Abstract
Antimicrobial resistance (AMR) is a global health concern, with natural ecosystems acting as reservoirs for resistant bacteria. We assessed AMR in Escherichia coli isolated from two wild sloth species in Costa Rica. E. coli from two-toed sloths (Choloepus hoffmanni), a species with greater mobility and a broader diet, showed resistance to sulfamethoxazole (25%), tetracycline (9.4%), chloramphenicol (6.3%), ampicillin (6.3%), trimethoprim (3.1%), and ciprofloxacin (3.1%), which correlated with the presence of resistance genes (tet(A), tet(B), blaTEM-1B, aph(3”)-Id, aph(6)-Id, sul2, qnrS1, floR and dfrA8). E. coli from three-toed sloths (Bradypus variegatus) showed 40% resistance to sulfamethoxazole despite no detected resistance genes, suggesting a regional effect. A significant negative correlation was found between AMR and distance to human-populated areas, highlighting anthropogenic impact on AMR spread. Notably, E. coli isolates from remote areas with no human impact indicate that some ecosystems remain unaffected. Preserving these areas is essential to protect environmental and public health.
Similar content being viewed by others
Introduction
Antimicrobial resistance (AMR), stemming from the widespread use of antibiotics, constitutes modern medicine’s foremost challenge. Many antimicrobial-resistant bacteria (ARB) emerge and disseminate in animal contexts, and a correlation between anthropogenic activities and AMR levels has been demonstrated1,2. Extensive studies have been conducted regarding this premise on humans, domestic animals, and livestock. Antibiotic consumption in animal production has reached up to 70% of global usage, and for decades, they have been administered as growth promoters or prophylactically3. In recent years, implementing regulatory changes to control AMR in many countries has drastically reduced livestock antibiotic consumption and AMR levels4. However, the development and spread of ARBs and antimicrobial resistance genes (ARGs) are still possible through the manure and wastewater of farms and slaughterhouses5. AMR remains one of the most significant challenges in livestock farming and can become a source of ARGs for humans and other animal bacteria, including those from wildlife. For its surveillance, a few bacteria have been considered sentinels, such as Escherichia coli, a common, ubiquitous bacterial species in most homeothermic species. It is easy to cultivate, highly survivable in the environment, and readily acquires AMR mechanisms. Moreover, it has been described as a good indicator of the selective pressure imposed by antimicrobial exposure in animals and, as a pathogen, E. coli stands out as one of the significant contributors to the increasing deaths associated with AMR in recent years (approximately 25% of death related to AMR in 2019 were attributable to E. coli)6,7.
Wildlife is supposed to interact little with human activities and, therefore, is less exposed to anthropogenic AMR8. For this reason, studies assessing the level of interaction between ecologically significant wildlife species and human activity are indispensable for comprehending the extent of anthropization in ecosystems and the dynamics of pathogenic agents and AMRs. Besides, an increasing number of species considered urban wildlife have closer contact with human activities, as they obtain unlimited resources to live, feed, and reproduce from cities9. In this sense, species such as raccoons (Procyon lotor) or tapirs (Tapirus bairdii) are more prone to AMR acquisition than non-urban wildlife species and can serve as a nexus between cities and ecosystems in the ARB spread8,10,11. Assessing the AMR dynamics in wildlife species is crucial to understanding their role in ecosystems and controlling ARB’s origin, reservoir, and spread. However, studies about AMR in wildlife are scarce, and its repercussions in natural ecosystems remain poorly understood12.
In this context, Costa Rica is known as one of the most biodiverse countries on the planet. It is recognized as a global biodiversity hotspot due to its rich variety of plant and animal species13. Previous studies have also identified AMR in isolates of rescued wildlife at a local rescue center14. Many Costa Rican species, such as Hoffmann’s two-toed sloth (Choloepus hoffmanni) and the three-toed sloth (Bradypus variegatus), are threatened by human activities like deforestation, habitat fragmentation, hunting, and pollution15. These sloths are iconic indigenous species with critical ecological roles. The ecological differences between the two sloth species are notable, particularly in their feeding habits. Two-toed sloths are primarily folivores and have a more varied diet that includes flowers, shoots, and fruits. and move in extensive areas16,17, while three-toed sloths are strictly folivores, relying on various arboreal leaves and moving very little from a specific deep forest area18,19. During the past decades, the increase in human activities has led to the rise of sloth habitat disturbance and the closer interaction between those species20.
In the present study, we analyzed the ARGs of E. coli populations using the two species of sloths from Costa Rica as indicators. The main objective of this research was to understand the impact of anthropogenic influence and ecological space on the bacterial and genomic composition of the E. coli populations in both sloth species. For this purpose, we assessed the resistome of two-toed and three-toed sloths from Costa Rica.
