Abstract
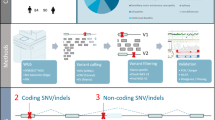
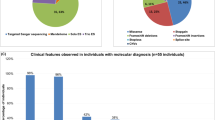
Structural variants contribute to genetic variability in human genomes and they can be presented in population-specific patterns. We aimed to understand the landscape of structural variants in the genomes of healthy Indian individuals and explore their potential implications in genetic disease conditions. For the identification of structural variants, a whole genome sequencing dataset of 1029 self-declared healthy Indian individuals from the IndiGen project was analysed. Further, these variants were evaluated for potential pathogenicity and their associations with genetic diseases. We also compared our identified variations with the existing global datasets. We generated a compendium of total 38,560 high-confident structural variants, comprising 28,393 deletions, 5030 duplications, 5038 insertions, and 99 inversions. Particularly, we identified around 55% of all these variants were found to be unique to the studied population. Further analysis revealed 134 deletions with predicted pathogenic/likely pathogenic effects and their affected genes were majorly enriched for neurological disease conditions, such as intellectual disability and neurodegenerative diseases. The IndiGenomes dataset helped us to understand the unique spectrum of structural variants in the Indian population. More than half of identified variants were not present in the publicly available global dataset on structural variants. Clinically important deletions identified in IndiGenomes might aid in improving the diagnosis of unsolved genetic diseases, particularly in neurological conditions. Along with basal allele frequency data and clinically important deletions, IndiGenomes data might serve as a baseline resource for future studies on genomic structural variant analysis in the Indian population.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
269,00 € per year
only 22,42 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data sets supporting the findings of this article are included within the article and as Supplementary Data. The variant file for the final SV callset is made available for download on the IndiGenomes database website. https://clingen.igib.res.in/indigen/.
References
Collins RL, Brand H, Karczewski KJ, Zhao X, Alföldi J, Francioli LC, et al. A structural variation reference for medical and population genetics. Nature. 2020;581:444–51.
Keegan NP, Wilton SD, Fletcher S. Breakpoint junction features of seven DMD deletion mutations. Hum Genome Var. 2019;6:39.
Cusin V, Clermont O, Gérard B, Chantereau D, Elion J. Prevalence of SMN1 deletion and duplication in carrier and normal populations: implication for genetic counselling. J Med Genet. 2003;40:e39.
Kurtovic-Kozaric A, Mehinovic L, Stomornjak-Vukadin M, Kurtovic-Basic I, Catibusic F, Kozaric M, et al. Diagnostics of common microdeletion syndromes using fluorescence in situ hybridization: single center experience in a developing country. Bosn J Basic Med Sci. 2016;16:121–5.
Sudmant PH, Rausch T, Gardner EJ, Handsaker RE, Abyzov A, Huddleston J, et al. An integrated map of structural variation in 2504 human genomes. Nature. 2015;526:75–81.
Ramaswamy S, Jain R, El Naofal M, Halabi N, Yaslam S, Taylor A, et al. Middle Eastern Genetic Variation Improves Clinical Annotation of the Human Genome. J Pers Med [Internet]. 2022;12. Available from: https://doi.org/10.3390/jpm12030423
Genome of the Netherlands Consortium. Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat Genet. 2014;46:818–25.
Gautam P, Jha P, Kumar D, Tyagi S, Varma B, Dash D, et al. Spectrum of large copy number variations in 26 diverse Indian populations: potential involvement in phenotypic diversity. Hum Genet. 2012;131:131–43.
Mastana SS. Unity in diversity: an overview of the genomic anthropology of India. Ann Hum Biol. 2014;41:287–99.
Sharma SK, Kalam MA, Ghosh S, Roy S. Prevalence and determinants of consanguineous marriage and its types in India: evidence from the National Family Health Survey, 2015–2016. J Biosoc Sci. 2021;53:566–76.
Reich D, Thangaraj K, Patterson N, Price AL, Singh L. Reconstructing Indian population history. Nature. 2009;461:489–94.
