Abstract
Translocation-associated sarcomas (TAS) are rare, phenotypically heterogeneous, with predisposition for young adults. We aimed to investigate the clinical impact of germline pathogenic/likely pathogenic (P/LP) variants in a diverse group of TAS and to conduct a comprehensive comparative analysis of clinicopathologic features, genomic alterations, and survival outcomes. A retrospective cohort of 426 TAS patients with both tumor and germline DNA sequencing was investigated for clinical actionability of P/LP variants, and potential impact on current screening guidelines and clinical interventions. Twenty-eight patients (6.6%) carried Tier 1 germline P/LP variants (moderate to high penetrance autosomal dominant (AD) variants), while 27 (6.3%) patients carried Tier 2 variants (monoallelic autosomal recessive or low penetrance AD variants). Compared to Tier 2, Tier 1 patients were more commonly of European ancestry and had a higher frequency of first- and second-degree relatives with cancer history. Notably, the frequency of both tiers variants was lower among pediatric patients compared to older patients and differed across TAS histologies, with the highest observed in solitary fibrous tumors. All germline P/LP variants were monoallelic, dispersed across multiple genes, and enriched in DNA damage repair pathways. There was no association between the germline P/LP variants and somatic genomic profile, nor any survival impact when stratified by histotype. Our findings highlight the incidence of clinically significant germline P/LP variants in TAS is lower in pediatric patients, questioning current sarcoma genetic screening guidelines and supporting germline testing for all TAS patients. Significant interventions were triggered in 46% of Tier 1 (n = 13), including platinum-based chemotherapy and PARP inhibitors in two BRCA1/2 patients.
Similar content being viewed by others
Introduction
The increased rate of tumor-normal sequencing and evaluation of germline variants in cancer predisposition genes has brought opportunities for identifying patients with previously unknown cancer predisposing syndromes, aiding in therapeutic stratification1,2. Translocation associated sarcomas (TAS) are rare, phenotypically heterogenous, and occurring preferentially in young adults. The clinical benefit of germline testing in this group of sarcomas patients is not well established. Traditionally, the criteria for selecting sarcoma patients for genetic testing include the age at diagnosis, family cancer history, tumor’s pathologic characteristics, and other factors outlined in clinical practice guidelines3. According to the American College of Medical Genetics (ACMG), the guidelines to refer a patient with sarcoma to genetic counseling includes those patients with one additional Li-Fraumeni syndrome tumor (brain tumor, breast cancer, adrenocortical tumor, leukemia, bronchoalveolar cancer and colorectal cancer) in the same person or in 2 close relatives, with one diagnosis at age <45 years or patients with sarcoma diagnosis at age <18 years3. However, studies have shown that current criteria for germline testing, primarily involving common cancers, often result in about half of the patients with pathogenic and likely pathogenic (P/LP) variants going undiagnosed4,5. Furthermore, Ballinger et al. demonstrated that family patterns are unreliable indicators of underlying genotypes in patients with sarcoma, thereby, questioning the relevance of the proposed clinical criteria in the context of affordable genetic testing6. Prior studies have demonstrated a high frequency of P/LP variants in patients with various sarcomas, ranging from 6.6 to up to 28.0%6,7,8,9,10. Specific hereditary cancer predisposition syndromes carrying germline P/LP variants in tumor suppressor genes have been shown to be associated with increased risk of developing certain sarcoma, predominantly sarcomas with complex genome. Such sarcomas include osteosarcoma in Li-Fraumeni and hereditary retinoblastoma syndromes10, leiomyosarcoma in hereditary retinoblastoma syndrome8, and malignant peripheral nerve sheath tumor (MPNST) in neurofibromatosis-1 (NF-1). With the exception of few studies addressing the genetic predisposition in Ewing sarcoma, the genetic underpinnings of TAS remain largely unexplored6,11.
We set out to report our experience and examine the prevalence and spectrum of germline P/LP variants among patients with a wide range of common TAS subtypes treated at our tertiary cancer center. Additionally, we performed a comparative clinicopathologic, genomic and survival analysis of patients with and without germline P/LP variants to further define their impact.
Results
Cohort summary
A total of 426 patients with TAS who consented for MSK-IMPACT and secondary germline testing were identified in our archives between January 2015 and February 2024. The cohort consisted of 255 males (59.9%) and 171 females (40.1%) with a median age of 22 years (6 months-75 years). The majority of the patients were of European ancestry, constituting 47.6% (n = 203). This was followed by patients with admixed ancestry at 16.7% (n = 71), Ashkenazi Jewish European ancestry at 13.6% (n = 58), African ancestry at 7.6% (n = 32), East Asian ancestry at 4.3% (n = 19), South Asian ancestry at 3.6% (n = 15) and Native American at 0.7% (n = 3). Ancestral information was unavailable for 5.8% of the patients.