Results
Bacterial identification and AMR phenotypic evaluation
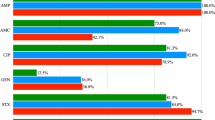
All the sloths were positive for confirmed E. coli, recovering a total of 47 isolates: 32 from two-toed sloths and 15 from three-toed sloths. AST revealed low levels of AMR in E. coli isolates from both sloth species. In two-toed sloth isolates, resistance to SMX (25%, 8/32), TET (9.4%, 3/32), CHL (6.3%, 2/32), AMP (6.3%, 2/32), TMP (3.1%, 1/32), and CIP (3.1%, 1/32) was found, whilst only SMX resistance was observed in three-toed sloths (40%, 6/15) (Fig. 1; Supplementary Data 1).
The tree reflects the phylogenetic relationships among the isolates and includes associated data on phylogroups, STs, AMR profiles, ARG content (Resfinder Database), and plasmid replicons identified (Plasmidfinder and MOB-Suite). * Replicon detected only with MOB-suite. Tree created with FastTree and edited with iTOL and Microreact.org.
Moreover, two of the two-toed sloth isolates were identified as MDR, comprising 25% of the resistant isolates (2/8) and 6.3% of the total isolates (2/32). One exhibited resistance to five antibiotic classes (SMX-CIP-TET-CHL-AMP), while the other showed resistance to four classes (SMX-TMP-TET-AMP). Notably, MDR isolates were obtained from two-toed sloths, both adult males, and collected 3.20 km apart, with a time interval of 32 days.
In contrast, 24 E. coli isolates demonstrated susceptibility to all 14 antimicrobials, constituting 75% of the total two-toed sloth isolates tested (24/32).
E. coli population structure
The 47 E. coli isolates exhibited a significant diversity, with 39 distinct Sequence Types (STs) identified across all samples (Fig. 1). In two-toed sloths, 27 different STs were recovered from 32 samples, while in three-toed sloths, there were 12 STs from 15 samples. Five new STs were identified and submitted to Enterobase for ST assignation (four isolates from ST12527, ESC_WA6892AA, ESC_YA9347AA, ESC_YA9349AA, and ESC_ZA0040AA; ST13059, ESC_ZA4827AA; ST13060, ESC_YA9348AA; ST13061, ESC_YA9350AA; and ST13061, ESC_YA9351AA).
Regarding population diversity, it was observed that 40% (6/15) of the isolates obtained from three-toed sloths belonged to the novel STs, underscoring the richness and uniqueness of the E. coli population in this species. The ST12527 described in this study was found in four isolates of three-toed sloths in entirely separate locations, but it was not found in the two-toed sloth. This species also identified two other isolates, attributed as ST13060 and ST13061. In contrast, for the two-toed sloth, only 6.3% (2 out of 32) of the STs were newly discovered (ST13259 and ST13062), while the remaining STs had already been described in previous studies. Based on the core genome, the single-nucleotide polymorphism (SNP) distance between the isolates ranged from 42 to 105,412 SNPs. Notably, there were only 42 SNPs between isolates BB1587 and BB1589 (ST12527), even though the three-toed sloths sampled were from two locations, 4.72 km apart and separated by 17 days. A similar observation was made with isolates BB1583 and BB1584 (ST12527), where only 52 SNPs differentiated them. These strains were isolated from two three-toed sloths (an adult and a baby, respectively) rescued from two geographical locations 3.95 km apart and admitted to the JCR one day apart (on 10/10 and 11/10, respectively). This novel ST appeared to be highly conserved in this sloth species.
The analysis revealed distinct E. coli phylogroups in the two-toed sloth, encompassing phylogroups A, B1, B2, D, F, and G. Conversely, the three-toed sloth exhibited a different phylogroup distribution, featuring B1, B2, D, E, and G (Fig. 1).
Antimicrobial resistance genes and plasmid content
Resistome analysis revealed a core set of intrinsic resistance genes exhibiting high prevalence across the studied population. Specifically, 41 genes were identified in ≥80% of the isolates, with a subset of 37 genes detected universally across all 47 strains (Supplementary Data 1). This core resistome comprised key components of efflux systems (e.g., mdtABCDEF, emrAB, acrB, acrD, acrA, acrF, acrS, acrE, tolC, mdfA, mdtP) and global regulatory elements (eptA, yojI, CRP, H-NS, cpxA, baeR, baeS), as well as the beta-lactamase gene ampC.
However, most isolates (90.6%; 29/32) did not exhibit acquired resistance genes. ARGs were identified in 9.4% (3/32) of the isolates from the two-toed sloth, encompassing a variety of genes including those for tetracyclines (tet(A), tet(B)), beta-lactams (blaTEM-1B), aminoglycosides (aph(3”)-Id, aph(6)-Id), sulfonamides (sul2), quinolones (qnrS1), amphenicols (floR), and diaminopyrimidines (dfrA8).
Unexpectedly, no acquired ARGs were identified in the isolates from the three-toed sloth, which did not correlate with the phenotypic resistance profiles exhibited against sulfamethoxazole. The identity for detecting any acquired gene related to sulfonamide in the ResFinder database decreased to 30% (default parameters ResFinder were 90% identity and 60% coverage). However, no sul genes were found in almost all these isolates, except for isolate BB1574, where the sul2 gene was identified with 100% identity and 100% coverage when compared against the ResFinder database.
