adj. P < 0.05 was used as a parameter.
Immune cell score
Gene set variation analysis (GSVA) was used to quantify by applying a single-sample gene set enrichment analysis (ssGSEA) score on a set of genes contained in the pathway.
Identification of key genes
The CD8+ T cell related differential genes were intersected with the differential genes computed by GSE54514 taking the intersection. And R package “clusterProfiler(v4.12.6)” was used for KEGG enrichment analysis of these DEGs15,16,17,19.
Mouse model of CLP-induced sepsis
Male C57BL/6 mice, aged 3–4 weeks, were routinely fed and kept at 22 °C for 12 h with light and dark cycles. Mice with sepsis were subjected to CLP 1 week after adaptation. There were 22 mice in the sepsis group and 6 mice in the sham group. After abdominal anesthesia (Zoletil, 40 mg/kg), CLP was performed on all the mice (except for the Sham group), and the abdominal cavity was closed before subcutaneous injection of normal saline (37 °C, 50 mL/kg) for resuscitation.
The mice used in this study were obtained from the Laboratory Animal Center of Zhejiang (Hangzhou, China). We used carbon dioxide (CO2) inhalation equipment to carry out euthanasia. We placed the mice in a chamber during euthanasia, gradually supplying CO2 to raise its concentration. The euthanasia personnel were required to observe the procedure and pause for a minimum of 1 min once they detected no movement, visible breathing, or heartbeat. The trials received approval from the Institutional Animal Care and Use Committee, ZJCLA (No. ZJCLA-IACUC-20010069). Animal experiments were complied with the ARRIVE guidelines and all methods were performed in accordance with the relevant guidelines and regulations.
Determination of the mold results
24 h after CLP surgery, we collected lung and kidney tissue from the mice. The tissue samples were preserved in a solution containing 4% paraformaldehyde, embedded in paraffin, and finally sliced into 5-micrometer-thick sections. The sections were stained with H&E to examine morphological damage under a microscope.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Twenty-four hours after CLP surgery, the mice were categorized into two different groups based on their mortality: the survivor group and the nonsurvivor group. We used cardiac blood collection of 0.5 mL, and PBMCs were extracted using Ficoll solution (Solarbio Life Sciences, Beijing, China). Then, RNA was extracted from the PBMCs via the Trizol method for qRT-PCR. Supplementary Table S1 shows the primers used.
Results
Single-cell transcriptome profiling of PBMCs
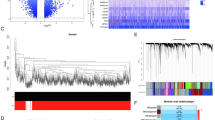
he scRNA-seq data for PBMCs were acquired from the GSE167363 database in the GEO database. A total of 13566, 4573 and 5547 cells were obtained from Hcs and survivors and nonsurvivors of sepsis, respectively, for scRNA analysis. Then, the t-SNE algorithm was used to reduce the dimensionality of the 30 initial PCs, and cluster analysis was performed on all cells (Fig. 1A-B). Based on our PCA and t-SNE results, PBMCs were classified into multiple clusters and annotated as dendritic cells, B cells, activated CD4 + T cells, natural killer (NK) cells, naive T cells, cytotoxic CD8+ T cells, platelet cells, or monocytes via marker genes14 (Fig. 1C).
Cell proportion analysis
Figure 2A shows the percentage of cells expressing characteristic markers and their scaled relative expression values in distinct cell clusters. The results were as follows: (1) monocytes characterized by high expression of S100A4 and CST3; (2) platelet cells specifically expressing the cell marker PPBP; (3) cytotoxic CD8+ T cells highly expressing CD8A; (4) naive T cells with high expression of IL7R; (5) NK cells specifically expressing the markers GNLY, NKG7, GZMA, and KLRF1; (6) activated CD4 + T cells expressing CD4, FOXP320; (7) B cells specifically expressing the cell marker MS4A1; and (8) dendritic cells with high expression of LYZ. The proportions of cell subsets were significantly different among the healthy controls and survivors and nonsurvivors of sepsis (Fig. 2B), indicating heterogeneity and consistency among these sepsis samples.
