Abstract
Epigenetic responses to cannabis use could link cannabis use to health problems. We examined the DNA-methylation profiles of long-term cannabis users in midlife, re-evaluating a set of 246 cannabis-associated methylation markers that were previously identified in other studies. Data were from the Dunedin Study, a five-decade longitudinal study of a birth cohort (analytic n = 787). Peripheral whole blood was drawn when the cohort was age 45, and DNA methylation was assayed using the EPIC 850 K BeadChip. Analyses compared long-term cannabis users with non-users and, for a benchmark comparison, long-term tobacco users. Results showed that long-term cannabis use was associated with sixteen of the previously published 246 cannabis-related methylation markers. Methylation markers that were associated with long-term cannabis use were also associated with long-term tobacco use. However, after adjusting for long-term tobacco use and other covariates, long-term cannabis use was robustly associated with hypomethylation of nine markers: cg05575921, cg21566642, cg03636183, cg21161138, cg01940273, cg17739917, cg05086879, cg02978227, cg23079012. Cannabis-related hypomethylation was associated with higher gene expression in the Dunedin Cohort, suggesting meaningful biological associations. A comparison of long-term cannabis users with cannabis quitters revealed that quitters showed less extreme DNA hypomethylation. Long-term cannabis use could affect the epigenome similarly to tobacco use, possibly at least partly though smoke inhalation. Cannabis cessation, like tobacco cessation, may reverse altered DNA methylation.
Similar content being viewed by others
Introduction
Long-term cannabis use has been linked to a number of health problems, including respiratory problems, cardiovascular diseases, cognitive decline, and mental illness [1]. Although some associations are inconsistent, evidence suggests that exposure to cannabinoids such as delta-9-tetrahydrocannabinol (THC) may pose risks to health. For example, cannabis administration studies have shown that THC, the main psychoactive constituent of cannabis, has acute effects on the vasculature [2] and on mental health symptoms [3]. Moreover, it is well known that smoking has negative effects on various aspects of health, and most people who use cannabis smoke it [4, 5]. These findings support the biological plausibility of effects of long-term cannabis use on some aspects of health.
Epigenetic responses to substance use, which can influence gene expression without altering genetic sequence, are increasingly recognized as mechanisms linking substance use to adverse health outcomes [6]. DNA methylation is one epigenetic mechanism, and it involves the addition or removal of methyl groups to DNA, usually at cytosine-guanine dinucleotides (CpG sites) [7]. Of all the substances, tobacco smoking has most consistently been associated with differential DNA methylation, notably hypomethylation of a CpG site located in the aryl hydrocarbon receptor repressor (AHRR) gene -- cg05575921 [8]. Hypomethylation of this site has been found in some studies to predict the development of lung cancer [9]. Importantly, hypomethylation of cg05575921 is linked to tobacco smoking and not non-combustible tobacco use [8], suggesting that it is exposure to the harmful chemicals in smoke, as opposed to nicotine, that causes the hypomethylation. The inhaled combustion products for cannabis and tobacco are largely similar [10], raising the question: do people who smoke cannabis show similar patterns of differential methylation to those who smoke tobacco?
The DNA methylation profiles of tobacco users have been fairly well characterized, but few studies have reported on the DNA methylation profiles of cannabis users. This represents a significant knowledge gap, especially given cannabis legalization and increases in cannabis use [11,12,13,14,15]. We identified eight studies published by August 2024 that reported on cannabis-related DNA methylation for up to eight hundred and fifty thousand CpG sites (Table S1) [16,17,18,19,20,21,22,23]. The studies generally found that cannabis use was associated with differential methylation of a small number of CpG sites after adjusting for multiple testing. However, the studies disagreed on which sites and on whether tobacco smoking accounted for cannabis associations. Moreover, some studies relied on small, unrepresentative samples. Most studies used relatively crude measures of cannabis use. For example, two studies distinguished only between those who had ever used versus never used cannabis [17, 23]. No studies examined whether cannabis cessation could restore DNA methylation profiles to that of non-users.
We aimed to redress these limitations in a representative birth cohort followed prospectively to midlife. We capitalized on the study’s strong and repeated assessments of cannabis use over a span of thirty years to identify, at midlife, a group of long-term cannabis users. Long-term cannabis users are of special clinical interest because long-term cannabis use is associated with more health, cognitive, and social problems compared with occasional, recreational use [1, 24,25,26,27]. Moreover, midlife is of interest because it is the developmental period when physical health decline becomes apparent [28] and when chronic diseases tend to onset [29].
This study addressed five questions. First, do previously reported cannabis-methylation associations replicate in a group of middle-age long-term cannabis users from an independent cohort? We elected a replication approach, as opposed to conducting an exploratory epigenome wide association study (EWAS), because EWAS requires very large samples to detect what are likely small-effect associations across hundreds of thousands of sites. We selected for analysis a set of 246 previously published cannabis-associated CpG sites (Table S2).