Furthermore, analysis of non-synonymous mutations in Bakta-annotated coding sequences between resistant isolates and closest sensitive isolates (BB1559 vs BB1569, BB1584 and BB1587 vs BB1583, and BB1591 vs BB1563; Fig. 1) did not reveal any significantly affected metabolic pathway or previously described genetic mutation common to all resistant isolates that would account for the observed AMR phenotypic values.
In contrast, the scenario concerning plasmid content exhibited stark differences. In the case of the two-toed sloth, plasmids were identified in 59.4% (19/32) of the isolates. Meanwhile, a higher percentage was observed in the three-toed sloth, precisely 93.3% (14/15) of the isolates. The main plasmid replicons detected in both species isolates were ColRNAI, IncFIB, and IncFII.
Biological and geographic factors
Statistical analysis using a GLM revealed varying degrees of influence of biological factors (sloth species, age, and sex) and spatial variables on phenotypic AMR levels in the isolates. Most biological factors were not statistically significant, indicating that they do not exert strong individual effects on phenotypic AMR. Specifically, sloth species (p = 0.8471), sex (p = 0.5574), and age (p = 0.0749 for babies and p = 0.6737 for juveniles) did not present significant associations with AMR levels.
In contrast, the proximity to district centroids or minimum distance to district centroids was statistically significant (β = −0.0017, p = 0.0277). This result indicates that sloths located closer to district centroids are more likely to exhibit phenotypic AMR. The negative coefficient suggests that proximity to these centroids—used as proxies for areas of human activity—is associated with an increased presence of AMR determinants.
The overall model fit was assessed using residual deviance (Residual deviance = 39.777 on 31 degrees of freedom) and the Akaike Information Criterion (AIC = 71.777), indicating an acceptable balance between model complexity and explanatory power.
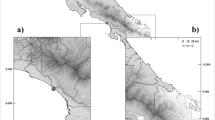
Figure 2 illustrates the spatial relationship between sloth sampling points, phenotypic AMR levels, and areas of varying human population density across Limón, Costa Rica. High population density areas, marked by hatching, align with urbanized zones and serve as indicators for human activity. At the same time, district centroids provide standardized reference points to measure sloth proximity to these regions. Higher AMR levels are observed near densely populated areas like Talamanca and coastal regions, emphasizing the impact of human activity.
Point size represents the number of individuals sampled, and the color indicates the number of antibiotics for which phenotypic AMR was detected (# of Resistance), from 0 (green) to 7 (red). The map highlights regions with larger human populations and includes centroids with a 2000-unit radius. Sloth populations are predominantly clustered along the coastal area.
Discussion
The present pilot study described phenotypic AMR in various isolates from sloth species inhabiting the southern region of Costa Rica. Despite the importance of AMR and the ecological niches of sloths in Costa Rica, only one study has been previously published on this subject20. Interestingly, phenotypic AMR levels detected in the present study are comparatively lower than those reported by Fernandes et al.20. This variance can likely be attributed to the sampling strategy, which exclusively focused on wild animals, excluding individuals with potential human contact. Despite this, two-toed sloths showed phenotypic AMR to several antibiotics previously identified, including AMP, TET, CIP, and CHL20. However, we uncovered resistance to TMP and SMX, constituting the first documented report of these resistance profiles in sloths. Notably, among two-toed sloths, only two isolates were characterized as MDR (6.3%, 2/32). At the same time, Fernandes et al.20 detected a higher percentage of MDR in E. coli (10%, 4/40) and one extensively drug-resistant (XDR) isolate from a sloth previously treated in a wildlife rescue center20. Conversely, the only phenotypic AMR identified in three-toed sloths was SMX. This discrepancy might signify a distinction between the species. It is important to note that over 2/3 of the rectal samples were from two-toed sloths. Beyond dietary differences, the spatial ecology of the two sloth species also differs, possibly linked to variations in their microbiome or AMR8. Moreover, two-toed sloths spend about 20% of their time on the forest floor, compared to 12% for three-toed sloths16,21 exposing them to a more diverse bacterial environment. This highlights their distinct ecological roles and dietary niches, which could increase the probability of ARB acquisition16,21. These sloths, considered keystone species for ecosystem conservation, play a crucial role in maintaining vegetal biodiversity and mutualism with moths and algae22.
Furthermore, our study delved into the genomic profiles of different E. coli strains, an aspect previously overlooked in this wildlife species. The considerable diversity among the various E. coli isolates within the same environment is striking. Overall, we identified 39 distinct STs, representing a wide range of variations with low repetition, particularly within the same ecological niche and geographical ___location. Five new STs were also described, highlighting the lack of data from this ecogeographical context. Five STs were isolated twice (ST155, ST127, ST939, ST720, ST394), and one was isolated from four different three-toed sloths (ST12527), each from different locations, sexes, and ages. This ST appeared highly conserved, as evidenced by the limited SNP distances between the isolated bacteria. Both isolates, with only 42 SNPs of difference, belonged to ST12527 (BB1587 and BB1589) in the three-toed sloths and were found in a 17-day frame in two different locations. Although rapid bacterial evolution or environmental pressure could influence these circumstances, in this case, it is more likely due to a common sharing area or a direct or indirect bacterial transmission between individuals. These findings suggest that similar E. coli strains are present in this wildlife species, even across different geographical areas, pointing to a bacterial homogeneity probably caused by a highly conserved lifestyle. Regarding the distribution of phylogroups, a few isolates belonging to the A, E, and F phylogroups are the only differences. Therefore, our results suggest ST differences but a similar phylogroup composition that is preserved.
