Abstract
In recent years, numerous studies have explored the relationship between atmospheric conditions and respiratory viral infections. However, these investigations have faced certain limitations, such as the use of modestly sized datasets, a restricted geographical focus, and an emphasis on a limited number of respiratory pathogens. This study aimed to develop a nationwide respiratory virus infection risk prediction model through machine learning approach. We utilized the CRFC algorithm, a random forest-based method for multi-label classification, to predict the presence of various respiratory viruses. The model integrated binary classification outcomes for each virus category and incorporated air quality and meteorological data to enhance its accuracy. The data was collected from 31 regions in China between 2016 and 2021, encompassing pathogen detection, air quality indices, and meteorological measurements. The model’s performance was evaluated using ROC curves, AUC scores, and precision-recall curves. Our model demonstrated robust performance across various metrics, with an average overall accuracy of 0.76, macro sensitivity of 0.75, macro precision of 0.77, and an average AUC score of 0.9. The SHAP framework was employed to interpret the model’s predictions, revealing significant contributions from parameters such as age, NO2 levels, and meteorological conditions. Our model provides a reliable tool for predicting respiratory virus risks, with a comprehensive integration of environmental and clinical data. The model’s performance metrics indicate its potential utility in clinical decision-making and public health planning. Future work will focus on refining the model and expanding its applicability to diverse populations and settings.
Similar content being viewed by others
Introduction
Respiratory viruses constitute a major cause of acute respiratory infections (ARI), prevalent in clinical settings and significantly contributing to global morbidity and mortality rates. For example, Respiratory Syncytial Virus and Influenza A has been identified as the main culprits behind severe acute respiratory infections in children1,2. For other viruses like human rhinoviruses (HRVs), this is less well established. Co-infection with multiple viruses refers to the presence of two or more different viruses within the same host at the same time. This phenomenon is relatively common in nature and poses a significant threat to public health3.
The COVID-19 pandemic has demonstrated the substantial impact of human respiratory virus infections, leading to a spectrum of respiratory symptoms and severe illnesses, resulting in a substantial surge in global morbidity, mortality, and economic losses4. Common respiratory viruses include influenza virus (IV), human coronaviruses (HCoV), respiratory syncytial virus (RSV), parainfluenza virus (PIV), adenovirus (ADV), human metapneumovirus (hMPV), and bocavirus (HBoV)5. Among these, IV, RSV, PIV, and hMPV are major pathogens associated with acute lower respiratory infections, imposing a significant disease burden, especially in young children and older adults. According to the World Health Organization (WHO), the influenza virus causes approximately 3 to 5 million severe cases and approximately 290,000 to 650,000 respiratory deaths worldwide6. RSV leads to an estimated 33.8 million lower respiratory infections globally each year, with 3.4 million necessitating hospitalization and at least 66,000 deaths among children under 5, with a higher prevalence in developing countries7.
The epidemiology of respiratory viruses varies not only by region, season, and year, but also appears to be influenced by environmental factors such as temperature, humidity, precipitation, air currents, and ventilation8. Additionally, air pollution has been identified as a risk factor for respiratory virus infections, with Nitrogen Oxides (NOX), Sulfur Dioxide(SO2), Fine Particulate Matter with a diameter of 2.5 micrometers or less (PM2.5), and Particulate Matter with a diameter of 10 micrometers or less (PM10) associated with increased RSV incidence9. Hence, continuous monitoring and understanding of the transmission patterns and epidemiological characteristics of respiratory viruses in a given region are crucial for controlling acute respiratory infections.
Given the diverse range of viral species causing respiratory infections, accurate identification of the causative agent is essential for effective treatment. However, conventional clinical examination methods may not promptly and accurately identify the virus, necessitating virus-specific testing. Detecting respiratory infection viruses typically involves sputum culture, blood tests, and molecular biology techniques. These methods have certain limitations, including time delays10 and technical requirements11.
The landscape of respiratory pathogen monitoring and forecasting is constantly evolving, and there is a pressing need for more sophisticated models that can accurately predict the trends of respiratory diseases. Hence, the monitoring and forecasting of respiratory viruses merit profound consideration as a way to assist researchers and clinicians in creating precise diagnostic approaches. In order to counteract the deficiency in predictive capabilities for respiratory diseases in China and to bolster clinical diagnostic efforts, we employed a chained random forest classifier (CRFC) model on a comprehensive dataset of respiratory virus tests to forecast outcomes of respiratory virus detection12,13. By integrating air quality and meteorological data, our model not only accurately predicts the occurrence of respiratory virus infections but also contributes significantly to the development of public services in China.