Of the 426 patients, 55 (12.9%) harbored germline P/LP variants conferring cancer predisposition. The germline P/LP variants occurred in 28 males (50.9%) and 27 females (49.1%) with a median age of 29 years (7–75 years). Twenty-eight patients (6.6%) carried a Tier 1 germline P/LP variant, while 27 patients (6.3%) harbored a Tier 2 P/LP variants. Tier 1 variants occurred in 18 males (64.3%) and 10 females (35.7%) with a median age of 33 years (13–67), while tier 2 variants occurred in 10 males (37%) and 17 females (63%) with a median age of 27 years (7–75). Among Tier 1 patients, European ancestry was the most prevalent (n = 10, 36%), followed by Ashkenazi Jewish European ancestry (n = 5, 18%). In contrast, Tier 2 patients predominantly had Ashkenazi Jewish European ancestry (n = 13, 48%), with European ancestry being the second most common (n = 7, 26%). Among patients with negative germline P/LP testing, there were 227 males (61.2%) and 144 females (38.8%), with a significantly lower median age of 21 years (range 0.5–71 years, Wilcoxon rank sum test with continuity correction, p-value = 0.003, Supplementary Table 3). The most commonly reported ancestry in this group was European at 50% (n = 186), followed by admixed ancestry at 17.2% (n = 64). One patient carried two germline P/LP variants, while all other patients had only a single germline P/LP variant. The clinical characteristics of the study cohort are summarized in Table 1.
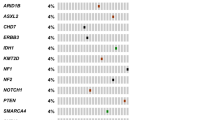
Three of the 28 patients (10.7%) with Tier 1 variants had prior history of malignancy including breast cancer, colon cancer and prostate cancer. Additionally, one patient with NF1 germline mutation had clinical manifestation of the disease including multiple neurofibromas. On the other hand, only one patient with Tier 2 variants had prior history of malignancy (prostate cancer and lymphoma). Among patients with Tier 1 variants, 12 (42.9%) had at least one first-degree relative, and 17 (60.7%) had at least one second-degree relative with history of cancer. In contrast, 10 (37%) and 11 (40.1%) patients with Tier 2 variants had at least one first- and second-degree relatives with cancer, respectively. Overall, forty patients with germline P/LP variants (73%) had at least one first- or second-degree relatives with malignancy. Common cancers were the predominant malignancies among first- and second-degree relatives, accounting for 86.3% and 92.9% of the cancers, respectively. Two patients with Tier 2 variants had first degree relatives with sarcoma; the first is a patient with Ewing sarcoma who had a sibling diagnosed with ARMS diagnosed at 5 years of age and the second is a patient with BCOR-altered sarcoma with a parent with bone sarcoma. Figure 1 is an oncoplot displaying the germline P/LP variants among the TAS cohort. Clinicopathologic characteristics of patients with germline P/LP variants are summarized in Table 2.
Oncoplot displaying P/LP variants among TAS along with clinicopathologic characteristics of the involved patients. DSRCT desmoplastic small round cell tumor, RMS rhabdomyosarcoma, EHE epithelioid hemangioendothelioma, CCS clear cell sarcoma, IMT inflammatory myofibroblastic sarcoma, LGFMS low grade fibromyxoid sarcoma, LPS liposarcoma, DFSP dermatofibrosarcoma protuberans, H&N head and neck, DOD died of disease, AWD alive with disease, NED no evidence of disease.
To compare the frequency of Tier 1 and Tier 2 variants across different age groups, we categorized the patients based on their age at diagnosis into three groups: pediatric (≤18 years), young adults (19–40 years), and older adults (>40 years). The frequency of Tier 1 variants among the pediatric category was significantly lower (3.6%) compared to patients older than 18 years (8.5%) (Fisher exact test p-value = 0.046). However, the frequency of Tier 2 variants among the pediatric category was not significantly different compared to patients older than 18 years (p-value = 0.84). Similarly, the frequency of Tier 1 and Tier 2 variants in patients ≤40 was lower (5.75% for both) compared to 10.3% in patients >40 years (both tiers). However, the difference was not statistically significant (Fisher exact test p-value > 0.05) (Supplementary Table 4A, B).
The frequency of Tier 1 and Tier 2 variants differed across various histologic subtypes of TAS, with the highest frequency observed in SFT for both tiers (16.6% and 25%, respectively), while the lowest frequencies were found in SEF and ASPS at 0% for both tiers. In addition to SFT, EHE and SS exhibited the highest frequency of Tier 1 variants, whereas EHE and ARMS had the highest frequency of Tier 2 variants. Figure 2 displays the number of patients with germline Tier 1 or Tier 2 variants in TAS. Across ES, DSRCT, SS, ARMS, and EHE, patients with Tier 1 variants had a higher mean age compared to those with negative P/LP; however, the difference was only statistically significant among ARMS patients (p < 0.05) (Supplementary Fig. 1). In ARMS, there was no statistically significant difference in the frequency of germline P/LP variants in regard to fusion partners [Fisher exact test p-value = 1.0, PAX 3 (3/26) and PAX7 (1/6)].