Second, how do long-term cannabis users compare with lifelong non-users and long-term tobacco users? We compared long-term cannabis users not only with non-users but also with long-term tobacco users, because many people who use cannabis also smoke tobacco and because the methylation profiles of long-term tobacco users are well characterized. Therefore, tobacco findings provide important context for interpreting cannabis findings. Analyses took two complementary approaches: (1) group comparisons of long-term cannabis users with lifelong non-users and long-term tobacco users, and (2) tests of dose-response associations, using, as the exposures, continuously-distributed quantitative measures of the extent of persistence of regular cannabis use and persistence of tobacco dependence over a lifetime. We elected to use a group comparison approach because it is the approach used in case-control studies. We elected to test dose-response associations because these tests are statistically powerful, and dose-response associations would be expected if associations were causal. Consistent findings across the two approaches would bolster confidence in cannabis-methylation associations.
Third, is differential DNA methylation among long-term cannabis users robust to control for confounders? We selected a set of plausible confounders and included them as covariates in tests of dose-response associations: childhood socioeconomic deprivation; low childhood self-control; family history of substance dependence; and persistence of tobacco, alcohol, and illicit drug use.
Fourth, do any of the cannabis-related methylation markers represent meaningful biological associations? Analyses examined correlations between cannabis-related methylation marker levels and whole-genome gene expression. Fifth, does cannabis cessation reverse differences in DNA methylation profiles? Analyses compared cannabis and tobacco quitters with non-users and with long-term cannabis users and long-term tobacco users. Our approach to addressing these questions is summarized in Fig. 1. Replicated findings of cannabis-related differential DNA methylation would suggest alterations in DNA methylation that could have important implications for later health.
Methods
Participants
Participants are members of the Dunedin Longitudinal Study, a representative birth cohort (N = 1,037; 91% of eligible births; 52% male) born April 1972-March 1973 in Dunedin, New Zealand (NZ), who were eligible based on residence in the province and who participated in the first assessment at age 3 years. As adults, the cohort’s distributions match the range in national surveys of same-age New Zealanders on educational attainment, body mass index, smoking, physical activity, and physician visits [30, 31]. Study participants are primarily of New Zealand European ethnicity; 8.6% reported Māori ethnicity at age 45.
Assessments were carried out at birth and ages 3, 5, 7, 9, 11, 13, 15, 18, 21, 26, 32, 38, and most recently (completed April 2019) 45 years. Of the original 1037 cohort participants, 997 were still alive at age 45; of these, 938 (94.1%) participated in the age-45 assessments [474 men (50.5%)]. Participants assessed at age 45 did not differ significantly from other living participants in terms of childhood SES, childhood IQ, or history of psychopathology (Fig. S1).
Ethics approval and consent to participate
Participants gave written informed consent. Study protocols were approved by the NZ Health and Disability Ethics Committee (Protocol: 2017-1211). All methods were performed in accordance with the relevant guidelines and regulations.
Measures
Measures are described here and more details are shown in Table S3.
Age-45 DNA methylation
At age 45, DNA extracted from peripheral whole blood was assayed using the EPIC 850 K BeadChip. Analyses are restricted to cohort members who did not self-identify as Māori at the time of the first blood draw and who consented to phlebotomy and whose DNA methylation passed quality control (n = 818).
As previously described [32], whole blood was collected in 10 mL K2EDTA tubes from 90% (N = 824) of participants at age 45. DNA was extracted from the buffy coat using standard procedures [33, 34]. ~500 ng of DNA from each sample was treated with sodium bisulfite using the EZ-96 DNA Methylation kit (Zymo Research, CA, USA). DNA methylation was quantified using the Illumina Infinium HumanMethylationEPIC BeadChip (“Illumina EPIC BeadChip”) run on an Illumina iScan System (Illumina, CA, USA) at the Molecular Genomics Core at the Duke Molecular Physiology Institute. All sample processing and array profiling was performed as a single experiment (i.e. at the same time).
Data were processed using ‘wateRmelon’ (v1.26.0; [35]) and were normalized using the ‘methylumi’ (v2.30.0; [36]) Bioconductor package from the R statistical programming environment and subjected to quality control (QC) analyses. Samples were removed if the average detection p-value was >=0.05. To confirm genetic identity of the DNA samples, we assessed genotype concordance between SNP probes on the EPIC array and data generated using Illumina OmniExpress12v1.1 genotyping BeadChips. Principal components analysis was performed on the full, normalized dataset and the first two components plotted [37]. Samples formed two major clusters separating on the 1st component, which corresponded to recorded sex. This was used to confirm sex assignment. Samples from 818 age-45 participants passed our QC pipeline.