Results
Baseline characteristics
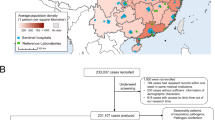
After integrating the pathogen detection, air quality, and meteorological data, a total of 528,471 entries were obtained. After selecting the pathogens of interest with positive test results, 19,161 entries between 2016-01-09 and 2021-12-22 were used for model training and testing. Furthermore, we calculated the total number of hospitals contributing pathogen data in each province and created a geographical distribution map of China to visualize the hospital locations in Fig. 1. The dataset was then split into training and test sets using the Iterative Stratification method14 which ensures an even distribution of labels across the splits by considering each label separately. For detailed inclusions, see Table 1 and “Methods”. The baseline characteristics of the training set and the test set are shown in Table 1.
In both the training and test sets, the median age of the test subjects is 2 years old. The interquartile range (IQR) is 3.17 in the training set and 3.07 in the test set. In terms of sex, males are in the majority, accounting for 60% in the training group and 59.7% in the test group, while females make up 39.3% and 39% respectively, making the male count about 1.5 times that of the females. In both the training and test sets, the number of people tested on non-holiday days is approximately four times higher than on holiday days. In both the training set and the test set, the average grade of AQI is good, and the values of all other key pollutants have not exceeded the national required concentration limits.
Box plots presented all differential continuous variables between four groups (IV, ADV, RSV, and others) in Fig. 2. AQI exhibits distinct median values for each category, with ‘IV’ showing a median of 55.04, ‘ADV’ at 50.80, ‘RSV’ at 34.11, and ‘others’ at 51.98. The IQR indicates the spread of the middle 50% of the data, with ‘RSV’ having the smallest variation and ‘IV’ the largest. In the case of CO, the IQR suggests a closer distribution of CO levels, particularly for ‘RSV’. NO2 levels have ‘IV’ with a median of 37.31 and the largest IQR, pointing to a broader spread of values. ‘ADV’ and ‘others’ have similar medians, while ‘RSV’ shows the lowest median and IQR, indicating more consistently lower levels. O3 levels are generally higher, with ‘ADV’ and ‘others’ showing notably higher medians and IQRs, suggesting a wider range of values and potential for higher peaks in ozone concentration. For PM10, ‘IV’ has a higher median and IQR, indicating more variable and generally higher levels compared to other categories. PM2.5 follows a similar trend but with lower overall median values. SO2 levels are comparatively lower, with ‘RSV’ showing the lowest median and IQR, indicating less variability and consistently lower SO2 levels. Air temperature shows a wide range of median values across categories, with ‘RSV’ having the highest and ‘IV’ the lowest. The IQR for ‘IV’ suggests a significant spread in temperatures. Dew point temperature also varies, with ‘RSV’ again showing the highest median and IQR, indicating not only higher median values but also a greater variability. Wind direction, presented as a degree measure, has the smallest IQR for ‘RSV’, suggesting less variability in wind direction within this category. ‘IV’ and ‘ADV’ have similar medians, while ‘others’ show a slightly higher spread. Wind speed is relatively consistent across categories, with medians ranging from 25.47 to 33.33 and IQRs indicating a moderate spread of values. The ‘Age’ category indicates a median patient age of 25.47 for ‘IV’, 32.13 for ‘ADV’, 33.33 for ‘RSV’, and 26.09 for ‘others’. The IQR shows a narrow age range for ‘RSV’, suggesting a more concentrated age group among these patients, while ‘ADV’ has a broader IQR, indicating a wider distribution of ages. It is worth noting that compared to other factors, NO2 exhibits a relatively large difference within the respiratory virus categories, suggesting it may have an impact on the model. Therefore, we conducted a Kruskal-Wallis H test on NO2, calculated all possible pairwise comparisons (Supplementary Table 1). The results of the pairwise comparisons between IV, ADV, RSV, and others all showed significant differences, with p-values less than 0.05. The finding was consistent with previous research15, suggesting that changes in NO2 concentrations are associated with the risk of respiratory virus infection.