Variant detection
42P/LP variants (total variants, n = 56) in 30 genes were detected in our cohort. The inheritance patterns of the involved genes were as follows: 17 genes exhibited autosomal dominant inheritance (Tier 1), 9 genes exhibited autosomal recessive inheritance (Tier 2), and 4 genes exhibited either autosomal dominant or recessive inheritance (Tier 1). Among both tiers, pathway enrichment analysis using the KEGG database revealed significant enrichment in several DNA repair-related pathways, including homologous recombination, Fanconi anemia pathway, DNA mismatch repair and base-excision repair. Variants involved in DNA damage repair pathway accounted for 79% (22/28) and 32% (9/28) of the variants among Tier 1 and Tier 2, respectively. In addition, enrichment in platinum drug resistance, breast, colorectal and endometrial cancer were noted. Supplementary Fig. 2A, B displays KEGG pathway enrichment analysis of Tier 1 and Tier 2P/LP variants among TAS.
MSK-IMPACT findings
The tumor mutational burden (median TMB < 10 mt/MB) and median fraction genome altered (FGA) were low across all the cases. A comparative study across different sarcomas histotypes showed no significant differences in TMB and FGA between tumors with Tier 1, Tier 2 variants and those without (ANNOVA and paired p-values > 0.05) (Supplementary Fig. 3A, B). All tumors were microsatellite stable (MSS) except for four cases, which were indeterminate: three of these had negative germline P/LP variants, and one had a positive germline P/LP variant (synovial sarcoma with germline CHEK2 mutation). The latter showed retained expression of MMR proteins on immunohistochemical stains. No sarcomas were microsatellite high (MSI-H) including those who had germline mutations in MMR genes. There was no additional somatic second hit mutation in any of the genes affected by P/LP germline variants across all cases. We have also conducted a comparison of the somatic mutational profiles between TAS patients with and without germline P/LP variants, focusing on common recurrent somatic mutations. There was no significant difference between Ewing sarcoma cases with Tier 1 or Tier 2 and those without germline P/LP variants regarding the rates of known recurrent somatic mutations in STAG2, TP53 and CDKN2A/2B (Fig. 3). In addition, among the recurrent somatic mutations in DSRCT, the rates of the common somatic mutations (ARID1A, TP53, TERT, CRLF2, ATM and FGFR4) were not statistically significant between patients with Tier 1 or Tier 2 and those with no germline pathogenic variants (p-value > 0.05) (Supplementary Fig. 4). Similar findings were noted among SS and ARMS regarding rates of respective recurrent somatic mutations (p-value > 0.05) (Supplementary Figs. 5 and 6). Mutation tumor signatures could not be assessed by MSK-IMPACT due to the low number of mutations seen, as expected, in TAS.
Loss of heterozygosity (LOH)
Among cases harboring germline P/LP variants, LOH was observed in only 4 cases (7.2%). The LOH events were noted in two Tier 1 variant (NF1 in a synovial sarcoma case and BRCA1 in a solitary fibrous tumor) and in two Tier 2 variants (NTHL1 in a desmoplastic small round cell tumor (DSRCT) case, and an APC variant in a BCOR-altered sarcoma case).
Survival studies
Survival comparative studies showed no significant difference in overall survival between patients harboring germline P/LP Tier 1 or Tier 2 and those without in all studied histologic subtypes: ES, DSRCT, SS, SFT, ARMS and EHE (p-value > 0.05). Kaplan–Meier survival curves of patients with Tier 1 and Tier 2 variants and those without germline P/LP variants across the various histologic subtypes are displayed in Supplementary Fig. 7A–F.
Clinical implication of germline testing results
All patients with positive germline P/LP variants were advised to undergo a clinical genetic consultation. Of the 55 patients, 39 (70.9%) underwent genetic consultation and were advised on suitable screening options. Genetic testing was offered to the families of all patients. Significant interventions were given to patients with Tier 1 variants including BRCA1, BRCA2, MLH1, PMS2, MSH6, TP53, YAP1 and CHEK2 mutations (n = 13). In two patients with BRCA1 and BRCA2 variants, discovery of the mutations led to initiation or consideration of poly adenosine diphosphate-ribose polymerase (PARP) inhibitors and/or platinum-based chemotherapy. The first patient was diagnosed with metastatic malignant SFT and had a germline BRCA1 mutation. Although there was an initial response to the PARP inhibitors, nthe medication was discontinued due to subsequent unavailability. The second patient, diagnosed with metastatic alveolar rhabdomyosarcoma, was offered PARP inhibitors or platinum-based chemotherapy after the discovery of a germline BRCA2 mutation. However, the patient was lost to follow-up.