To permit control for technical variation, we used residualized DNA methylation values for the principal components (PCs) using DNA methylation beta values for the 186 QC probes on the EPIC BeadChip. A total of 32 PCs were retained explaining 90% of the variation in QC probes. To control for cell type composition, we also included white cell-type counts measured using flow cytometry (Sysmex Corporation, Japan) in whole blood samples taken concurrently with the DNA sample.
Substance use
At ages 18, 21, 26, 32, 38, and 45, participants were interviewed about their substance use using the Diagnostic Interview Schedule [38, 39], and past-year substance-use dependencies were assessed following Diagnostic and Statistical Manual of Mental Disorders criteria. This information was used to define exposures for group comparisons and for tests of dose-response associations.
Groups for comparison
Groups comprised cohort members who met criteria at age 45 for (i) long-term cannabis use, (ii) long-term tobacco use, (ii) lifelong non-use of cannabis and tobacco, (iv) former cannabis use, (v) former tobacco use (Fig. S2). Subsets of cohort members who met criteria for these pre-registered groups were selected as described below and as previously published [24, 40, 41] to mimic groups from case-control studies. If a cohort member did not meet criteria for a group, they were not included in group comparisons (n = 332), but they were included in tests of dose-response associations. Table S4 shows background characteristics and substance use for the cohort and for each group described below.
Long-term cannabis users (n = 74; 68% male) used cannabis weekly or more frequently in the past year at age 45, or were dependent on cannabis at age 45, and also used weekly or more frequently at one or more previous assessment waves. Of these, 28% used cannabis before age 18; 74% met criteria for cannabis dependence at one or more waves; and 89% used regularly (4+ days per week) at one or more waves. Age-45 cannabis consumption was a median of 300 days in the past year, with 62% using 4+ days per week. Most long-term cannabis users had a history of tobacco dependence (84%) and smoked tobacco daily at age 45 (66%).
Long-term tobacco users (n = 57; 40% male) smoked tobacco daily at age 45 and also smoked daily at one or more previous waves; were free from cannabis at age 45; and had no history of weekly cannabis use or dependence.
Lifelong cannabis/tobacco non-users (n = 189; 40% male) never used cannabis, never used tobacco daily, and never had a diagnosis of any substance-use disorder.
Cannabis quitters (n = 50; 60% male) did not use cannabis at age 45 but previously either were diagnosed with cannabis dependence or used cannabis regularly (4+ days per week). Most had a history of tobacco dependence (60%) and a quarter smoked tobacco daily at age 45.
Tobacco quitters (n = 148; 45% male) did not use tobacco at age 45 but were previously diagnosed with tobacco dependence. Nearly a third of tobacco quitters had a history of cannabis dependence (29%), but few of them used cannabis regularly at age 45 (5%).
Quantitative exposures for tests of dose-response associations
We report on two previously published quantitative exposures [24, 40]: persistence of regular cannabis use and persistence of tobacco dependence.
Persistence of regular cannabis use (i.e., 4+ days per week) comprised participants who (i) never used cannabis (n = 244), (ii) used but never regularly (n = 450), (iii) used regularly at one study wave from age 18–45 years (n = 44), (iv) two waves (n = 26), (v) three waves (n = 26), and (vi) 4+ waves (n = 27).
Persistence of tobacco dependence comprised participants who (i) never smoked tobacco (n = 411), (ii) smoked tobacco daily at one or more assessment waves but were never diagnosed with tobacco dependence (n = 114), (iii) were diagnosed at one wave (n = 92), (iv) two waves (n = 81), (v) three waves (n = 52), and (vi) four or more waves (n = 67).
Covariates
The following covariates were included in all statistical tests: sex, methylation array control probe principal components indexing technical variation, and white blood cell counts. White blood cell counts (neutrophils, lymphocytes, monocytes, eosinophils, and basophils) were included because there can be differences in patterns of methylation between different cell types, and people differ in the distribution of these cells in blood. The following additional covariates were included in tests of dose-response associations due to their association with cannabis use and/or DNA methylation: childhood socioeconomic status [42], low childhood self-control [43], family history of substance dependence [44], and persistent use of other substances (tobacco, alcohol, other illicit drugs) [45, 46].
Statistical analyses
DNA methylation data were not normally distributed and showed evidence of outliers, influential observations, and, for some methylation markers, non-normally distributed residuals. Robust regression is appropriate when data contain outliers and influential observations. Further, research has shown that non-normally distributed residuals do not bias epigenome-wide association findings in terms of false negatives or false positives [47].
We used robust regression to compare groups on age-45 CpG sites, with groups represented by dummy codes. We used robust regression to test dose-response associations between quantitative exposures (persistence of regular cannabis use, persistence of tobacco dependence) and age-45 CpG sites. In tests of dose-response associations, covariates were added sequentially: sex, methylation array control probe principal components, and white blood cell counts (Model 1); aforementioned covariates plus childhood SES, low childhood self-control, and family history of substance dependence (Model 2); and aforementioned covariates plus persistent use of other substances (Model 3).