A AQI; B CO; C NO2; D O3; E PM10; F PM2.5; G SO2; H air temperature; I dew point temperature; J wind direction; K wind speed; L age.The central box spans from the first quartile (Q1) to the third quartile (Q3), representing the interquartile range (IQR), which contains the middle 50% of the data. A line inside the box marks the median (Q2), which is the middle value of the dataset. Lines extend from the edges of the box to the minimum and maximum values that are not considered outliers. These lines are called whiskers. Data points that fall outside the whiskers are plotted as individual points and are considered outliers. They are typically defined as points that are more than 1.5 times the IQR below Q1 or above Q3. The ends of the whiskers represent the minimum and maximum values within the non-outlier range.
Analyzing features included in models
Twelve selected continuous variables included in the CRFC model exhibited a correlation relationship as shown in Fig. 3. The heatmap displays the correlation coefficients between pairs of variables, with darker shades indicating stronger positive or negative correlations. The correlation coefficients range from -1 (perfect negative correlation) to 1 (perfect positive correlation), with 0 indicating no correlation. The Correlation Heatmap displays a matrix of correlation coefficients that reveal the strength and direction of relationships between different environmental variables. Notably, there is a very strong positive correlation between PM10 and PM2.5, as well as between AQI and PM2.5, indicating that these particulate matters are closely linked to overall air quality. The perfect correlation between Air Temperature and Dew Point Temperature suggests they are highly interdependent measures of atmospheric conditions. On the other hand, Wind Speed shows minimal correlation with most variables, suggesting it may be less influential in the dynamics of air quality. Additionally, the moderate negative correlation between AQI and Wind Direction could imply that specific wind patterns may be associated with poorer air quality, while the negative correlation between SO2 and Age is also of moderate strength, indicating a potential inverse relationship.
The closer a color is to red end of the spectrum, the stronger the positive correlation it indicates. Similarly, the closer it is to blue end of the spectrum, the stronger the negative correlation it indicates. AQI: Air Quality Index; CO: Carbon Monoxide; NO2: Nitrogen Dioxide; PM10: Particulate Matter with a diameter of 10 micrometers or less; PM2.5: Fine Particulate Matter with a diameter of 2.5 micrometers or less; SO2: Sulfur Dioxide.
To demonstrate the importance and correlation of features in the multi-label classification model CRFC in the prediction of respiratory virus infections, we used the SHAP method to interpret our CRFC model. The mean absolute SHAP value was chosen to gain insight into the impact of features and virus types on respiratory virus prediction. Figure 4 shows the mean absolute SHAP values of the top 20 features of the 100 testing samples. The features are listed in descending order of importance, with the most influential features at the top. Year, patients’ ages, province, NO2 and air temperature are among the top five factors that affect the CRFC model’s results. Overall, year, age, and province have the most significant impact on predicting ADV infections. Compared to other viruses, patients’ ages have the greatest influence on predicting susceptibility to IV infections, while other factors have a relatively smaller impact on predicting IV infections. The year has the most significant effect on ADV and RSV.
The x-axis represents the average impact on model output magnitude and the y-axis represents the feature variables used to build the model. Different colors represent different viruses, each mapped onto the feature variables, with the more prominent feature variables having a greater impact on the model. On a single feature variable, the longer the color band, the greater the contribution value.
Building upon the variable importance insights gleaned from the previous SHAP analysis, SHAP plots were generated to specifically examine feature impacts for each respiratory virus class. As shown in Fig. 5, age emerged as one of the top five most influential factors for predicting all four virus categories. Age and SO2 also featured prominently in the top five for IV. Age and air temperature exhibited high importance for ADV classification. Air temperature was identified as among the key predictors for RSV occurrence. Notably, when considering all pathogen types collectively, age, air temperature, NO2 and SO2 surfaced as particularly crucial variables overall for distinguishing respiratory virus classification. These SHAP findings help validate the real-world influence of certain atmospheric and host-related attributes on specific viral transmission dynamics.
Figure 6 shows the SHAP dependence plots for key variables in respiratory virus prediction. Figure 6A reveals that patient age above 20 serves as an inflection point, correlating with an increased probability of influenza infection. Figure 6B indicates wind direction larger than 150°emerges as a threshold where likelihood of ADV detection decreases. To the contrary of Fig. 6C,D, patients over 20 years significantly reduced vulnerability to “others” viruses. Additionally, the risk of “other” viruses increases when the wind direction exceeds 150°, suggesting a potential protective wind-related mechanism against these pathogens below this threshold.