Discussion
In this study, we examined a large cohort of translocation-associated sarcomas (TAS), utilizing a clinically designed approach to identify underlying germline P/LP variants in cancer-predisposing genes. To our knowledge, this is the first study specifically focused on investigating this genomic cohort. The overall frequency of germline P/LP variants in TAS patients was 12.9%, while the frequency of significant clinically actionable autosomal dominant variants (Tier 1) was 6.6%. This overall frequency of P/LP variants among TAS was lower compared to their frequency among patients with advanced cancers studied within our institution (205/1040; 19.7%)5. The frequency of germline P/LP variants was variable among different histotypes, ranging from 0% in SEF and ASPS up to 16.6% and 25% for Tier 1 and Tier 2, respectively in SFT. While no prior comprehensive study has specifically focused on this group of sarcomas, the frequencies of germline P/LP variants in certain histologic subtypes within our cohort align with previous reports. For instance, after excluding monoallelic MUTYH variants to adjust for prior reported studies, the frequency of germline P/LP variants in our Ewing sarcoma subset was 13.3% (12/90), aligning with the 13.1% frequency reported by Brohl et al. (23/175)12. On the other hand, the overall frequency of germline P/LP variants in ARMS was 13.1% (5/38), exceeding the 3% (2/67) reported in the discovery cohort and the 5.3% (2/38) reported in the secondary cohort of FOXO1-fusion positive ARMS7. Despite the limited number of cases in the literature, germline P/LP variants in SFT have been documented, with one series reporting a frequency as high as 25% (1/4)9. Additionally, Prejac et al. identified a rare TP53 germline pathogenic variant in a 36-year-old female who developed malignant SFT and subsequent breast cancer13. Notably, 9 of the 12 SFTs in our cohort were classified as malignant (4/5 with germline P/LP variants and 5/7 with negative germline testing). The difference in germline P/LP variant frequencies between the current and the reported studies is likely due to the small sample size in both cohorts, selection bias (with advanced diseases and referral center patients being tested), and genetic background variability among patients. Despite earlier research indicating a trend of younger age among patients with germline P/LP variants6,7,12, our study observed a higher median age in patients with these variants (including both tiers) compared to those without. However, this age difference was only statistically significant among ARMS when analyzed within each histologic subtype. Moreover, we observed a lower frequency of germline P/LP variants among younger patients, particularly for Tier 1 variants. This finding challenges the current ACMG recommendation to test patients with one sarcoma under 18 years old and supports germline testing in all patients with TAS.
Our study demonstrated that pathogenic germline mutations in TAS are monoallelic and are not recurrent in a single gene; rather, these mutations are dispersed across multiple genes, exhibiting similar functional clustering. Significant enrichment was observed in DNA damage repair pathways, with the highest frequency in HR pathway, followed by BER (base excision repair), MMR (DNA mismatch repair), FA (Fanconi anemia), and NER (nucleotide excision repair) pathways. These findings align with previous reports showing DNA damage repair gene enrichment in Ewing sarcoma6,11,12, although other TAS were largely underexplored. Recent studies showed that susceptibility to sarcoma can arise not only from monogenic high-risk variants but also from combined effect of multiple less penetrant variants, which together contribute to sarcoma risk through a polygenic risk model6. Specifically, it has been suggested that polygenic effects and monoallelic germline variants in DNA damage repair genes may predispose patients to TAS subtypes6. Subsequently, this was shown by Gillani et al. who demonstrated that the recurrent enrichment of heterozygous pathogenic germline variants in FANCC substantiates its role in increasing the risk for certain individuals with Ewing sarcoma11. They suggested that monoallelic germline variants in FA genes might also elevate the risk for other translocation-associated cancers. In addition, using parent-proband trios, Gillani et al. demonstrated that pathogenic germline mutation in DNA damage repair variants found in individuals with Ewing sarcoma are inherited from their parents through autosomal inheritance11. These moderate penetrance risk variants play a substantial role in increasing risk for developing Ewing sarcoma but are most likely not sufficient to cause the disease in isolation11. The enrichment of germline P/LP mutations in DNA damage repair genes among our TAS cohort is in keeping with their observation. Ultimately, additional studies involving large cohorts of patients and controls are necessary to determine if there is a genuine functional association between this class of mutations and the development of fusion-driven cancers.
Rare germline P/LP TP53 variants in Ewing sarcoma have been documented in the literature12,14, including within our cohort (1/15). However, comparative analyses showed that the frequency of these variants is significantly lower in Ewing sarcoma compared to other pediatric sarcoma subtypes, predominantly sarcoma with complex genomes, such as osteosarcoma and rhabdomyosarcoma11. This observation was further supported by the fact that Ewing sarcoma is not commonly found in families with Li-Fraumeni syndrome15. Except for the single Ewing sarcoma case harboring a germline TP53 variant, germline mutations in cancer predisposition genes known to be associated with sarcomas, predominantly sarcomas with complex genome, such as TP53, RB1 and DICER1, were not observed across our TAS cohort.