Prior to analyses, methylation probes were standardized on the full age-45 cohort (N = 818, M = 0.00, SD = 1.00). Standardized mean differences between groups can be interpreted as effect sizes, with 0.2, 0.5, 0.8 reflecting small, medium, and large effects, respectively [48]. Results present nominal p-values and indicate which CpG sites survived Bonferroni correction. All statistical tests are two-sided.
Results
A comparison of long-term cannabis users, long-term tobacco users, and non-users
Groups were compared on the 246 CpG sites shown in previous studies to be associated with cannabis use.
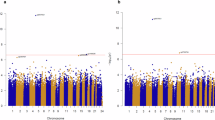
Long-term cannabis users showed both hypomethylation and hypermethylation at age 45 relative to lifelong cannabis/tobacco non-users, but mostly hypomethylation (Fig. 2A). Of 246 CpG sites, 35 differed between long-term cannabis users and non-users at α = 0.05, and 17 at α adjusted for 246 tests (Table 1).
The x axis is the mean difference between long-term cannabis users and cannabis/tobacco non-users (Panel A), long-term tobacco users and cannabis/tobacco non-users (Panel B), and long-term cannabis users and long-term tobacco users (Panel C) on the 246 CpG sites at age 45. Mean differences were estimated using robust regression and were adjusted for sex, control probe principal components indexing technical variation, and white blood cell counts. Negative mean differences indicate hypomethylation relative to the comparison group, and positive mean differences indicate hypermethylation. The y-axis is the negative log base 10 p-value, and these were truncated at a value of 100. The dashed vertical line is the reference line that represents no difference in means between groups. Panel A=Long-term Cannabis Users (N = 73) vs. Cannabis/Tobacco Non-users (N = 182). Panel B=Long-term Tobacco Users (N = 56) vs. Cannabis/Tobacco Non-users (N = 182). Panel C=Long-term Cannabis Users (N = 73) vs. Long-term Tobacco Users (N = 56). The colors indicate statistical significance: Blue= p < 0.00020 (significant at Bonferroni-adjusted threshold for 246 tests). Red= p < 0.05. Gray= p ≥ 0.05.
Long-term tobacco users showed both hypomethylation and hypermethylation at age 45 compared with lifelong cannabis/tobacco non-users, but mostly hypomethylation (Fig. 2B). Of 246 CpG sites, 36 differed between long-term tobacco users and non-users at α = 0.05, and 17 at α adjusted for 246 tests (Table 1).
Long-term cannabis users and long-term tobacco users generally did not differ in terms of age-45 DNA methylation (Fig. 2C). Of 246 CpG sites, 17 differed between long-term cannabis users and long-term tobacco users at α = 0.05, and 3 at α adjusted for 246 tests (Table 1).
Tests of dose-response associations for persistence of regular cannabis use and persistence of tobacco dependence
Greater persistence of regular cannabis use from age 18–45 was associated with differential DNA methylation of 52 of 246 CpG sites at α = 0.05, and 17 at α adjusted for 246 tests (Table 2, Model 1). Associations were largely unchanged after adjusting for childhood SES, low childhood self-control, and family history of substance dependence, with 45 of the 52 significant CpG sites from Model 1 remaining differentially methylated at α = 0.05, and 15 at α adjusted for 52 tests (Table 2, Model 2). A number of associations decreased substantially after additionally adjusting for persistence of tobacco, alcohol, and illicit drug use. Nonetheless, 19 of the 45 significant CpG sites from Model 2 remained differentially methylated at α = 0.05 after adjusting for all covariates, and 9 of these were significant at α adjusted for 45 tests (Table 2, Model 3).
Greater persistence of tobacco dependence was associated with differential methylation of 57 of the 246 CpG sites at α = 0.05, and 22 at α adjusted for 246 tests (Table 3, Model 1). Associations were largely unchanged after adjusting for childhood SES, low childhood self-control, and family history of substance dependence, with 47 of the 57 significant CpG sites from Model 1 remaining differentially methylated at α = 0.05, and 22 at α adjusted for 57 tests (Table 3, Model 2). Associations were only slightly attenuated after adjusting for persistence of cannabis, alcohol, and illicit drug use, with 37 of the 47 significant CpG sites from Model 2 remaining differentially methylated at α = 0.05 after adjustment for all covariates, and 20 of these were significant at α adjusted for 47 tests (Table 3, Model 3).