Potential cell-cell interactions assessment in patients with sepsis
To characterize the intercellular communication networks of the normal controls and the survivors and nonsurvivors of sepsis, a dataset of human ligand‒receptor pairs was used to construct a dense intercellular communication network (Fig. 3A-C). Our results highlighted that most cell‒cell interactions were between cytotoxic CD8+ T cells and monocytes in the healthy samples. A comparison investigation was performed to examine the intercellular interactions between the survivors and nonsurvivors of sepsis (Fig. 3D). The red edge indicates much stronger intercellular interactions in the nonsurvivors of sepsis, while the blue edge indicates stronger cell‒cell communication in the survivors of sepsis. These results showed that intercellular communication among cytotoxic CD8+ T cells was reduced in sepsis survivors compared to sepsis nonsurvivors. Figure 4 illustrates the ligand‒receptor or signaling pathways mediating intercellular communication in the normal controls and in the survivors and nonsurvivors of sepsis.
Circle network diagram illustrating important potential cell-cell interactions pathways in HCs. (A) Healthy samples; (B) survived samples; (C) Non-survived samples; (D) Network representing the difference in potential cell-cell interactions between survived samples and non-survived samples, with the red edge indicating stronger intercellular interactions in the sepsis nonsurvivors and the blue edge representing stronger potential cell-cell interactions in the sepsis survivors.
Enrichment of pathways in cytotoxic CD8+ T cells
To observe the effect of cytotoxic CD8+ T cells on sepsis, we used GSEA to analyze the enrichment of the KEGG pathways in the survivors and nonsurvivors of sepsis. The results showed that the ribosome, spliceosome, and RNA degradation pathways were highly enriched in cytotoxic CD8+ T cells from the survivors of sepsis. Notably, the ribosome, NK cell-mediated cytotoxicity, graft versus host disease and antigen processing and presentation pathways were highly enriched in cytotoxic CD8+ T cells in the nonsurvivors of sepsis (Fig. 5).
DEGs in the GSE167363 and GSE54514 datasets
To further clarify the molecular mechanism by which cytotoxic CD8+ T cells affect the survival of patients with sepsis, we identified DEGs in cytotoxic CD8+ T cells between the survivors and nonsurvivors of sepsis by FindMarker (min.pct = 0.2, logfc.threshold = 0.25), and a total of 582 DEGs were identified (Fig. 6A). Moreover, DEGs between sepsis survivors and nonsurvivors in the GSE54514 dataset were analyzed with the limma package. With the criteria of |logFC| > 0.5 and P < 0.05, 210 DEGs were obtained (110 upregulated DEGs and 100 downregulated DEGs) (Fig. 6B). DEGs that were simultaneously upregulated or downregulated in both cytotoxic CD8+ T cells and the GSE54514 dataset are shown in the Venn diagram. Finally, 12 DEGs were obtained (3 upregulated DEGs and 9 downregulated DEGs) (Fig. 6C-D) (Table 1).
(A) Heatmap showing DEGs in cytotoxic CD8+ T cells between sepsis survivors and sepsis nonsurvivors (GSE167363). (B) Volcano plot showing DEGs between the survivors and nonsurvivors of sepsis in the GSE54514 dataset. Genes upregulated or downregulated by more than 0.5-fold are shown in red. (C, D) Venn diagram of the overlapping upregulated (C) and downregulated (D) DEGs based on the DEGs identified in the Cytotoxic CD8+ T cell (GSE167363) and GSE54514 datasets.
KEGG enrichment analysis
Then, these 12 overlapping DEGs were subjected to KEGG enrichment analysis. The pathway that was shown to be significantly enriched was cell adhesion molecules. Among the 12 overlapping DEGs, ITGB2, SELL and ICAM3 were enriched in the pathway of cell adhesion molecules (Fig. 7A; Table 2), with downregulated expression in the nonsurvivors (Fig. 7B).
Correlation of ITGB2, SELL and ICAM3 expression with immune infiltration and cytotoxic CD8+ T cells
29 immune-associated gene sets representing distinct types, functions, and pathways of immune cells were analyzed by ssGSEA21, and the Wilcoxon test was used to assess the disparity between sepsis survivors and nonsurvivors (Fig. 8A). Then, we examined the relationships between the expression levels of ITGB2, SELL, and ICAM3 and the level of infiltration by CD8+ T cells. The abundance of CD8+ T cells was negatively correlated with the expression of ITGB2, SELL, and ICAM3 (Fig. 8B-D).