AThe SHAP plots for age in Group IV; B The SHAP plots for wind direction in Group ADV; C The SHAP plots for age in Group others; D The SHAP plots for wind direction in Group others. The blue dots represent the smaller values in the X-axis, and the vertical axis represents the SHAP values, where positive values indicate a positive impact on testing positive for a certain virus. The red dots represent the bigger values in the X-axis, and the vertical axis represents the SHAP values, where negative values indicate a negative impact on testing positive for a certain virus.
SHAP can not only be used to explain the overall predictive performance of a model but also to make predictions for a specific group of people as needed. The SHAP explanation plots for a specific group of people are shown in Fig. 7. After inputting information such as gender, age, and province of the population, we can generate the SHAP force plots, as detailed in Fig. 7. The SHAP Force Plot illustrates the individual level within the model CRFC. What decision each model makes and how much each attribute contributes to the decision are detailed in the SHAP force plots. The red color block represents an increased risk of infection, and blue represents a decreased risk. The longer the color band, the greater the contribution value. The text on the graph represents the demographic information entered for the population. The impact of each variable is proportionate to its bar length. The variable wind_speed, NO2, wind_direction and PM2.5 can enhance model output in the given “other” whereas the wind_speed, NO2 and SO2 can decrease prediction in the given “ADV”.
Model performance
To boost overall classification performance, classifier chains with random forest models as base learners were employed. Classifier chains treat previous virus label prediction results as input features, each random forest predicts whether each sample is associated with each virus label.
In our implementation of CRFC using this approach, the model CRFC was run 50 times to evaluate consistency. Encouragingly, CRFC achieved a mean overall accuracy of 0.76, along with mean of macro sensitivity of 0.75, mean of macro precision of 0.77, mean of macro f1-score of 0.76, mean of AUC of 0.90.
For one run (overall accuracy of 0.76, macro sensitivity of 0.75, macro precision of 0.77, macro f1-score of 0.76, macro AUC score of 0.90), further performance metrics for each virus in detailed are shown in Table 2, Table 3 and Fig. 8.
The true positive number for IV, ADV, RSV, and others are 235, 1527, 596 and 546, respectively. The true negative number for IV, ADV, RSV, and others are 3508, 1557, 2780 and 2723. The false positive number for IV, ADV, RSV, and others are 46, 454, 98 and 331. The false negative number for IV, ADV, RSV, and others are 44, 294, 358 and 232.
As for the performance evaluation metrics, for IV, the accuracy is 0.98, indicating that 98% of the predictions are correct. For ADV, it’s 0.80. RSV has an accuracy of 0.88, and for others, it is 0.85. The IV has a sensitivity of 0.84, meaning it correctly identifies 84% of the positive cases. ADV also has a sensitivity of 0.84, RSV has 0.62, and others have a sensitivity of 0.7. The IV has an impressive specificity of 0.99, indicating it correctly identifies 99% of the negative cases. ADV has a specificity of 0.77, RSV has 0.97, and others have 0.89. The IV has a precision of 0.84, ADV is at 0.77. RSV has 0.86, and others have 0.62. The IV has an f1- score of 0.84. ADV is slightly lower at 0.80, RSV has 0.72, and others have 0.66. The AUC is a performance measurement for classification problems at various threshold settings. It represents the degree or measure of separability. The higher the AUC, the better the model is at distinguishing between the two classes. IV has an AUC score of 0.99. ADV has a score of 0.9, RSV has 0.87, and others have 0.85 (Fig. 8A).
Figure 8B illustrates the ROC and Precision-Recall curves for classifier chains. The classes include IV, ADV, RSV, and Others. The Precision-Recall curves indicate the following AP values: Class IV = 0.92; Class ADV = 0.88; Class RSV = 0.80; The Others class = 0.69.
These curves are essential tools for evaluating the performance of classifiers, offering a visual representation of the trade-offs involved in choosing different thresholds for classification. A higher AUC/AP value indicates superior classifier performance.
Performance comparison between CRFC and other prediction models
To comprehensively evaluate the effectiveness of our proposed method, Classifier Chain with Random Forest, we conducted extensive comparisons with additional prediction models, including Decision Tree, KNN, Logistic Regression, Neural Network, and SVM, utilizing both Binary Relevance16 and Classifier Chain methods. The results, summarized in Table 4, demonstrate that Classifier Chain with Random Forest consistently outperforms the other models across key metrics. Specifically, it achieved the highest Accuracy (0.76) and Macro Recall (0.75), underscoring its superior performance and robustness in predicting respiratory pathogen categories. These findings highlight the effectiveness of the proposed method in comparison to a diverse range of alternative models.