Summary of group comparisons and tests of dose-response associations
Table S5 summarizes the cannabis-related CpGs from tests of group comparisons and tests of dose-response associations. Of the 246 candidate CpG sites that we sought to replicate, 35 replicated in tests comparing long-term cannabis users with non-users and 52 replicated in tests of persistence of regular cannabis use, using comparable covariates across the two approaches (sex, control probe principal components indexing technical variation, white blood cell counts) and using α = 0.05. After adjusting for multiple testing, those numbers reduced to 17 (Table 1) and 17 (Table 2, Model 1) CpG sites, respectively, with 16 CpGs in common across the two approaches. When dose-response associations were additionally adjusted for substantive covariates, including childhood SES, low childhood self-control, family history of substance dependence, and persistence of tobacco, alcohol, and illicit drug use, nine CpGs remained associated with persistence of regular cannabis use at α adjusted for multiple testing (Table 2, Model 3: cg05575921, cg21566642, cg03636183, cg21161138, cg01940273, cg17739917, cg05086879, cg02978227, cg23079012). By comparison, there were 17 tobacco-related CpG sites that survived adjustment for multiple testing in group comparisons and were robust to covariate-adjustment and adjustment for multiple testing in tests of dose-response associations. The 9 cannabis-related CpG sites that replicated in tests of group comparisons and covariate-adjusted tests of dose-response associations were the focus of the next two analyses: (1) a test of associations between methylation sites and gene expression, and (2) a test of whether quitters showed less extreme hypomethylation than long-term cannabis users.
Association between DNA methylation and gene expression levels
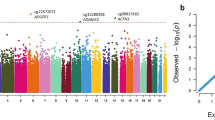
To test whether the nine cannabis-related methylation sites represent meaningful biological associations, we examined correlations between methylation site levels and whole-genome gene expression (N = 45,374 probesets). Methods are described in Supplemental Text. Gene expression was measured when study members were 38 years old. Therefore, these analyses used gene expression and DNA methylation data collected concurrently at age 38. Because DNA methylation data were assayed using different arrays at age 38 and age 45, three of the nine cannabis-related CpG sites at age 45 were not present in the age-38 data. Of the remaining six CpG sites (cg05575921, cg21566642, cg03636183, cg21161138, cg01940273, cg23079012), all were associated with long-term cannabis use at age 38, like at age 45 (Table S6), and all showed statistically significant negative associations with gene expression levels after adjustment for multiple testing (Fig. S3). Specifically, across the six cannabis-related CpG sites, hypomethylation was associated with higher expression of AHRR, P2RY6, LRRN3, GPR15, and DSC2. Two sites were at the AHRR locus (cg05575921 and cg21161138).
A comparison of cannabis and tobacco quitters with non-users and long-term users
Cannabis quitters showed a pattern of DNA methylation that was intermediate between long-term cannabis users and lifelong non-users (Fig. 3A, Table S7).
Panel A Cannabis quitters versus cannabis/tobacco non-users and long-term cannabis users. Panel B Tobacco quitters versus cannabis/tobacco non-users and long-term tobacco users. Means are crude means, standardized (M = 0.00, SD = 1.00) on the representative cohort. Therefore, means of 0.00 reflect the representative cohort norm. Error bars are 95% confidence intervals. Statistical tests used robust regression, with covariates (sex, principal components indexing technical variation, and white blood cell counts). Asterisks (*) indicate that the comparison with the quitter group was statistically significant at α adjusted for multiple testing (cannabis = 9 tests; tobacco = 17 tests).
Likewise, tobacco quitters showed a pattern of DNA methylation that was intermediate between long-term tobacco users and lifelong non-users (Fig. 3B, Table S8).
Further, longer cannabis quit length was dose-dependently associated with increases in methylation at age 45, after adjusting for all covariates (Table S9). Specifically, whereas long-term cannabis users who had not quit showed hypomethylation at age 45, former cannabis users showed less extreme hypomethylation with each additional increment in quit length (i.e., longer quit length was positively associated with methylation).
Discussion
This study characterized the DNA methylation profiles of long-term cannabis users in midlife, re-evaluating a set of 246 methylation sites shown in previous research to be associated with cannabis use. Key findings are as follows. First, 16 sites of cannabis-related differential DNA methylation were replicated across our comparisons of long-term cannabis-user versus non-user groups and our tests of dose-response associations, the latter of which used prospectively-assessed persistence of regular cannabis use from age 18–45 as the continuously-measured exposure. Second, when tests of dose-response associations were adjusted for a number of potential confounders (childhood socioeconomic deprivation; low childhood self-control; family history of substance dependence; and persistence of tobacco, alcohol, and illicit drug use), persistence of regular cannabis use remained robustly associated with hypomethylation of nine sites: cg05575921, cg21566642, cg03636183, cg21161138, cg01940273, cg17739917, cg05086879, cg02978227, cg23079012. Third, the nine sites were not specific to long-term cannabis users; long-term tobacco users showed hypomethylation of those same sites. Fourth, cannabis-related methylation sites showed biologically meaningful associations with gene expression. Fifth, the group of cannabis quitters showed less extreme DNA hypomethylation than long-term cannabis users, suggesting that cessation of cannabis use has the potential to reverse altered DNA methylation.