To elucidate the roles and importance of ITGB2, SELL and ICAM3, we determined the expression of ITGB2, SELL and ICAM3 in cytotoxic CD8+ T cells from survivors and nonsurvivors of sepsis. As shown in Fig. 8E, downregulated ITGB2, SELL and ICAM3 were significantly correlated with sepsis survivors and nonsurvivors. Combined with the above findings, these results suggested that cytotoxic CD8+ T cells with low expression of ITGB2, SELL and ICAM3 were more likely to have adverse effects on the survival of patients with sepsis than were those with high expression of the above genes.
(A) ssGSEA of immune cells between survivors and nonsurvivors (GSE54514, Include 26 sepsis survivors and 9 sepsis nonsurvivors). (B-D) The correlation diagrams showing the correlation between CD8+ T cells and the expression of ITGB2, SELL and ICAM3 (GSE54514). (E) The expression of ICAM3, ITGB2 and SELL in cytotoxic CD8+ T cells between survivors and nonsurvivors of sepsis (GSE167363, Include 2 cases of survivors and nonsurvivors).
Mouse model of CLP-induced sepsis
Determination of the mouse model of CLP-induced sepsis
To verify the above results, a mouse model of CLP-induced sepsis was constructed. 24 h after CLP surgery, lung and kidney tissues were collected for H&E staining. Figure 9A shows that the lungs of CLP mice were infiltrated with an enormous amount of inflammatory cells, and some alveolar cavities were fused into pulmonary bullae with interstitial hyperemia and edema. Figure 9B shows a high level of leukocyte infiltration in the renal interstitium of CLP mice and hyperemia of the glomerular capillaries. These results suggested that the mouse model of CLP-induced sepsis was successfully constructed.
The mRNA expression of ITGB2, SELL and ICAM3 in the CLP mouse model
After the mouse model of CLP-induced sepsis was successfully generated, the PBMCs of all 22 model mice were obtained, including 15 from the group of survivors and 7 from the group of nonsurvivors. Then, we detected the expression levels of ITGB2, SELL, and ICAM3 by qRT-PCR. Figure 10 shows that, in comparison to those in survivors, the expression levels of ITGB2 and ICAM3 were substantially lower in nonsurvivors, while there was no significant difference in the expression of SELL between the two groups.
Discussion
Sepsis is a systemic inflammatory syndrome caused by infectious diseases and is closely associated with the pathophysiological changes that occur in various systems and organs22. Recent studies have shown that sepsis is closely related to immunity23,24. Previous studies have shown that sepsis is associated with severe and sustained immunosuppression, immune dysfunction is considered to be the core mechanism of sepsis, and immune response disorders are susceptibility factors for secondary infection and increased mortality25. Therefore, in this study, scRNA-seq data from human PBMCs were used for cluster analysis, and the results were annotated as dendritic cells, B cells, activated CD4 + T cells, NK cells, naive T cells, cytotoxic CD8+ T cells, platelet cells, and monocytes. Subsequently, we performed a cell proportion analysis, and significant changes were revealed in the proportions of immune cells among normal controls, as well as survivors and nonsurvivors of sepsis, which further confirmed the crucial role of immune cells in sepsis pathogenesis.
On this basis, we constructed an intercellular communication network to investigate the correlations among these immune cells. The intercellular communication of cytotoxic CD8+ T cells was more significant in sepsis nonsurvivors than in sepsis survivors, which implies that enhanced cytotoxic CD8+ T-cell communication may be significantly associated with reduced survival in sepsis patients. As a type of specific T cell, CD8+ T cells can secrete a variety of cytokines to participate in the immune response and resist the invasion of some foreign pathogens26. Some studies have shown that increased interaction between platelets and CD8+ T cells is strongly linked to an unfavorable prognosis in sepsis patients27. However, studies on the mechanism of action of cytotoxic CD8+ T cells in sepsis and the correlation of cytotoxic CD8+ T cells with the prognosis of sepsis patients are lacking. It may be due to the fact that studies related to sepsis and immunity have focused on immune homeostasis, so there are relatively few CD8+ T cell-related studies28. In this study, we suggested that the intercellular interactions of cytotoxic CD8+ T cells are closely associated with the prognosis of sepsis patients. Subsequently, cytotoxic CD8+ T cells were chosen for further analysis to investigate the key genes associated with cytotoxic CD8+ T cells in sepsis.