Discussion
In recent years, numerous studies have investigated the relationship between atmospheric conditions and respiratory viral infections. However, these studies exhibit limitations, including small sample sizes in datasets, restricted geographical coverage, and a focus on only a few types of respiratory pathogens. For instance, Jin’s work proposed a model based on meteorological factors and machine learning to forecast influenza outbreaks, but it only utilized a relatively small dataset of influenza virus information from a single city in China between 2012 and 201617. Similarly, Steindór’s study applied a respiratory symptom classification model to predict whether patients would need to see a doctor, but the data was limited to 6 viruses and 17,177 entries from 2016 to 201818. Compared to the previous study, our prediction method demonstrates superiority. Previous research has shown that meteorological conditions have a significant impact on the seasonal activity of respiratory viruses19. Therefore, we incorporate meteorological data into our predictions, extending applicability to a wider range of areas. A previous study in Bogotá, Colombia, utilized a Naive Bayes classifier and the receiver operating characteristic curve to predict the onset of RSV outbreaks20, whereas our model is based on a multi-label classification with a random forest, capable of predicting various viral infections. In this study, we developed the CRFC, an ensemble model comprising four chained random forest models, which demonstrated accurate prediction of respiratory virus types using air pollutants and environmental data.
In the analysis of variable importance, the year ranks first in predicting the likelihood of respiratory virus infection, a result that may be attributed to our data encompassing the years 2020 and 2021, which coincide with the COVID-19 pandemic. Due to the severity and urgency of the COVID-19 outbreak, many medical institutions and laboratories may have prioritized the expansion of their testing capabilities for SARS-CoV-2, including increasing diagnostic equipment and training of professionals. This concentrated use of resources, while enhancing the detection capacity for SARS-CoV-2, may have also led to a reduced focus and capability for detecting other respiratory viruses21. Age ranks as the second most influential factor, aligning with the consensus that children are more prone to respiratory infections22. NO2 is also a key factor affecting the model, which may be related to the fact that NO2 exposure can affect the function and immune response of respiratory epithelial cells23. At the same time, different concentrations of NO2 may also have different effects on respiratory epithelial cells15. Furthermore, the province of residence ranks third in influence, with the susceptibility of individuals to viruses potentially being related to climate and air quality, as well as economic conditions and the density of urban populations24.
The primary advantage of the CRFC model lies in its generalizability. As the study is based on nationwide respiratory virus detection data, the extensive scope and comprehensive representation of viral types allow the model to be applicable to diverse geographical regions and populations, contributing to its potential utility in varied healthcare settings. The second notable advantage of the CRFC model is its practicability. By exclusively utilizing the basic personal information of patients, air pollution data, and meteorological factors, while excluding clinical data, the model becomes user-friendly and easy to implement across various healthcare settings. Additionally, the CRFC model exhibits a certain level of interpretability. Through the application of SHAP25, the impacts of air pollutants and meteorological factors on respiratory viruses were thoroughly analyzed. This innovative aspect enables an in-depth investigation of the associations between respiratory virus risks and air pollution and meteorological factors, providing accurate insights into the underlying parameters of the prediction model.
Despite its strengths, the present study does possess a limitation. Due to the specific business characteristics of Guangzhou KingMed Diagnostics Group Co., Ltd., the tests were conducted in China, potentially making the model more applicable for China. However, its application and generalizability to other regions may require further validation and adaptation. The data in our study was collected using the PCR method, which has certain limitations in detection and is prone to potential false positives26, as well as risks of decreased sensitivity and increased variability in results27. Additionally, the current sample size in our study is insufficient and we need to study a greater variety of pathogens. These are areas that we plan to address in the future.
In conclusion, the CRFC model exhibited notable advantages in terms of generalizability, practicability, and interpretability. Its application can be valuable in a broader context, but further considerations are necessary for regional adaptability and the inclusion of clinical data to enhance the accuracy of predicting respiratory virus infections.