It is worth noting that most of the previously published markers of cannabis-associated differential DNA methylation were not replicated in this study. One factor might be our focus on cannabis use that is long-term, a difference from several prior studies that reported on less sustained use. However, our record can be compared to the literature where among the 246 previously published cannabis-related CpG sites, only one was reported by more than one study: cg05575921 [16, 20, 22] (Table S2). Another factor may be test-retest reliability. Our team previously documented the highly variable, but on average low, test-retest reliability of ~450,000 CpG sites assayed once using the 450 K BeadChip and once using the EPIC BeadChip, in the same DNA sample at the same time (mean intraclass correlation [ICC] = 0.21) [49]. Using that database (https://osf.io/83ucs/), we obtained the test-retest reliability of the 246 CpG sites that were the focus of the current study. Of the 246 sites, 156 had test-retest data because they were present on both the 450k and EPIC BeadChips, and their average ICC was only 0.28 (SD = 0.30, range = −0.13, 0.98). Sites that do not yield the same value if re-tested have limited utility. Importantly, the sites with better test-retest reliability showed the largest effect sizes in our tests of dose-response associations for persistence of regular cannabis use (r = 0.32, p < 0.0001) and persistence of tobacco dependence (r = 0.35, p < 0.0001). One possibility for future research, which may improve reproducibility, is to pre-screen for reliable CpG sites [49]. Another possibility is to use epigenetic measures that are enriched for reliable CpG sites [49]. For example, with one exception [50], several recent studies have shown that cannabis use is associated with accelerated epigenetic aging [51,52,53], using epigenetic clocks that correlate with physiological deterioration and a broad range of health indices.
Despite the relatively low replication rate, nine cannabis-related methylation sites were robust to adjustment for a range of covariates, and six of those sites with available gene expression data (cg05575921, cg21566642, cg03636183, cg21161138, cg01940273, cg23079012) were related to gene expression in a biologically meaningful way. Specifically, cannabis-related hypomethylation was associated with higher expression of the following genes: AHRR, P2RY6, LRRN3, GPR15, and DSC2. These genes are implicated in the metabolism of environmental toxins; immune system function and inflammatory response; and neurodevelopment, synaptogenesis, and neuroplasticity [54,55,56,57,58]. Higher expression is linked with some cancers (AHRR, DSC2) [59, 60], poorer prognosis of lung cancer (P2RY6) [55], and cardiovascular diseases (GPR15, DCS2) [61, 62]. Most of these genes have been implicated in tobacco smoking. Our findings are consistent with previous studies documenting that the cannabis-related methylation sites reported here are related to tobacco smoking and lung cancer (Table S10). However, unlike tobacco, cannabis use is not clearly associated with lung cancer [1] and is inconsistently associated with elevated risk of cardiovascular events [63].
The high degree of overlap between the methylation profiles of long-term cannabis users and long-term tobacco users has implications for the specific mechanisms underlying cannabis-related differential methylation. Although our findings showed that some associations between long-term cannabis use and DNA methylation were robust to adjustment for long-term tobacco use, it is difficult to disentangle cannabis effects from tobacco effects because most people who use cannabis also use tobacco. One possibility is that our analyses of long-term cannabis use did not fully account for tobacco effects, despite covariate adjustment, and tobacco use explains the cannabis-related differential methylation. However, another possibility is that cannabis and tobacco may independently affect DNA methylation. Importantly, one of the nine cannabis-associated sites from our study, cg05575921, has been shown to be associated specifically with tobacco smoking and not vaping or use of non-combustible tobacco products [8], which suggests smoking as a potential common mechanism. We found that long-term cannabis users and long-term tobacco users showed hypomethylation of cg05575921, but the effect was smaller for long-term cannabis users. The smaller effect for long-term cannabis users, and lack of a clear association between cannabis use and smoking-related health outcomes such as lung cancer, may be explained by the less intensive patterns of use typical of cannabis relative to tobacco. As cannabis use intensity increases [15], associations with lung cancer and other diseases could emerge.
A key question is whether THC exposure might also be a mechanism by which cannabis alters methylation. Rodent studies have shown that THC administration alters DNA methylation [64]. However, if THC were a mechanism underlying cannabis-related methylation in the present study, it is perhaps surprising that there were few methylation sites that were unique to cannabis. In this regard, it is worth noting that THC and nicotine affect shared brain pathways, including pathways that underlie reward, mood, and cognition. For example, despite binding to different brain receptors, THC and nicotine result in altered neurotransmission (e.g., increased dopamine release) [65], which could affect the epigenome in similar ways [66]. Moreover, the endocannabinoid and nicotinic systems appear to interact, with one system modulating the other [65]. Strikingly, epidemiological studies have documented that cannabis and tobacco relate similarly to a number of health outcomes, including psychosis [67], cognitive impairment [68], MRI-assessed brain structure [41], and periodontal disease [69]. This epidemiological evidence underscores the possibility that cannabis and tobacco use share biological pathways to health outcomes.