Acquired ARGs are widespread, extending even into wildlife populations8. The widespread occurrence of acquired genes in our study strongly suggests that intrinsic resistance within these strains is likely maintained through clonal propagation and constitutes a stable component of their core resistome. It is crucial to recognize that animals with minimal human interaction may still harbor acquired ARGs23. The AMR genes found in the samples, including tet(A), tet(B), blaTEM-1B, aph(3”)-Id, aph(6)-Id, sul2, qnrS1, floR, and dfrA8, were entirely concordant with the observed phenotypic resistance in the two-toed sloth isolates. This observation demonstrates the inherent ability of E. coli to acquire and maintain resistance determinants, which might result from the adaptation to environmental antibiotic pressures. These findings are supported by the country’s significant prevalence of ARGs, as indicated by elevated antibiotic residue levels24. On the contrary, there was a lack of acquired SMX-resistance genes in all the isolates that presented resistance against that antibiotic in the three-toed sloth and eight isolates in the two-toed sloth. This absence of acquired ARGs suggests the presence of unidentified mechanisms of SMX resistance that are not conferred by sul-like genes. Notably, our analysis identified multiple intrinsic drug-resistance efflux pumps that could contribute to the observed SMX resistance phenotype. Importantly, intrinsic efflux pump genes, even in the absence of genetic mutations, can drive resistance phenotypes through overexpression or increased activity, often triggered by environmental stressors or regulatory changes (e.g., upregulation of marA or soxS regulators). Such systems not only confer resistance to antibiotics but also enhance tolerance to biocides, detergents, and other toxic compounds25. However, comparative genomic analysis revealed no genetic divergence in these efflux pump systems between clonally related resistant and susceptible isolates. Furthermore, none of the protein variants identified between closely related isolates with different SMX-resistance phenotypes analyzed could be associated with metabolic mechanisms of resistance to sulfonamides described before, such as detoxification routes, p-aminobenzoic acid hyperproduction, or sulfonamide-resistant dihydropteroate synthetase mutations26. These findings suggest that the observed SMX resistance disparities between closely related clonal isolates may arise from transcriptional variations (e.g., differential expression of efflux pump regulators) or post-translational modifications affecting pump activity, rather than genetic alterations in the efflux pump genes themselves27. Since this undefined SMX resistance was identified across different STs and phylogroups, the observed phenotype could reflect either a species-wide regulatory feature in E. coli (e.g., conserved stress response pathways) or a horizontally transferable mechanism if the implicated regulators are linked to mobile genetic elements. Experimental validation through transcriptomic profiling (e.g., RNA-seq) and efflux pump inhibition assays would help elucidate these putative regulatory drivers, highlighting the need to characterize novel resistance mechanisms in understudied ecological niches.
Surprisingly, in the present study, a large number of plasmid replicons were found in isolates from three-toed sloths (93.3%), coincidentally, the species with no acquired ARGs. A similar situation was observed in the two-toed species, but in a lower percentage (59.4%). Acquiring and maintaining plasmids might entail a high fitness cost for the bacteria, which can result in slower colony growth28. Therefore, plasmid maintenance could have a biological function in this ecological niche and confer significant advantages to the bacteria, such as beneficial metabolic functions29. Furthermore, most isolates, independently of ST or phylogroup, shared the same plasmid replicons, strongly suggesting horizontal transfer of these elements and their potential adaptive genetic traits. Likewise, the broad presence of these specific plasmids could favor the rapid acquisition, maintenance, and dissemination of newly acquired genetic material, including ARGs. The relationship between AMR and population density was further supported by statistical analysis, which found a significant association between phenotypic AMR levels and proximity to district centroids (p = 0.0277). Although the use of district centroids as proxies for human population centers has inherent limitations, the spatial structure of human settlements along the southern Costa Rican coast, characterized by a relatively continuous, linear urban development along the main roadways, supports the validity of this approximation in our study context. While centroids serve as a useful proxy, the observed alignment of high AMR levels with hatched areas suggests that population density plays a key role in driving the patterns of AMR in sloths. The significant negative correlation between minimal distance to the district and AMR levels suggests that factors associated with urban centers, such as higher antibiotic usage, increased population density, and greater access to healthcare, might be pivotal in shaping resistance patterns.
These findings are particularly relevant to the impact of the human-wildlife interface on AMR since urban environments could accelerate the development and spread of resistant strains due to increased antibiotic exposure and higher transmission rates1. The lack of significant associations with more specific geographic factors (like cantons or districts) suggests a complex interplay of environmental, ecological, and socio-economic factors in shaping resistance. These findings highlight the need to incorporate population density and urbanization metrics into future models to elucidate better the anthropogenic activity’s effect on wildlife health and AMR metrics in future analyses.