Material and methods
Data sources
The pathogen detection information was collected from Guangzhou KingMed Diagnostics Group Co., Ltd., one of the pioneers and leaders of the Independent Clinical Laboratories (ICL) industry in China. The data was collected nationwide from 2016 to 2021, encompassing a range of respiratory pathogens including HBoV, IV, ADV, RSV, rhinovirus (RhV), PIV, hMPV, and HCoV. Among these pathogens, HBoV, RhV, PIV, hMPV, and HCoV were categorized as the “others” group. The air quality were collected from a public dataset covering daily observations across the country from 2016 to 2021 in the National Urban Air Quality Real time Release Platform (https://air.cnemc.cn:18007). The air quality data included Air Quality Index (AQI), Ozone Concentration (O3), Nitrogen Dioxide (NO2), Carbon Monoxide (CO), PM10, PM2.5, SO2. The meteorological data was collected in National Centers for Environmental Information (https://www.ncei.noaa.gov/) including air temperature, dew point temperature, wind direction, and wind speed. The exposure of participants was assessed based on administrative divisions of the hospital addresses, that is, we matched the regions with environmental data through their latitude and longitude, and then utilized the average environmental data from the past 7 days for each participant for further study.
The research data included pathogen detection information for 654,830 individuals, 8,059,519 meteorological data entries, and 3,319,259 air quality data entries. Our data includes 31 regions across China, encompassing Anhui Province, Shanghai Municipality, Yunnan Province, Inner Mongolia Autonomous Region, Beijing Municipality, Jilin Province, Sichuan Province, Tianjin Municipality, Ningxia Hui Autonomous Region, Shandong Province, Shanxi Province, Guangdong Province, Guangxi Zhuang Autonomous Region, Xinjiang Uyghur Autonomous Region, Jiangsu Province, Jiangxi Province, Hebei Province, Henan Province, Zhejiang Province, Hainan Province, Hubei Province, Hunan Province, Gansu Province, Fujian Province, Tibet Autonomous Region, Guizhou Province, Liaoning Province, Chongqing Municipality, Shaanxi Province, Qinghai Province, and Heilongjiang Province.
By associating these data with the hospital address and date of testing for the detected cases, we aim to classify different respiratory virus categories under the obtained air quality and meteorological conditions. After data integration and cleaning, we ultimately obtained 19,161 entries whose test results are positive and which included information on testing personnel, pathogen detection results, meteorological data, and air quality data.
The selection of these data sources was based on their wide temporal and geographical coverage, enabling the acquisition of representative sample sets to better understand the transmission and infection characteristics of viruses. We adhered to the principles of data legality and privacy protection to ensure the security and reliability of the data used.
Study design
We employed a multi-label ensemble learning framework to construct a national respiratory virus infection risk prediction model, enabling real-time and precise assessment of viral transmission trends and potential risks. The model was built using a random forest-based multi-label chained ensemble learning algorithm called CRFC to perform binary classification for each category of respiratory virus. Ultimately, the four category-specific classification results were integrated to obtain the final prediction for each case in detail. Additionally, by incorporating air quality and meteorological data, we aim to establish a more accurate and comprehensive model, thereby providing the necessary support for clinical decision-making. The Receiver Operating Characteristic (ROC) curves with Area Under the Curve (AUC) score and Precision-Recall (PR) curves for classifier chains, were used to showcase the performance metrics for different classes of respiratory viruses. All of our data cleaning and statistical work was completed using Python version 3.8.19. The utilized Python libraries encompass “sklearn”, “imblearn”, “skmultilearn”, “shap”. The flowchart of model construction was shown in Fig. 9.
Model construction
Based on the study design, we constructed a classifier chains framework with random forest as the base model, utilizing air pollutants and environmental factors to predict the specific type of RTI virus affecting the patient. To generate a training dataset, we randomly sampled 80% of the cases, while the remaining 20% formed a balanced testing dataset. To mitigate sampling bias, we repeated the workflow 50 times. Notably, the Python package “iterative_train_test_split28” was utilized to split the data, ensuring that the sample distribution in both the training and test sets is similar to that of the entire dataset. In our model, we incorporated age, sex, year, month, week, day, day of week, season, and holiday as covariates in addition to air pollutants and environmental factors.