Quitting cannabis may at least partially reverse cannabis-related alterations to the epigenome. Cannabis quitters showed DNA methylation profiles that were intermediate between long-term cannabis users and non-users. Tobacco quitters showed the same pattern. The finding for tobacco has been shown before [70, 71] and is consistent with research documenting that tobacco cessation improves health outcomes [72]. The finding for cannabis is new and suggests that cannabis cessation may improve health outcomes.
This study has limitations. First, findings are based on a single New Zealand (NZ) cohort born in the 1970s when cannabis was much less potent (i.e., characterized by lower THC concentrations) than it is today [73, 74]. Relatedly, the heaviest cannabis users in the Dunedin cohort had used cannabis on an almost daily basis for years, but they comprised only ~6% of the cohort. Recent data from the US show that the number of daily cannabis users is increasing [15]. If long-term frequent use of high-potency cannabis underlies cannabis effects on methylation, then our study may have underestimated cannabis-methylation associations. In terms of how NZ currently compares with the US and Europe, medical cannabis is legal in NZ but non-medical use (i.e., recreational use) is not, similar to many American states. The NZ medical cannabis market has shifted toward high-THC products, and the potency of medical and illegal market cannabis products in NZ is similar to the US [75]. The rate of disability adjusted life years attributable to cannabis use disorder is higher in NZ than in the US and UK [76].
Second, the DNA is from blood, and research is needed to determine if findings replicate in other tissues (e.g., lung, which is obtained much more invasively than blood). Third, the study cannot address the mechanisms of cannabis-associated hypomethylation. Most people who use cannabis smoke it [4, 5], and although smoking is one plausible mechanism, research is needed to ascertain whether users who primarily vape or ingest cannabis also show differential DNA methylation.
Fourth, this paper focused on cannabis use. We included tobacco comparisons because tobacco associations provide important context for interpreting cannabis associations. However, there are tobacco-associated DNA methylation markers that we do not report on here, because we selected previously published cannabis-associated methylation markers for replication. Nonetheless, all 17 tobacco-associated markers of differential DNA methylation that replicated across tests of group comparisons and tests of dose-response associations are reported as tobacco-associated in the EWAS catalog in the same direction as we report here. Fifth, in group comparisons, small N’s may have reduced statistical power (n = 49–182). However, our dose-response tests had ample power (n = 787). Finally, this study is observational and cannot determine causality. Nonetheless, we found evidence of dose-response associations, which bolsters biological plausibility of a causal effect. Moreover, dose-response tests addressed confounding by adjusting for factors known to characterize long-term cannabis users: socioeconomically deprived circumstances, poor childhood self-control, family history of substance dependence, and persistent tobacco, alcohol, and illicit drug use. The field generally lacks tests of dose-response associations. Our findings represent a unique contribution.
This study has several implications. First, long-term cannabis use could affect DNA methylation in similar ways to tobacco, possibly at least partly though the shared mechanism of smoke inhalation. Second, research is needed to ascertain whether cannabis-related hypomethylation and associated alterations in gene expression result in the development of disease. Third, at a time when cannabis and cannabinoids are increasingly touted and used for salutary effects, it is important to recognize that the epigenome may be responsive to long-term cannabis use regardless of whether use is for medical or non-medical reasons. Finally, cannabis cessation could reverse altered DNA methylation.
Data availability
Dunedin Study data is available via managed access at https://dunedinstudy.otago.ac.nz/for-investigators/policy-statement-code-of-practice.
Code availability
All scripts used in the analyses are available here: https://github.com/MoffittCaspiLab/Meier_MolecularPsychiatry_2025.
References
Campeny E, López-Pelayo H, Nutt D, Blithikioti C, Oliveras C, Nuño L, et al. The blind men and the elephant: systematic review of systematic reviews of cannabis use related health harms. Eur Neuropsychopharmacol. 2020;33:1–35.
Ghasemiesfe M, Ravi D, Casino T, Korenstein D, Keyhani S. Acute cardiovascular effects of marijuana use. J Gen Intern Med. 2020;35:969–74.
Hindley G, Beck K, Borgan F, Ginestet CE, McCutcheon R, Kleinloog D, et al. Psychiatric symptoms caused by cannabis constituents: a systematic review and meta-analysis. Lancet Psychiatry. 2020;7:344–53.
Hindocha C, Brose LS, Walsh H, Cheeseman H. Cannabis use and co‐use in tobacco smokers and non‐smokers: prevalence and associations with mental health in a cross‐sectional, nationally representative sample of adults in great Britain, 2020. Addiction. 2021;116:2209–19.