Secondary somatic mutations in germline P/LP variants appear to be uncommon among TAS. Although other histologic subtypes were not thoroughly studied in the literature, this finding was previously noted by Zhang et al.14 and Brohl et al.12 across their Ewing sarcoma cases. Additionally, LOH in germline P/LP variants was infrequent and was only noted in four cases in our cohort. The genes involved were APC in a case of BCOR altered sarcoma, NTHL1 in a case of DSRCT, NF1 in a case of SS and BRCA1 in a solitary fibrous tumor. LOH in sarcomas are expected to occur in tumor suppressor genes such as NF1, TP53 or RB1, through the loss of the wild-type allele. While LOH, specifically copy neutral LOH, was demonstrated to be the most prevalent type of second hit in tumors from individuals with constitutional NF1 including MPNST, glioma and gastrointestinal stromal tumor, LOH of the NF1 gene in SS has not been reported in the literature to date. LOH of the APC gene is a common form of allelic imbalance that has been observed in many types of cancer including colorectal cancer16, renal cell carcinoma17, gastric cancer18, endometrial cancer19 and squamous cell carcinoma20. To our knowledge, LOH in the APC gene in BCOR-altered sarcoma has not been reported in the literature, and its significance remains unclear. Moreover, the significance of LOH in genes not typically associated with sarcoma, such as NTHL1, is also undetermined. All five patients with germline P/LP variants in MMR genes had microsatellite-stable tumors, suggesting that these variants likely constitute incidental findings. Despite the incidental nature of these variants, identifying these clinically significant variants was essential for the patients’ clinical care.
Up to 75% of TAS patients with germline P/LP in our cohort had at least one first degree or second degree relative with cancer. This number might be under-represented, as family history was collected from patients’ charts and clinical genetics encounters (subset of patients). Prior studies have demonstrated an increased risk of cancer among patients with Ewing sarcoma and their relatives21,22. The most frequent malignancies in first-degree relatives were brain, lung, gynecologic, and prostate cancers. In second-degree relatives, common cancers included breast cancer, nonmelanoma eye cancer, and MPNST21. One report indicated that the EWSR1::FLI1 transcription induces R-loops and impedes BRCA1-mediated repair in Ewing sarcoma which could explain the observed increased risk of breast cancer23. Interestingly, Ewing sarcoma was not detected among the first-, second-, or third-degree relatives of patients with Ewing sarcoma21. In this study, we observed a high incidence of common carcinomas, particularly breast, prostate, lung, and renal cancers, among first- and second-degree relatives of patients with TAS, while the incidence of sarcomas remained low. Although this increase might be partly due to the natural rise in cancer incidence among older individuals, these findings underscore the importance of regular cancer screening and vigilant monitoring for relatives of patients with TAS to facilitate early detection and timely intervention.
The somatic genomic profiles of the four studied TAS histotypes (ES, DSRCT, SS and ARMS) did not show significant differences between tumors with and without germline mutations. These findings are consistent with those reported by Brohl et al., who observed no significant differences in somatic mutations in STAG2, CDKN2A, and TP53 between Ewing sarcoma patients with germline P/LP variants and those without12. Data regarding other TAS histologies is limited in the literature. Furthermore, we demonstrated that TMB and FGA are not significantly different between the translocation associated sarcomas with and without germline mutations. In addition, our comparative survival studies revealed no significant difference between TAS with germline P/LP variants and those without, when stratified by histologic subtypes. These findings are consistent with those reported in the literature7,12.
The identification of germline P/LP variants in patients with TAS remains critical as it not only prompts genetic counseling for patients and their family members, but also allows for potential targeted therapeutic strategies. For instance, as in two of our patients, the presence of germline mutations in DNA repair pathways may render tumors more susceptible to platinum-based chemotherapy and PARP inhibitors24,25,26,27,28.
In conclusion, we present our experience on pathogenic variants among TAS, reporting a frequency of 6.6% clinically significant germline P/LP variants in this group. Moreover, we observed a lower frequency of germline P/LP variants, among younger patients, particularly for Tier 1 variants, thus, questioning the current recommendations for genetic screening in patients with sarcoma which might miss a considerable number of patients with germline P/LP variants and support germline testing in all patients with TAS. Further research is needed to determine if these interventions will improve outcomes for both the patients and their family members.
Methods
Patient cohort
The study was approved by the institutional review board (#12-245). The cohort comprised of patients with common TAS subtypes [Ewing sarcoma (ES), desmoplastic small round cell sarcoma (DSRCT), synovial sarcoma (SS), alveolar rhabdomyosarcoma (ARMS), solitary fibrous tumor (SFT), epithelioid hemangioendothelioma (EHE), low grade fibromyxoid sarcoma (LGFMS), BCOR-altered sarcoma, myxoid liposarcoma (MLPS), CIC-rearranged sarcoma, clear cell sarcoma (CCS), inflammatory myofibroblastic tumor/ epithelioid inflammatory myofibroblastic sarcoma (IMT/EIMS), dermatofibrosarcoma protuberans (DFSP), and sclerosing epithelioid fibrosarcoma (SEF)] at Memorial Sloan Kettering Cancer Center (MSKCC) between January 2015 and February 2024, who underwent tumor and normal DNA sequencing using MSK-IMPACT (Integrated Mutation Profiling of Actionable Cancer Targets) and provided an additional consent for germline analysis. Patients were selected and referred for germline analysis upon the discretion of their treating physicians. The electronic medical records were searched for clinical data including the patient’s age, sex and anatomic site of primary tumor. Personal and family history were gathered from the patient’s medical charts including the clinical genetic encounters. The gene fusions in all cases were confirmed by fluorescence in situ hybridization (FISH) and/or targeted RNA/DNA sequencing. RNA sequencing was performed using the Archer FusionPlex (Archer, Boulder, CO), an anchored multiplex PCR-based assay consisting of a custom amplicon-based next generation sequencing (NGS) that targets specific exons in 129 genes, applying a standard protocol29.