Model evaluation
The resultant model predicted the respiratory virus type affecting the patient within the training dataset, and we assessed its performance with the test dataset using various metrics, including overall accuracy, macro recall, macro precision, macro f1-score, and macro AUC score. Accuracy is the ratio of correctly predicted observations to the total observations. It measures the overall correctness of the model. The equation was as follows:
Here, TP which means true positives was the number of samples that can be correctly predicted as positive by the model. TN (True Negatives) was the number of samples that can be correctly predicted as negative by the model. The recall (sensitivity) measures the proportion of actual positives that are correctly identified. The value of recall was calculated using the following formula:
In this formula, TP means the same as mentioned above. FNc was the number of false negatives for class c by the model, which are instances that are actually positive but were predicted as negative. The recall was particularly important in cases where the cost of missing a positive instance was high. The precision was the ratio of correctly predicted positive observations to the total predicted positives. It was the ability of the model to not label a negative instance as positive, which can be reflected as:
Also, TP means the same as mentioned above. FP (False Positive) referred to the instances where a model incorrectly predicts a negative sample as a positive one. The f1- Score was the harmonic mean of precision and recall, providing a single measure that balances both metrics. It was particularly useful when the class distribution is uneven. The equation was as follows:
The Precisionc was the precision for class c, which was the ratio of true positives to the total predicted positives. Recallc is the recall for class c, which was the ratio of true positives to the actual positives. The trained model was then employed to predict the status of patients in the test dataset, and its performance was again evaluated using the same metrics.
We used ROC and PR curves to illustrate the model’s performance. These curves are essential tools for evaluating the performance of classifiers, offering a visual representation of the trade-offs involved in choosing different thresholds for classification. The ROC curves represent the trade-off between the true positive rates and the false positive rates for each class, along with the AUC scores. The PR curves represent the trade-off between precision and recall for each class, along with the Average Precision (AP) scores. A higher AUC/AP score indicates superior classifier performance. In our implementation of CRFC using this approach, the model CRFC was run 50 times to evaluate consistency. At the same time, we also conducted extensive comparisons with additional prediction models, including Decision Tree, KNN, Logistic Regression, Neural Network, and SVM, utilizing both Binary Relevance16 and Classifier Chain methods.
Interpretable machine learning model
In this study, we employed the interpretable SHAP29 machine learning framework to explore the impact mechanism of air pollution and meteorological factors on respiratory viruses.
To begin with, based on the concepts of cooperative game theory, we calculated the contributions of parameters in the multi-label classifier chain to determine their importance, thus identifying the air pollutants and meteorological factors that significantly influence the detection results of respiratory viruses.
Furthermore, we utilized an additive feature attribution method to interpret the model’s predictions as linear functions of binary variables and perform probability transformations. By presenting the interaction effects between different parameters and their collective impacts on viral risk in terms of probability values, we aimed to unveil the comprehensive influence of various factors on the outcomes. In addition to showing the overall impact of various factors on the model, SHAP is also utilized to illustrate the effects on a particular group of individuals.
Data availability
The datasets used and/or analyzed during the current study will be available from the corresponding authors on reasonable requests.
References
Moesker, F. M. et al. Viruses as sole causative agents of severe acute respiratory tract infections in children. PLoS One 11, e0150776 (2016).
Geretti, A. M. The expanding role of common respiratory viruses in human disease. J. Med. Microbiol. 52, 443–445 (2003).
Alizon, S. et al. Multiple infections and the evolution of virulence. Ecol. Lett. 16, 556–567 (2013).
Wang, D. et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. JAMA 323, 1061–1069 (2020).
Huang, X. B. et al. Epidemiological characteristics of respiratory viruses in patients with acute respiratory infections during 2009–2018 in southern China. Int. J. Infect. Dis. 98, 21–32 (2020).
WHO. Influenza(Seasonal). 2023; Available from: https://www.who.int/news-room/fact-sheets/detail/influenza-(seasonal).
Borchers, A. T. et al. Respiratory syncytial virus—a comprehensive review. Clin. Rev. Allergy Immunol. 45, 331–379 (2013).
Pica, N. & Bouvier, N. M. Environmental factors affecting the transmission of respiratory viruses. Curr. Opin. Virol. 2, 90–95 (2012). (1879-6265 (Electronic)).
Loaiza-Ceballos, M. A.-O. et al. Viral respiratory infections and air pollutants. Air Quality, Atmosphere & Health, 2021.
Somerville, L. K. et al. Molecular Diagnosis of Respiratory Viruses. Pathology 47, 243–249 (2010).
Martins, L. A. et al. A-302 Agreement Assessment Between Molecular and Rapid Tests in Respiratory Viruses Detection. Clinical Chemistry, 2023.
Shelton, C. R., Huie W., & Kan K. F. Chained Boosting. In Advances in Neural Information Processing Systems 19: Proceedings of the 2006 Conference (eds Schölkopf B., Platt J. & Hofmann T.) (The MIT Press, 2007).