Subsequently, GSEA was performed on the cytotoxic CD8+ T cells from both the sepsis survivors and nonsurvivors. The enrichment functions and pathways were revealed to be inconsistent between these two groups, which also confirmed the heterogeneity of cytotoxic CD8+ T cells in sepsis patients with different prognoses. Next, the DEGs of cytotoxic CD8+ T cells were screened from both the sepsis survivors and nonsurvivors and intersected with the DEGs from the survivors and nonsurvivors of sepsis in the GSE54514 dataset. Finally, 12 DEGs were obtained, and KEGG enrichment results showed that cell adhesion molecules were most significantly correlated with these DEGs. Cell adhesion molecule (CAM) is a collective term referring to various molecules that facilitate cellular interactions and adhesion, either between cells or between cells and the extracellular matrix (ECM). It serves as the molecular foundation for a range of significant physiological and pathological processes29. It has been confirmed in the literature that CAM affects sepsis and its complications30. Some studies have shown that FAM46C can antagonize cardiac dysfunction caused by sepsis by downregulating CAM and inhibiting apoptosis. It has also been found that silencing or inhibiting PFKFB3 can significantly downregulate the expression of ICAM-1 and vascular cell adhesion molecule-1 (VCAM-1) in sepsis, thus improving acute lung injury induced by sepsis31. Moreover, a large number of studies have recognized the diagnostic and predictive value of ICAM-1 in sepsis through bioinformatics analysis32,33. Therefore, we selected the downregulated genes of ITGB2, SELL, and ICAM3 that are enriched in the pathway of cell adhesion molecules for further study.
To further verify the scientific nature of the above results, a mouse model of CLP-induced sepsis was constructed, and PBMCs were obtained from both survivors and nonsurvivors of sepsis 24 h after surgery for qRT‒PCR. ITGB2 and ICAM3 expression levels were significantly lower in nonsurvivors than in survivors, but there was no significant difference in the SELL between the two groups.
ITGB2 has been reported to affect the progression of sepsis by participating in neutrophil recruitment, and defects in this gene can lead to defects in leukocyte adhesion in sepsis34. Unfortunately, the correlation between the expression of ITGB2 and the infiltration of immune cells other than neutrophils during the progression of sepsis has not been widely studied. For example, effective interactions between T cells and their targets depend on T-cell receptor-mediated ITGB2 activation35. Similarly, the interaction between ITGB2 and cytotoxic CD8+ T cells in the pathogenesis of sepsis has not been fully studied. In addition, ICAM3 belongs to the family of intercellular adhesion molecules and is the ligand of the leukocyte adhesion protein LFA-1 (integrin αL/β2)36. Some studies indicate that ICAM3 is critical for immune cell interactions and T lymphocyte activation37,38. Numerous studies have proposed that ICAM3 mediates inflammatory signaling, thereby promoting cancer cell stemness39. Dendritic cell-specific ICAM3-grabbing nonintegrin (DC-sign) inhibition plays a protective role in sepsis-associated organ injury and systemic inflammation40. However, the relationship between ICAM3 and cytotoxic CD8+ T cells in sepsis has not been elucidated.
ITGB2, also known as integrin β2, is considered a key regulator of neonatal sepsis and is essential for the progression of this condition41. In this study, we supposed that ITGB2 and ICAM3 are strongly associated with cytotoxic CD8+ T cell intercellular communication and are associated with reduced survival in patients with sepsis. Targeting ICAM3 and ITGB2 may affect the prognosis of sepsis patients by affecting the intercellular communication of cytotoxic CD8+ T cells.
However, there are still many defects in this study. First, this study is based on single-cell sequencing data, which tends to have fewer samples. Then we only validated the expression levels of key genes in CLP mice with sepsis, not in a larger clinical sequence. Second, the mechanism of action of cytotoxic CD8+ T cells in sepsis has not been thoroughly explored, and the predictive significance of key genes for the prognosis of sepsis needs to be further verified. Last, there are few applications in clinical practice, and we may be able to use efficient clinical subtypes to recognize sepsis and achieve personalized treatment goals42.
Conclusion
This study suggested that ITGB2 and ICAM3 could serve as biomarkers to predict survival rates in sepsis patients, primarily by decreasing intercellular communication among cytotoxic CD8+ T cells. This research demonstrated a close association between the levels of ITGB2 and ICAM3 and reduced cellular interactions, thereby contributing to improved patient survival rates. Moreover, this discovery highlights a potential therapeutic target for improving sepsis prognosis. Targeted regulation of these molecules may lead to new treatment strategies, optimizing the therapeutic outcomes for sepsis patients and ultimately boosting their survival rates and quality of life.