Nakatsuka N, Moorjani P, Rai N, Sarkar B, Tandon A, Patterson N, et al. The promise of discovering population-specific disease-associated genes in South Asia. Nat Genet. 2017;49:1403–7.
Jain A, Bhoyar RC, Pandhare K, Mishra A, Sharma D, Imran M, et al. IndiGenomes: a comprehensive resource of genetic variants from over 1000 Indian genomes. Nucleic Acids Res. 2021;49:D1225–32.
Mahmoud M, Gobet N, Cruz-Dávalos DI, Mounier N, Dessimoz C, Sedlazeck FJ. Structural variant calling: the long and the short of it. Genome Biol. 2019;20:246.
Cameron DL, Di Stefano L, Papenfuss AT. Comprehensive evaluation and characterisation of short read general-purpose structural variant calling software. Nat Commun. 2019;10:3240.
Sarwal V, Niehus S, Ayyala R, Kim M, Sarkar A, Chang S, et al. A comprehensive benchmarking of WGS-based deletion structural variant callers. Brief Bioinform [Internet]. 2022;23. Available from: https://doi.org/10.1093/bib/bbac221
Kosugi S, Momozawa Y, Liu X, Terao C, Kubo M, Kamatani Y. Comprehensive evaluation of structural variation detection algorithms for whole genome sequencing. Genome Biol. 2019;20:117.
Geoffroy V, Herenger Y, Kress A, Stoetzel C, Piton A, Dollfus H, et al. AnnotSV: an integrated tool for structural variations annotation. Bioinformatics. 2018;34:3572–4.
Layer RM, Chiang C, Quinlan AR, Hall IM. LUMPY: a probabilistic framework for structural variant discovery. Genome Biol. 2014;15:R84.
Chen X, Schulz-Trieglaff O, Shaw R, Barnes B, Schlesinger F, Källberg M, et al. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics. 2016;32:1220–2.
Chiang C, Layer RM, Faust GG, Lindberg MR, Rose DB, Garrison EP, et al. SpeedSeq: ultra-fast personal genome analysis and interpretation. Nat Methods. 2015;12:966–8.
Jeffares DC, Jolly C, Hoti M, Speed D, Shaw L, Rallis C, et al. Transient structural variations have strong effects on quantitative traits and reproductive isolation in fission yeast. Nat Commun. 2017;8:14061.
Riggs ER, Andersen EF, Cherry AM, Kantarci S, Kearney H, Patel A, et al. Technical standards for the interpretation and reporting of constitutional copy-number variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen). Genet Med. 2020;22:245–57.
Zhang D, Hu Q, Liu X, Zou K, Sarkodie EK, Liu X, et al. AllEnricher: a comprehensive gene set function enrichment tool for both model and non-model species. BMC Bioinforma. 2020;21:106.
Karolchik D, Hinrichs AS, Kent WJ. The UCSC genome browser. Curr Protoc Bioinforma. 2009;Chapter 1:Unit1.4.
Halldorsson BV, Eggertsson HP, Moore KHS, Hauswedell H, Eiriksson O, Ulfarsson MO, et al. The sequences of 150,119 genomes in the UK Biobank. Nature. 2022;607:732–40.
Bose P, Hermetz KE, Conneely KN, Rudd MK. Tandem repeats and G-rich sequences are enriched at human CNV breakpoints. PLoS One. 2014;9:e101607.
Wright CF, Fitzgerald TW, Jones WD, Clayton S, McRae JF, van Kogelenberg M, et al. Genetic diagnosis of developmental disorders in the DDD study: a scalable analysis of genome-wide research data. Lancet. 2015;385:1305–14.
Amberger JS, Bocchini CA, Scott AF, Hamosh A. OMIM.org: leveraging knowledge across phenotype-gene relationships. Nucleic Acids Res. 2019;47:D1038–43.