MSK-IMPACT testing and variant calling
Detailed descriptions of MSK-IMPACT workflow and data analysis, a hybridization capture-based targeted DNA NGS assay for solid tumors, were described previously30. All mutational and copy number calls were generated by the standard MSK-IMPACT pipeline30. Copy number amplification and deletion are defined as gains and losses of gene-level copy number greater than two-fold in the tumor relative to pooled FFPE normal based on NGS. Germline analysis consisted of 76, 88 or 90 genes on the MSK-IMPACT panel associated with hereditary cancer predisposition, including all cancer predisposing genes identified by ACMG guidelines (Supplementary Table 1)31,32,33. Variants were interpreted based on ACMG criteria by molecular genetic pathologists34. Variants with variant allele fraction (VAF) less than 25% for single nucleotide variants (SNV) and 15% for indels and variants with less than 20× coverage were filtered. Variants of unknown significance were not included in the study and were not reported in the clinical reports.
In this study, we categorized P/LP variants into two tier groups. Tier 1 comprised of autosomal dominant variants with moderate to high penetrance, which confer a significant clinical actionability. Tier 2 included monoallelic autosomal recessive variants, and autosomal dominant variants with low penetrance (APC p.Ile1307Lys, FH p.Lys477dup35) which are associated with a low risk of clinically actionable disease. Variants with either autosomal dominant or recessive patterns were classified as Tier 1. Details of the variants included in each tier are summarized in Supplementary Table 2.
Pathway analysis of germline P/LP variants was performed using the “clusterProfiler”36 package version 4.13.0 (RRID:SCR_016884), leveraging the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. The analysis parameters were set to include the entire KEGG database for human genes, and we applied the Benjamini–Hochberg (BH) method to adjust p-values for multiple testing. Pathways with an adjusted p-value < 0.05 were considered significantly enriched.
Loss of heterozygosity and microsatellite instability
Loss of heterozygosity (LOH), defined as loss of the wild-type allele in the tumor at the locus of the germline mutation, was determined using segmented allele-specific copy number calls from FACETS tool, as previously published37. Briefly, FACETS tool uses aligned sequence bam files from NGS and performs analysis for joint segmentation of total-and allele-specific read counts and integer copy number calls corrected for tumor purity, ploidy and clonal heterogeneity to estimate LOH. Segments were classified as LOH if they have minor allele copy number of zero. The allele having LOH was identified by examining the VAF of germline variants in the tumor. Microsatellite instability (MSI) was assessed by MSI sensor, a computational algorithm that analyzes sequencing reads at designated microsatellite regions in tumor-normal pairs, reporting the percentage of unstable loci as a cumulative score38,39. Microsatellite instability high (MSI-H) was defined as >10% of loci on the MSK-IMPACT panel demonstrating microsatellite instability38. MSI at ≥3 to <10% was considered as indeterminate MSI and <3% was considered microsatellite stable.
Ancestry inference
Genetic ancestry was determined using data from the MSK-IMPACT clinical sequencing panel, as previously outlined40. Briefly, ADMIXTURE v1.341 (RRID:SCR_001263) was ran in supervised mode using the 1000 Genomes project42 cohort as a reference to infer ancestral proportions of African, Ashkenazi Jewish European, European, East Asian, Native American, and South Asian populations. Patients with an ancestral fraction greater than 0.8 for any single population were assigned the corresponding population label; those with no single population fraction exceeding 0.8 were classified as admixed.
Survival analysis
The clinical charts were manually reviewed, and date of initial diagnosis and survival status were documented. Survival analysis was performed using R (RRID:SCR_001905) packages “survminer” version 0.4.9 (RRID:SCR_021094) and “survival” version 3.2.13 (RRID:SCR_021137) by comparison of hazard ratios using log rank P testing and Kaplan–Meier curves.
Data availability
Anonymized clinical and Sequencing data generated by MSK-IMPACT will be publicly available at https://www.cbioportal.org/study/summary?id=soft_tissue_msk_2025.
Code availability
Code to generate the figures is available upon request from the corresponding author.
References
Huang, K. L. et al. Pathogenic Germline Variants in 10,389 Adult Cancers. Cell 173, 355–370.e314 (2018).
Jahn, A. et al. Comprehensive cancer predisposition testing within the prospective MASTER trial identifies hereditary cancer patients and supports treatment decisions for rare cancers. Ann. Oncol. 33, 1186–1199 (2022).
Hampel, H., Bennett, R. L., Buchanan, A., Pearlman, R. & Wiesner, G. L. A practice guideline from the American College of Medical Genetics and Genomics and the National Society of Genetic Counselors: referral indications for cancer predisposition assessment. Genet. Med. 17, 70–87 (2015).
Samadder, N. J. et al. Comparison of Universal Genetic Testing vs Guideline-Directed Targeted Testing for Patients With Hereditary Cancer Syndrome. JAMA Oncol. 7, 230–237 (2021).