Recognizing the constraints inherent in our study, future research endeavors could benefit from increased sample size and an active sampling approach. In our study, sampling was passive, as animals were selected based on their need for assistance from the rescue center. However, an active sampling strategy, where animals are actively sought out for inclusion, could provide a more comprehensive understanding of antimicrobial resistance dynamics in wildlife populations. Also, it is crucial to recognize the inherent constraints of wildlife studies, which are often influenced by practical complexities. Additionally, epidemiological studies provide a snapshot of the situation during sampling. Therefore, implementing a continuous sampling strategy could prove invaluable in monitoring the dynamics of AMR in wildlife over time. A limitation of this study is the moderate sample size (47 individuals), which, although substantial relative to the total number of rescued sloths annually in the region, may restrict the ability to detect finer-scale spatial associations or more subtle environmental effects. To be more precise in our results, it would have been ideal to collect at least three different strains from each sample. Thus, future studies with larger sample sizes and enhanced spatial resolution would be valuable in confirming and expanding upon the patterns observed here.
Our findings emphasize the critical role of ecological contexts in shaping bacterial populations and AMR dynamics in wildlife. Natural environments, characterized by minimal human interference, are associated with lower AMR prevalence and greater diversity in E. coli populations. Intriguingly, the distinct ecological behaviors of the two sloth species revealed a differential risk of gene acquisition: C. hoffmanni, with its greater mobility, varied diet, and frequent terrestrial interactions, exhibits a higher propensity for acquiring resistance genes compared to the predominantly arboreal and folivorous B. variegatus. Furthermore, our study highlights the heightened risk of AMR dissemination in regions where wildlife habitats intersect with human-populated areas. These findings emphasize the urgent need for research to unravel the complex bacterial wildlife populations and their interplay with anthropogenic pressures, informing strategies to mitigate AMR and safeguard biodiversity.
Methods
Sampling strategy and distribution
From September 2019 to January 2020, 47 individual rectal swab samples were collected from sloths rescued from different geographic locations on the southern Costa Rican coast: 32 two-toed sloths and 15 three-toed sloths. The geographic coordinates of the rescue points for the sloths were recorded, as well as data related to age, sex, and species. Some of the sloths tested were in protected natural areas such as Parque Nacional de Cahuita, where urbanization is forbidden. Others were located inside urban areas, living next to the human population: Puerto Viejo de Talamanca, Manzanillo, Limón, and Bribrí. All the animals were free-living and had no prior known contact with humans.
The sloths tested here were all patients from the Jaguar Rescue Center (JRC) in Costa Rica (https://www.jaguarrescue.foundation/en-us/) and had been rescued from the wild. Rectal swabs were collected upon arrival using sterile swabs with AMIES transport medium (Deltalab®, Barcelona, Spain), prior to any treatment or sedation, and without the use of anesthesia. Samples were immediately refrigerated at 4 °C until further processing (Fig. 3).
All animal handling and sample collection procedures adhered to Costa Rican legislation (ACLACDRFVS-PVS-002-2018) and the ethical guidelines of the JRC, following the Ministerio de Ambiente y Energía (MINAE) regulations. The study protocol was reviewed and approved by the Sistema Nacional de Áreas de Conservación (SINAC) and the Comisión Nacional para la Gestión de la Biodiversidad (CONAGEBIO) of Costa Rica, under resolution N°R-SINAC-PNI-ACLAC-039-2020 (Exp. Dig. N°M-PC-SINAC-PNI-ACLAC-035-2020).
Processing samples and AMR phenotypic evaluation
Each swab was streaked in MacConkey and Brilliance E. coli agar media (Oxoid, Thermo Scientific™, Massachusetts, United States) and incubated for 24 h at 37 ± 1 °C. One presumptive E. coli colony from each plate was selected and streaked into LB agar medium (Oxoid, Thermo Scientific™, Massachusetts, United States). After 24 h of incubation at 37 ± 1 °C, monoclonal cultures were obtained. Then, a subsequent subculture on blood agar was performed, and MALDI-TOF used the resulting pure colonies for bacterial species identification.
After confirming the species, an antimicrobial susceptibility test (AST) was performed according to the European Committee on Antimicrobial Susceptibility Testing30 guidelines, using the broth microdilution method in Sensititre EUVSEC® plates (Thermo Scientific™, Massachusetts, United States). The plates contain 14 antibiotics, including ampicillin (AMP; 1-64 mg/L), azithromycin (AZI; 2-64 mg/L), cefotaxime (FOT; 0.25-4 mg/L), ceftazidime (TAZ: 0.5-8 mg/L), chloramphenicol (CHL; 8-128 mg/L), ciprofloxacin (CIP; 0.015-8 mg/L), colistin (COL; 1-16 mg/L), gentamicin (GEN; 0.5-32 mg/L), meropenem (MERO; 0.03-16 mg/L), nalidixic acid (NAL; 4-128 mg/L), sulfamethoxazole (SMX; 8-1024 mg/L), tetracycline (TET; 2-64 mg/L), tigecycline (TGC; 0.25-8 mg/L), trimethoprim (TMP; 0.25-32 mg/L). Strains were classified as multidrug-resistant (MDR) when resistant to at least one agent in three or more antibiotic classes31.