Yuan B, Wang L, Liu P, Shaw C, Dai H, Cooper L, et al. CNVs cause autosomal recessive genetic diseases with or without involvement of SNV/indels. Genet Med. 2020;22:1633–41.
Lalani SR, Liu P, Rosenfeld JA, Watkin LB, Chiang T, Leduc MS, et al. Recurrent muscle weakness with rhabdomyolysis, metabolic crises, and cardiac arrhythmia due to Bi-allelic TANGO2 mutations. Am J Hum Genet. 2016;98:347–57.
Gupta D, Bijarnia-Mahay S, Saxena R, Kohli S, Dua-Puri R, Verma J, et al. Identification of mutations, genotype-phenotype correlation and prenatal diagnosis of maple syrup urine disease in Indian patients. Eur J Med Genet. 2015;58:471–8.
Huie ML, Shanske AL, Kasper JS, Marion RW, Hirschhorn R. A large Alu-mediated deletion, identified by PCR, as the molecular basis for glycogen storage disease type II (GSDII). Hum Genet. 1999;104:94–8.
Puri RD, Setia N, N V, Jagadeesh S, Nampoothiri S, Gupta N, et al. Late onset Pompe Disease in India - Beyond the Caucasian phenotype. Neuromuscul Disord. 2021;31:431–41.
Truty R, Paul J, Kennemer M, Lincoln SE, Olivares E, Nussbaum RL, et al. Prevalence and properties of intragenic copy-number variation in Mendelian disease genes. Genet Med. 2019;21:114–23.
Rice AM, McLysaght A. Dosage sensitivity is a major determinant of human copy number variant pathogenicity. Nat Commun. 2017;8:14366.
Aradhya S, Truty R. AB003. Prevalence of copy number and structural variants across Mendelian disorders. Ann Transl Med. AME Publishing Company; 2017;5:AB003–AB003.
India State-Level Disease Burden Initiative Neurological Disorders Collaborators. The burden of neurological disorders across the states of India: the Global Burden of Disease Study 1990–2019. Lancet Glob Health. 2021;9:e1129–44.
Nalls MA, Blauwendraat C, Vallerga CL, Heilbron K, Bandres-Ciga S, Chang D, et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet Neurol. 2019;18:1091–102.
GBD 2016 Parkinson’s Disease Collaborators. Global, regional, and national burden of Parkinson’s disease, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 2018;17:939–53.
Acknowledgements
Authors acknowledge the funding support from the Council of Scientific and Industrial Research (CSIR) India (Grant No. MLP1809, MLP1801, MLP2001). Authors acknowledge all the volunteers who participated in this study. MKD, AJ, AB, and BJ acknowledge the CSIR fellowship; MI acknowledges the ICMR fellowship; DS acknowledges Intel research fellowship; VG acknowledges the UGC fellowship. The funding body has no role in the design of the study and in writing the manuscript.
Author information
Authors and Affiliations
Contributions
SSB and V Scaria conceived the idea; MKD, AJ, RCB, VS, BJ, MI, DS, AB, and VG performed research; and MKD, SSB, and VS wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
This study was approved by the Institutional Human Ethics Committee (IHEC) of the CSIR-Institute of Genomics and Integrative Biology. The participants were explained about the informed consent process as per the approved IHEC guidelines.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
10038_2023_1131_MOESM1_ESM.docx
Data on benchmarking for variant detection tool, supplementary method for variant filtering and disease status of rare deletions affected genes in DDD and OMIM database
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Divakar, M.K., Jain, A., Bhoyar, R.C. et al. Whole-genome sequencing of 1029 Indian individuals reveals unique and rare structural variants. J Hum Genet 68, 409–417 (2023). https://doi.org/10.1038/s10038-023-01131-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-023-01131-7
This article is cited by
-
Whole genome sequencing of families diagnosed with cardiac channelopathies reveals structural variants missed by whole exome sequencing
Journal of Human Genetics (2024)
-
A Catalogue of Structural Variation across Ancestrally Diverse Asian Genomes
Nature Communications (2024)