Data availability
The datasets used and/or analyzed during the current study are publicly available from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/) (accession number: GSE167363, GSE54514).
References
Wu, Y. et al. Immunotherapy in the context of sepsis-induced immunological dysregulation. Front. Immunol. 15, 1391395 (2024).
Rudd, K. E. et al. Global, regional, and National sepsis incidence and mortality, 1990–2017: analysis for the global burden of disease Study. Lancet 395 (10219), 200–211 (2020).
Gu, Y. et al. Exploring the efficacious constituents and underlying mechanisms of Sini Decoction for sepsis treatment through network Pharmacology and multi-omics. Phytomedicine 123, 155212 (2024).
Zhang, Q. et al. Identification of hub genes for adult patients with sepsis via RNA sequencing. Sci. Rep. 12 (1), 5128 (2022).
Reyes, M. et al. An immune-cell signature of bacterial sepsis. Nat. Med. 26 (3), 333–340 (2020).
Zhang, X. et al. The potential immunological mechanisms of sepsis. Front. Immunol. 15, 1434688 (2024).
Wang, W. et al. The role of trained immunity in sepsis. Front. Immunol. 15, 1449986 (2024).
Guo, Y. et al. Spontaneous formation of neutrophil extracellular traps is associated with autophagy. Sci. Rep. 11 (1), 24005 (2021).
Park, S. Y. et al. Autophagy primes neutrophils for neutrophil extracellular trap formation during Sepsis. Am. J. Respir Crit. Care Med. 196 (5), 577–589 (2017).
Wang, L. X. et al. Sestrin2 protects against lethal sepsis by suppressing the pyroptosis of dendritic cells. Cell. Mol. Life Sci. 78 (24), 8209–8227 (2021).
Qiu, X. et al. Dynamic changes in human single-cell transcriptional signatures during fatal sepsis. J. Leukoc. Biol. 110 (6), 1253–1268 (2021).
Wen, M. et al. Single-cell transcriptomics reveals the alteration of peripheral blood mononuclear cells driven by sepsis. Ann. Transl Med. 8 (4), 125 (2020).
Parnell, G. P. et al. Identifying key regulatory genes in the whole blood of septic patients to monitor underlying immune dysfunctions. Shock 40 (3), 166–174 (2013).
Zhang, X. et al. CellMarker: a manually curated resource of cell markers in human and mouse. Nucleic Acids Res. 47 (D1), D721–d728 (2019).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28 (1), 27–30 (2000).
Kanehisa, M. Toward Understanding the origin and evolution of cellular organisms. Protein Science: Publication Protein Soc. 28 (11), 1947–1951 (2019).
Kanehisa, M. et al. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 51 (D1), D587–d592 (2023).
Wang, Y. et al. The shared biomarkers and pathways of systemic lupus erythematosus and metabolic syndrome analyzed by bioinformatics combining machine learning algorithm and single-cell sequencing analysis. Front. Immunol. 13, 1015882 (2022).
Wu, T. et al. ClusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innov. (Camb). 2 (3), 100141 (2021).
Guo, S. et al. Integrated analysis of single-cell RNA-seq and bulk RNA-seq unravels T cell-related prognostic risk model and tumor immune microenvironment modulation in triple-negative breast cancer. Comput. Biol. Med. 161, 107066 (2023).
He, Y. et al. Classification of triple-negative breast cancers based on Immunogenomic profiling. J. Exp. Clin. Cancer Res. 37 (1), 327 (2018).
Williams, B. et al. Sepsis-Induced coagulopathy: A comprehensive narrative review of pathophysiology, clinical presentation, diagnosis, and management Strategies. Anesth. Analg. 138 (4), 696–711 (2024).
Giamarellos-Bourboulis, E. J. et al. The pathophysiology of sepsis and precision-medicine-based immunotherapy. Nat. Immunol. 25 (1), 19–28 (2024).
Cajander, S. et al. Profiling the dysregulated immune response in sepsis: overcoming challenges to achieve the goal of precision medicine. Lancet Respir Med. 12 (4), 305–322 (2024).
Padovani, C. M. & Yin, K. Immunosuppression in sepsis: Biomarkers and specialized pro-resolving mediators. Biomedicines 12(1), 175 (2024).