Whole genome sequencing and genomic analysis
Whole Genome Sequencing and data processing by Illumina® were conducted in the Agricultural Technology Institute of Castilla and León (ITACyL) laboratory. First, genomic DNA was extracted and purified with the Wizard Genomic DNA purification kit (Promega Corp., Madison, WI, USA), following the manufacturer’s guidelines for gram-negative bacteria. Shortly, 101 bp paired-end reads (550 bp insert size) of the 46 genomic samples were generated on a HiSeq 2500 platform (Illumina Inc., San Diego, California, USA).
Raw reads quality control was done with FastQC32. Genome assembly was conducted with Unicycler v.0.5.133, while Bakta v1.7.034 was employed for genomic annotation, and pangenome analysis was performed using the Roary v3.13.035 pipeline. Snp_sites 2.5.136 and snp-dist v.0.8.237 were used to identify single-nucleotide polymorphisms (SNPs) and extract them from the core-genome alignment resulting from Roary, which were analyzed with FastTree 2.1.1138 to generate the phylogenetic tree. Sequence types (STs) were determined by multi-locus sequence typing (MLST)39, and novel STs were submitted to the Enterobase database40 to be assigned. ARGs and plasmids were identified using multiple tools and databases. ARGs were detected with ResFinder 4.141,42,43 and the Comprehensive Antibiotic Resistance Database (CARD)44. Plasmid content was assessed using PlasmidFinder 2.0.145 and MOB-suite46.
In addition, snippy v4.6.047 was used to identify genetic variations in coding sequences between phylogenetically close isolates with AMR phenotypic differences. Subsequently, BlastKOALA v 3.0 (KEGG Orthology And Links Annotation)48 was used to search for metabolic networks and enriched bacterial functions among the identified gene variants.
Spatial analysis of species distribution and urban proximity
A spatial dataset representing the ___location of each species was created to perform the Spatial Data Analysis, using the longitude and latitude coordinates of the areas where the animals were observed. This approach allowed for a detailed examination of species distribution in relation to standardized spatial reference points.
The geographical extent of the study covered the entire province of Limón, enabling analysis across multiple administrative levels (province, canton, and district). To ensure consistency and neutrality in the proximity analysis, the centroids of each district in Limón were calculated based on their geometric boundaries. Centroids were used as consistent, replicable reference points in spatial analysis because they provided a standardized proximity measure and a reasonable proxy for human activity near district centers. These centroids were selected over urban centers because they provide an objective, mathematically defined reference point not influenced by uneven population distribution or infrastructure ___location. Urban centers, while significant in human geography, vary in their size, definition, and ___location, potentially introducing biases or inconsistencies into the analysis.
To complement this analysis, a one-kilometer buffer was created by accounting for human influences near district boundaries, such as roads and settlements, ensuring a broader spatial context for analyzing interactions like human-animal hotspots. This buffer allowed us to identify species points near urban areas, providing additional spatial context to the analysis. For key districts, spatial grids were constructed, enabling a more granular examination of the relationship between species distribution and proximity to district centroids. These grids helped to map spatial patterns at a finer resolution, facilitating the identification of potential hotspots near areas of human activity.
We calculated the shortest distance from each species ___location to the nearest district centroid using a distance function. These centroid-based distances served as standardized metrics for assessing proximity, ensuring that the results were spatially unbiased and reproducible. While the analysis was grounded in these objective centroid-based distances, the buffer and grid-based approaches further contextualized the results, offering insights into the relationship between species distributions and areas of human influence, including urban centers.
Assessing the influence of spatial and biological factors
Spatial and biological variables associated with the phenotypic AMR levels (resistance) and MDR determination were assessed using a generalized linear model (GLM) with a binomial family. The model is defined as follows:
The model (formula 1) included categorical variables such as species (Sp; three-toed sloth or two-toed sloth), age categories (A; baby [0–12 months for two-toed sloths and 0–7 months for three-toed sloths], juvenile [1–3 years for two-toed sloths and 7 months–2 years for three-toed sloths], and adult [over 3 years for two-toed sloths and over 2 years for three-toed sloths]) and sex (Sx; male or female; Supplementary Data 1). Additionally, the model incorporated continuous variables, such as the minimal district distance (DD; formula 1). The latter was key to studying the influence of human activities on the presence of AMR determinants in sloth populations. To this end, geographical parameters (canton (C), district (D), and grid cells (GC)) were also incorporated as categorical predictors. The minimum distances between the rescue point of sloths and the centroids of the Costa Rican cantons were calculated, and their relationship with the level of bacterial phenotypic resistance was evaluated by logistic regression.