Becker, T. et al. Decision trees and random forests. Am. J. Orthod. Dentofac. Orthop. 164, 894–897 (2023).
Sechidis, K., Tsoumakas, G. & Vlahavas, I. On the stratification of multi-label data. Machine Learning and Knowledge Discovery in Databases (Springer Berlin Heidelberg, Berlin, Heidelberg, 2011).
Becker, S. & Soukup, J. M. Effect of nitrogen dioxide on respiratory viral infection in airway epithelial cells. Environ. Res. 81, 159–166 (1999).
Luaces, O. et al. Binary relevance efficacy for multilabel classification. Prog. Artif. Intell. 1, 303–313 (2012).
Ge, H. et al. How to determine the early warning threshold value of meteorological factors on influenza through big data analysis and machine learning. Comput Math. Methods Med 2020, 8845459 (2020).
Ellertsson, S. et al. Triaging patients with artificial intelligence for respiratory symptoms in primary care to improve patient outcomes: a retrospective diagnostic accuracy study. Ann. Fam. Med 21, 240–248 (2023).
Yusuf, S. et al. The relationship of meteorological conditions to the epidemic activity of respiratory syncytial virus. Epidemiol. Infect. 135, 1077–1090 (2007).
González-Parra, G., Querales, J. F. & Aranda, D. Prediction of the respiratory syncitial virus epidemic using climate variables in Bogotá, D.C. Revista Biomédica (2016).
Woolbert, M. E. et al. Sharp decline in rates of community respiratory viral detection among patients at the National Institutes of Health Clinical Center during the coronavirus disease 2019 (COVID-19) pandemic. Infect. Control Hospital Epidemiol. 44, 62–67 (2022).
Douros, K. et al. Many children aged two to five years have a persistent presence of respiratory viruses in their nasopharynx. Acta Paediatrica 105, e89–e92 (2016).
Frampton, M. W. et al. Nitrogen dioxide exposure: effects on airway and blood cells. Am. J. Physiol.-Lung Cell. Mol. Physiol. 282, L155–L165 (2002).
Beamer, P. I. et al. Spatial clusters of child lower respiratory illnesses associated with community-level risk factors. Pediatr. Pulmonol. 51, 633–642 (2016).
Lundberg, S. M. & Lee S.-I. A Unified Approach to Interpreting Model Predictions. In Neural Information Processing Systems. 2017. Curran Associates, Inc.
Hajia, M. Limitations of different PCR protocols used in diagnostic laboratories: a short review. Mod.-Med-Lab-J. 1, 1–6 (2018).
Hilscher, C., Vahrson, W. & Dittmer, D. P. Faster quantitative real-time PCR protocols may lose sensitivity and show increased variability. Nucleic Acids Res. 33, e182 (2005).
Szyma'nski, P. & Kajdanowicz, T. Proceedings of the first international workshop on learning with imbalanced domains: theory and applications. PMLR 74, 22–35 (2017).
Lundberg, S. M. & Lee S.-I. A Unified Approach to Interpreting Model Predictions. In Neural Information Processing Systems (2017).
Acknowledgements
This work is supported by the 2024 Guangzhou Agricultural and Social Development Science and Technology Special Project (No. 2024B03J1385).
Author information
Authors and Affiliations
Contributions
Shuting Shi, Haowen Lin and Leiming Jiang conducted the statistical analysis and interpretation of the results, ensuring the robustness of the findings. ChuiXu Lin, Yinghua Li, Pei Li, Zhiqi Zeng and Zifeng Yang contributed to the design of the study and provided critical feedback on the machine learning methodologies used. Shuting Shi and Haowen Lin prepared all figures and tables, and assisted in the writing of the results and discussion sections. Zhiqi Zeng and Zifeng Yang provided expertize in respiratory virus epidemiology. ChuiXu Lin and Yinghua Li oversaw the project, ensuring that all aspects of the research met the highest standards of scientific integrity. Each author has contributed significantly to the research and has given final approval to the version of the manuscript being submitted. Each author has confirmed that there are no known conflicts of interest associated with this publication.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Shi, S., Lin, H., Jiang, L. et al. Development of a respiratory virus risk model with environmental data based on interpretable machine learning methods. npj Clim Atmos Sci 8, 39 (2025). https://doi.org/10.1038/s41612-025-00894-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41612-025-00894-4