St Paul, M. & Ohashi, P. S. The roles of CD8(+) T cell subsets in antitumor Immunity. Trends Cell. Biol. 30 (9), 695–704 (2020).
Guo, L. et al. Platelet MHC class I mediates CD8+ T-cell suppression during sepsis. Blood 138 (5), 401–416 (2021).
Cao, M., Wang, G. & Xie, J. Immune dysregulation in sepsis: experiences, lessons and perspectives. Cell. Death Discov. 9 (1), 465 (2023).
Zhong, L., Simard, M. J. & Huot, J. Endothelial MicroRNAs regulating the NF-κB pathway and cell adhesion molecules during inflammation. Faseb J. 32 (8), 4070–4084 (2018).
Fang, Y. et al. The role of biomarkers of endothelial activation in predicting morbidity and mortality in patients with severe sepsis and septic shock in intensive care: A prospective observational study. Thromb. Res. 171, 149–154 (2018).
Wang, L. et al. Ablation of endothelial Pfkfb3 protects mice from acute lung injury in LPS-induced endotoxemia. Pharmacol. Res. 146, 104292 (2019).
Zonneveld, R. et al. Soluble adhesion molecules as markers for sepsis and the potential pathophysiological discrepancy in neonates, children and adults. Crit. Care. 18 (2), 204 (2014).
Li, X. J. et al. Accuracy of intercellular adhesion molecule-1 for diagnosing sepsis: A systematic review and meta-analysis protocol. Medicine (Baltim). 98 (24), e16019 (2019).
Yuki, K. & Hou, L. Role of Β2 integrins in neutrophils and sepsis. Infect. Immun. 88(6). (2020).
Dimitrov, S. et al. Gα(s)-coupled receptor signaling and sleep regulate integrin activation of human antigen-specific T cells. J. Exp. Med. 216 (3), 517–526 (2019).
Kristóf, E. et al. Novel role of ICAM3 and LFA-1 in the clearance of apoptotic neutrophils by human macrophages. Apoptosis 18 (10), 1235–1251 (2013).
Estecha, A. et al. RUNX3 regulates intercellular adhesion molecule 3 (ICAM-3) expression during macrophage differentiation and monocyte extravasation. PLoS One. 7 (3), e33313 (2012).
Montoya, M. C. et al. Role of ICAM-3 in the initial interaction of T lymphocytes and APCs. Nat. Immunol. 3 (2), 159–168 (2002).
Shen, W. et al. ICAM3 mediates inflammatory signaling to promote cancer cell stemness. Cancer Lett. 422, 29–43 (2018).
Chen, W. et al. DC-SIGN expression in intestinal epithelial cells regulates Sepsis-Associated acute intestinal injury via activating ERK1/2-NF-κB/P65 Signaling. Shock 52 (4), 434–442 (2019).
Bu, L. et al. Identification of key mRNAs and lncRNAs in neonatal sepsis by gene expression profiling. Comput. Math. Methods Med. 2020, 8741739 (2020).
Yang, J. et al. Identification of clinical subphenotypes of sepsis after laparoscopic surgery. Laparosc. Endoscopic Robotic Surg. 7 (1), 16–26 (2024).
Funding
This research was supported by grants from the Medical Science Research Foundation of Zhejiang Province (2022515073).
Author information
Authors and Affiliations
Contributions
Min Lei and Yaping Zhang designed this study, conducted bioinformatics analysis and wrote the main manuscript .Yijin Yu and text : Conceptualization, Investigation, Data curation, Funding acquisition, Formal analysis Writing – original draft.Yaping Zhang: Software, Formal analysis. Writing – review & editing.Yijin Yu: Formal analysis, Software . Gaojian Wang: Supervision. Conceptualization. Nianqiang Hu: Validation, Software, Visualization. Junran Xie: Supervision, Project administration.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics statement
Ethical approval was granted by the Institutional Animal Care and Use Committee, ZJCLA (No. ZJCLA-IACUC-20010069).
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Lei, M., Zhang, Y., Yu, Y. et al. ITGB2 and ICAM3 predict increased survival of sepsis with decreased intercellular communication in cytotoxic CD8+ T cells. Sci Rep 15, 12635 (2025). https://doi.org/10.1038/s41598-025-93685-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-93685-z