All the statistical analyses were performed using the software R (R Core Team version 4.1.3, Vienna, Austria)49, with a significance level of p < 0.05. The following R packages were utilized for statistical, visualization and spatial data analysis: tidyverse (version 1.3.1), tidymodels (version 0.1.4), psych (version 2.1.9), car (version 3.0-12), dplyr (version 1.0.7), readxl (version 1.3.1), terra (version 1.5-12), geodata (version 1.0.2), rnaturalearth (version 0.3.2), raster (version 3.5-15), CoordinateCleaner (version 2.0-19), sdmpredictors (version 0.2.9), fuzzySim (version 2.5), leaflet (version 2.1.1), mapmisc (version 1.2.8), cartogram (version 0.2.6), kableExtra (version 1.3.4).
Data availability
The data have been deposited with links to BioProject accession number PRJNA1219484 in the NCBI BioProject database.
References
Sacristán, I. et al. Antibiotic resistance genes as landscape anthropization indicators: using a wild felid as sentinel in Chile. Sci. Total Environ. 703, 134900 (2020).
Woolhouse, M., Ward, M., van Bunnik, B. & Farrar, J. Antimicrobial resistance in humans, livestock and the wider environment. Philos. Trans. R. Soc. Lond. B Biol. Sci. 370, 20140083 (2015).
Stevenson, P. Links between industrial livestock production, disease including zoonoses and antimicrobial resistance. Anim. Res. One Health 1, 137–144 (2023).
EFSA, E. C. D. C. The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2020/2021. EFSA J. 21, e07867 (2023).
Caneschi, A., Bardhi, A., Barbarossa, A. & Zaghini, A. The use of antibiotics and antimicrobial resistance in veterinary medicine, a complex phenomenon: a narrative review. Antibiotics 12, 487 (2023).
Nyirabahizi, E. et al. Evaluation of Escherichia coli as an indicator for antimicrobial resistance in Salmonella recovered from the same food or animal ceca samples. Food Control 115, 107280 (2020).
Murray, C. J. et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399, 629–655 (2022).
Ramey, A. M. & Ahlstrom, C. A. Antibiotic resistant bacteria in wildlife: perspectives on trends, acquisition and dissemination, data gaps, and future directions. J. Wildl. Dis. 56, 1–15 (2020).
Miranda, A. C. Mechanisms of behavioural change in urban animals: the role of microevolution and phenotypic plasticity. In Ecology and Conservation of Birds in Urban Environments (eds Murgui, E. & Hedblom, M.) 113–132 (Springer, 2017).
Rojas-Jiménez, J. et al. Pansusceptible Escherichia coli isolates obtained from faeces of free-ranging Baird’s tapirs (Tapirus bairdii) suggest low selective pressure for resistance determinants in the northwestern region of the Talamanca Mountain Range, Costa Rica. J. Glob. Antimicrob. Resist. 16, 140–143 (2019).
Worsley-Tonks, K. E. et al. Importance of anthropogenic sources at shaping the antimicrobial resistance profile of a peri-urban mesocarnivore. Sci. Total Environ. 764, 144166 (2021).
Laborda, P. et al. Wildlife and antibiotic resistance. Front. Cell. Infect. Microbiol. 12, 873989 (2022).
Holland, M. B. Mesoamerican biological corridor. In Climate and Conservation: Landscape and Seascape Science, Planning, and Action (eds Hilty, J. A., Chester, C. C. & Cross, M. S.) 56–66 (Island Press, 2012).
Fernandes, R. et al. Resistant Escherichia coli isolated from wild mammals from two rescue and rehabilitation centers in Costa Rica: characterization and public health relevance. Sci. Rep. 14, 8039 (2024).
IUCN. The IUCN Red List of Threatened Species. Version 2023-1. https://www.iucnredlist.org/ (accessed on 27 December 2023).
Hayssen, V. Choloepus hoffmanni (Pilosa: Megalonychidae). Mamm. Species 43, 37–55 (2011).
Sánchez-Chavez, A. P. Diet of Hoffmann’s two-toed sloth (Choloepus hoffmanni) in Andean forest. Mammalia 85, 515–524 (2021).
Urbani, B. & Bosque, C. Feeding ecology and postural behaviour of the three-toed sloth (Bradypus variegatus flaccidus) in northern Venezuela. Mamm. Biol. 72, 321–329 (2007).
Fernandez Giné, G. A., Mureb, L. S. & Cassano, C. R. Feeding ecology of the maned sloth (Bradypus torquatus): understanding diet composition and preferences, and prospects for future studies. Austral Ecol. 47, 1124–1135 (2022).
Fernandes, M. et al. Antimicrobial resistance and virulence profiles of Enterobacterales isolated from two-finger and three-finger sloths (Choloepus hoffmanni and Bradypus variegatus) of Costa Rica. PeerJ 10, e12911 (2022).
Vaughan, C. et al. Spatial ecology and conservation of two sloth species in a cacao landscape in Limón, Costa Rica. Biodivers. Conserv. 16, 2293–2310 (2007).
Borges, C. et al. Safeguarding sloths and anteaters in the future: Priority areas for conservation under climate change. Biotropica 55, 306–317 (2023).
Hwengwere, K. et al. Antimicrobial resistance in Antarctica: is it still a pristine environment?. Microbiome 10, 71 (2022).