Mandelker, D. et al. Mutation Detection in Patients With Advanced Cancer by Universal Sequencing of Cancer-Related Genes in Tumor and Normal DNA vs Guideline-Based Germline Testing. JAMA 318, 825–835 (2017).
Ballinger, M. L. et al. Monogenic and polygenic determinants of sarcoma risk: an international genetic study. Lancet Oncol. 17, 1261–1271 (2016).
Kim, J. et al. Pathogenic Germline Variants in Cancer Susceptibility Genes in Children and Young Adults With Rhabdomyosarcoma. JCO Precis. Oncol. 5, 75–87 (2021).
Kleinerman, R. A. et al. Risk of soft tissue sarcomas by individual subtype in survivors of hereditary retinoblastoma. J. Natl Cancer Inst. 99, 24–31 (2007).
Carvalho, N. A. et al. Prevalence and clinical implications of germline pathogenic variants in cancer predisposing genes in young patients across sarcoma subtypes. J. Med. Genet. 61, 61–68 (2023).
Mirabello, L. et al. Frequency of Pathogenic Germline Variants in Cancer-Susceptibility Genes in Patients With Osteosarcoma. JAMA Oncol. 6, 724–734 (2020).
Gillani, R. et al. Germline predisposition to pediatric Ewing sarcoma is characterized by inherited pathogenic variants in DNA damage repair genes. Am. J. Hum. Genet. 109, 1026–1037 (2022).
Brohl, A. S. et al. Frequent inactivating germline mutations in DNA repair genes in patients with Ewing sarcoma. Genet. Med. 19, 955–958 (2017).
Prejac, J., Dedić Plavetić, N., Gotovac Jerčić, K. & Borovečki, F. A first report of a rare TP53 variant associated with Li-Fraumeni syndrome manifesting as invasive breast cancer and malignant solitary fibrous tumor. World J. Surg. Oncol. 19, 254 (2021).
Zhang, J. et al. Germline Mutations in Predisposition Genes in Pediatric Cancer. N. Engl. J. Med. 373, 2336–2346 (2015).
Kratz, C. P. et al. Analysis of the Li-Fraumeni Spectrum Based on an International Germline TP53 Variant Data Set: An International Agency for Research on Cancer TP53 Database Analysis. JAMA Oncol. 7, 1800–1805 (2021).
Preston, J. L. & Stiffler, N. Epigenetic loss of heterozygosity of Apc and an inflammation-associated mutational signature detected in Lrig1(+/-)-driven murine colonic adenomas. BMC Cancer 20, 126 (2020).
Pećina-Slaus, N., Pavelić, K. & Pavelić, J. Loss of heterozygosity and protein expression of APC gene in renal cell carcinomas. J. Mol. Med. 77, 446–453 (1999).
Sanz-Ortega, J. et al. LOH at the APC/MCC gene (5Q21) in gastric cancer and preneoplastic lesions. Prognostic implications. Pathol. Res. Pr. 192, 1206–1210 (1996).
Moreno-Bueno, G. et al. Abnormalities of the APC/beta-catenin pathway in endometrial cancer. Oncogene 21, 7981–7990 (2002).
Rivero, E. R., Horta, M. C., Silva Guerra, E. N., Ferraz, A. R. & Nunes, F. D. Loss of heterozygosity of the APC gene in oral squamous cell carcinoma. Pathol. Res. Pr. 204, 793–797 (2008).
Abbott, D. et al. Increased risk for other cancers in individuals with Ewing sarcoma and their relatives. Cancer Med. 8, 7924–7930 (2019).
Ginsberg, J. P. et al. Long-term survivors of childhood Ewing sarcoma: report from the childhood cancer survivor study. J. Natl Cancer Inst. 102, 1272–1283 (2010).
Gorthi, A. et al. EWS-FLI1 increases transcription to cause R-loops and block BRCA1 repair in Ewing sarcoma. Nature 555, 387–391 (2018).
Farmer, H. et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434, 917–921 (2005).
Kristeleit, R. S., Miller, R. E. & Kohn, E. C. Gynecologic Cancers: Emerging Novel Strategies for Targeting DNA Repair Deficiency. Am. Soc. Clin. Oncol. Educ. Book 35, e259–e268 (2016).
Tung, N. M. et al. TBCRC 048: Phase II Study of Olaparib for Metastatic Breast Cancer and Mutations in Homologous Recombination-Related Genes. J. Clin. Oncol. 38, 4274–4282 (2020).
Litton, J. K. et al. Talazoparib in Patients with Advanced Breast Cancer and a Germline BRCA Mutation. N. Engl. J. Med. 379, 753–763 (2018).
Clark, C. A. & Yang, E. S. Therapeutic Targeting of DNA Damage Repair in the Era of Precision Oncology and Immune Checkpoint Inhibitors. J. Immunother. Precis. Oncol. 6, 31–49 (2023).
Zheng, Z. et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat. Med. 20, 1479–1484 (2014).
Cheng, D. T. et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J. Mol. Diagn. 17, 251–264 (2015).