Vargas-Villalobos, S. et al. A case study on pharmaceutical residues and antimicrobial resistance genes in Costa Rican rivers: A possible route of contamination for feline and other species. Environ. Res. 242, 117665 (2024).
Holden, E. R. & Webber, M. A. MarA, RamA, and SoxS as mediators of the stress response: survival at a cost. Front. Microbiol. 11, 828 (2020).
Then, R. L. Mechanisms of resistance to trimethoprim, the sulfonamides, and trimethoprim-sulfamethoxazole. Rev. Infect. Dis. 4, 261–269 (1982).
Li, X. Z. & Nikaido, H. Efflux-mediated drug resistance in bacteria: an update. Drugs 69, 1555–1623 (2009).
Prensky, H., Gomez-Simmonds, A., Uhlemann, A. C. & Lopatkin, A. J. Conjugation dynamics depend on both the plasmid acquisition cost and the fitness cost. Mol. Syst. Biol. 17, e9913 (2021).
Dai, D. et al. Long-read metagenomic sequencing reveals shifts in associations of antibiotic resistance genes with mobile genetic elements from sewage to activated sludge. Microbiome 10, 20 (2022).
EUCAST (European Committee on Antimicrobial Susceptibility Testing). (2021). Breakpoint Tables for Interpretation of MICs and Zone Diameters, Version 11. 2021. Available online: http://www.eucast.org (accessed on 15 March 2023).
Magiorakos, A. P. et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281 (2012).
Andrews, S. FastQC: A quality control tool for high throughput sequence data. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (2010).
Wick, R. R., Judd, L. M., Gorrie, C. L. & Holt, K. E. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13, e1005595 (2017).
Schwengers, O. et al. Bakta: rapid and standardized annotation of bacterial genomes via alignment-free sequence identification. Microb. Genom. 7, 000685 (2021).
Page, A. J. et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693 (2015).
Page, A. J. et al. SNP-sites: rapid efficient extraction of SNPs from multi-FASTA alignments. Microb. Genom. 2, e000056 (2016).
Seemann, T. Snp-dists. https://github.com/tseemann/snp-dists (2017).
Price, M. N., Dehal, P. S. & Arkin, A. P. FastTree 2 – Approximately maximum-likelihood trees for large alignments. PLoS ONE 5, e9490 (2010).
Jolley, K. A. & Maiden, M. C. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinforma. 11, 595 (2010).
Zhou, Z. et al. The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny and Escherichia core genomic diversity. Genome Res 30, 138–152 (2020).
Bortolaia, V. et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 75, 3491–3500 (2020).
Camacho, C. et al. BLAST+: architecture and applications. BMC Bioinforma. 10, 421 (2009).
Zankari, E. et al. PointFinder: a novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J. Antimicrob. Chemother. 72, 2764–2768 (2017).
Alcock, B. P. et al. CARD 2023: expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 51, D690–D699 (2023).
Carattoli, A. & Hasman, H. PlasmidFinder and in silico pMLST: Identification and typing of plasmid replicons in whole-genome sequencing (WGS). Methods Mol. Biol. 2075, 285–294 (2020).
Robertson, J. & Nash, J. H. E. MOB-suite: software tools for clustering, reconstruction and typing of plasmids from draft assemblies. Microb. Genom. 4, e000206 (2018).
Seemann, T. Snippy. https://github.com/tseemann/snippy (2015).
Kanehisa, M., Sato, Y. & Morishima, K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 428, 726–731 (2016).
R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria (2021).
Acknowledgements
This research was supported by the Antimicrobial Resistance Unit (ARU), Veterinary Faculty, University Complutense of Madrid, which provided financial and infrastructural support. We thank Marta Hernández (University of Valladolid, Spain), María Jesús Sanchez-Calabuig (University Complutense of Madrid, Spain), and all the members of the ARU Laboratory for their help and participation in this project. We also must consider the contribution of all the workers and volunteers of the Jaguar Rescue Center (JRC).
Author information
Authors and Affiliations
Contributions
C.C.-F., R.S. and E.G.-V. collected the samples. C.C.-F. and N.M. performed the laboratory analyses. C.C.-F. and M.P.-V. conducted the bioinformatic analyses. M.M.D.-N. carried out the statistical analysis. C.C.-F., B.M.-M., M.M.D.-N. and M.P.-V. wrote the manuscript. J.F.D.-B. supported the bioinformatic interpretation. B.G.-Z. and E.G.-V. provided funding. B.G.-Z. supervised the research. C.C.-F., M.M.D.-N. and B.G.-Z. conceived and designed the study. C.C.-F. and M.M.D.-N. prepared the data visualizations. All authors reviewed and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Calvo-Fernandez, C., Dolcet-Negre, M.M., Martin-Maldonado, B. et al. Human-wildlife ecological interactions shape Escherichia coli population and resistome in two sloth species from Costa Rica. npj Antimicrob Resist 3, 62 (2025). https://doi.org/10.1038/s44259-025-00134-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s44259-025-00134-y