Rahman, N. Realizing the promise of cancer predisposition genes. Nature 505, 302–308 (2014).
Kalia, S. S. et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet Med. 19, 249–255 (2017).
Cheng, D. T. et al. Comprehensive detection of germline variants by MSK-IMPACT, a clinical diagnostic platform for solid tumor molecular oncology and concurrent cancer predisposition testing. BMC Med. Genomics 10, 33 (2017).
Richards, S. et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 17, 405–424 (2015).
Zhang, L. et al. Fumarate hydratase FH c.1431_1433dupAAA (p.Lys477dup) variant is not associated with cancer including renal cell carcinoma. Hum. Mutat. 41, 103–109 (2020).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics 16, 284–287 (2012).
Shen, R. & Seshan, V. E. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. 44, e131 (2016).
Niu, B. et al. MSIsensor: microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics 30, 1015–1016 (2014).
Latham, A. et al. Microsatellite Instability Is Associated With the Presence of Lynch Syndrome Pan-Cancer. J. Clin. Oncol. 37, 286–295 (2019).
Arora, K. et al. Genetic Ancestry Correlates with Somatic Differences in a Real-World Clinical Cancer Sequencing Cohort. Cancer Discov. 12, 2552–2565 (2022).
Alexander, D. H., Novembre, J. & Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664 (2009).
Auton, A. et al. A global reference for human genetic variation. Nature 526, 68–74 (2015).
Acknowledgements
We gratefully acknowledge the members of the Molecular Diagnostics Service in the Department of Pathology and would like to acknowledge the Center Core grant (P30 CA008748) and the Marie-Josee and Henry R. Kravis Center for Molecular Oncology for use of MSK-IMPACT data. All authors report no conflict of interests related to this study.
Author information
Authors and Affiliations
Contributions
C.Saoud: Conceptualization, data curation, formal analysis, visualization, methodology, writing-original draft, writing-review and editing; J.K.Dermawan: Conceptualization, methodology, supervision, writing-review and editing; K. Arora: Data curation, formal analysis, W. Tap: Resources, methodology, writing-review and editing; D.Reed: Conceptualization, resources, methodology, writing-review and editing; E.K Slotkin: Conceptualization, resources, methodology, writing-review and editing; L.H.Wexler: Conceptualization, methodology, supervision, writing-review and editing; Y.R. Murciano-Goroff: Conceptualization, methodology, writing-review and editing; A. Latham: Conceptualization methodology, writing-review and editing; D.L.Mandelker: Conceptualization, resources, supervision, investigation, project administration, writing-review and editing; C.R.Antonescu: Conceptualization, resources, supervision, investigation, project administration, writing-original draft, writing-review and editing.
Corresponding author
Ethics declarations
Competing interests
Y.R.M.-G. reports travel, accommodation, and expenses from AstraZeneca and Loxo Oncology/Eli Lilly. She acknowledges honoraria from Virology Education and Projects in Knowledge (for a CME program funded by an educational grant from Amgen). She has been on an advisory board for Revolution Medicines, and consulted for AbbVie. She acknowledges associated research funding to the institution from Mirati Therapeutics, Bristol Myers Squibb/E.R. Squibb & Sons, Loxo Oncology at Eli Lilly, Elucida Oncology, Taiho Oncology, Hengrui USA, Ltd/Jiangsu Hengrui Pharmaceuticals, Luzsana Biotechnology, Endeavor Biomedicines, and AbbVie. She is an employee of Memorial Sloan Kettering Cancer Center, which has an institutional interest in Elucida. She acknowledges royalties from Rutgers University Press and Wolters Kluwer. She acknowledges food/beverages from Endeavor Biomedicines, and other services from Amgen, Loxo Oncology/ Eli Lilly, and AbbVie. Y.R.M.-G. acknowledges receipt of training through an institutional K30 grant from the NIH (CTSA UL1TR00457). She has received funding from a Kristina M. Day Young Investigator Award from Conquer Cancer, the ASCO Foundation, endowed by Dr. Charles M. Baum and Carol A. Baum. She is also funded by the Fiona and Stanley Druckenmiller Center for Lung Cancer Research, the Andrew Sabin Family Foundation, the Society for MSK, the Squeri Grant for Drug Development, and a Paul Calabresi Career Development Award for Clinical Oncology (NIH/NCI K12 CA184746) as well as through NIH/NCI R01 CA279264. E.K.S. reports advisory board for Inhibrx Pharmaceuticals. She acknowledges consultant work for Guidepoint Global. She acknowledges research funding from Eli Lilly, Asperas Pharma, and y-mAbs Pharmaceuticals. D.R. reports DSMC work for Springworks and Eisai. All other authors declare no Competing Financial or Non-Financial Interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Saoud, C., Dermawan, J.K., Arora, K. et al. Germline pathogenic variants in DNA repair pathways: a key feature in a significant subset of translocation-associated sarcomas. npj Precis. Onc. 9, 133 (2025). https://doi.org/10.1038/s41698-025-00925-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41698-025-00925-6