Wadsworth E, Craft S, Calder R, Hammond D. Prevalence and use of cannabis products and routes of administration among youth and young adults in Canada and the United States: a systematic review. Addict Behav. 2022;129:107258.
Szutorisz H, Hurd YL. Epigenetic effects of cannabis exposure. Biol Psychiatry. 2016;79:586–94.
Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38:23–38.
Andersen A, Reimer R, Dawes K, Becker A, Hutchens N, Miller S, et al. DNA methylation differentiates smoking from vaping and non-combustible tobacco use. Epigenetics. 2022;17:178–90.
Fasanelli F, Baglietto L, Ponzi E, Guida F, Campanella G, Johansson M, et al. Hypomethylation of smoking-related genes is associated with future lung cancer in four prospective cohorts. Nat Commun. 2015;6:10192.
Hoffmann D, Brunnemann K, Gori G, Wynder E. On the carcinogenicity of marijuana smoke. In: Runeckles VC, editor. Recent advances in phytochemistry. MA: Springer; 1975. pp. 63–81.
Cerdá M, Mauro C, Hamilton A, Levy NS, Santaella-Tenorio J, Hasin D, et al. Association between recreational marijuana legalization in the United States and changes in marijuana use and cannabis use disorder from 2008 to 2016. JAMA Psychiatry. 2020;77:165–71.
Bahji A, Kaur S, Devoe D, Patten S. Trends in Canadian cannabis consumption over time: a two-step meta-analysis of Canadian household survey data. Can J Addict. 2022;13:6–13.
Imtiaz S, Nigatu YT, Ali F, Douglas L, Hamilton HA, Rehm J, et al. Cannabis legalization and cannabis use, daily cannabis use and cannabis-related problems among adults in Ontario, Canada (2001–2019). Drug Alcohol Depend. 2023;244:109765.
Rubenstein D, McClernon FJ, Pacek LR. Trends in cannabis and tobacco co-use in the United States, 2002–2021. Addict Behav. 2024;158:108129.
Caulkins JP. Changes in self‐reported cannabis use in the United States from 1979 to 2022. Addiction. 2024;119:1648–52.
Osborne AJ, Pearson JF, Noble AJ, Gemmell NJ, Horwood LJ, Boden JM, et al. Genome-wide DNA methylation analysis of heavy cannabis exposure in a New Zealand longitudinal cohort. Transl Psychiatry. 2020;10:114.
Markunas CA, Hancock DB, Xu Z, Quach BC, Fang F, Sandler DP, et al. Epigenome‐wide analysis uncovers a blood‐based DNA methylation biomarker of lifetime cannabis use. Am Journal Med Genet B Neuropsychiatr Genet. 2021;186:173–82.
Clark SL, Chan R, Zhao M, Xie LY, Copeland WE, Aberg KA, et al. Methylomic investigation of problematic adolescent cannabis use and its negative mental health consequences. J Am Acad Child Adolesc Psychiatry. 2021;60:1524–32.
Wiedmann M, Kuitunen-Paul S, Basedow LA, Wolff M, DiDonato N, Franzen J, et al. DNA methylation changes associated with cannabis use and verbal learning performance in adolescents: an exploratory whole genome methylation study. Transl Psychiatry. 2022;12:317.
Nannini DR, Zheng Y, Joyce BT, Kim K, Gao T, Wang J, et al. Genome-wide DNA methylation association study of recent and cumulative marijuana use in middle aged adults. Mol Psychiatry. 2023;28:2572–82.
Carreras-Gallo N, Dwaraka VB, Cáceres A, Smith R, Mendez TL, Went H, et al. Impact of tobacco, alcohol, and marijuana on genome-wide DNA methylation and its relationship with hypertension. Epigenetics. 2023;18:2214392.
Garrett ME, Dennis MF, Bourassa KJ, Workgroup VM-AM, Hauser MA, Kimbrel NA, et al. Genome-wide DNA methylation analysis of cannabis use disorder in a veteran cohort enriched for posttraumatic stress disorder. Psychiatry Res. 2024;333:115757.
Fang F, Quach B, Lawrence KG, van Dongen J, Marks JA, Lundgren S, et al. Trans-ancestry epigenome-wide association meta-analysis of DNA methylation with lifetime cannabis use. Mol Psychiatry. 2024;29:124–33.
Meier MH, Caspi A, R. Knodt A, Hall W, Ambler A, Harrington H, et al. Long-term cannabis use and cognitive reserves and hippocampal volume in midlife. Am J Psychiatry. 2022;179:362–74.
Cerdá M, Moffitt TE, Meier MH, Harrington H, Houts R, Ramrakha S, et al. Persistent cannabis dependence and alcohol dependence represent risks for midlife economic and social problems: a longitudinal cohort study. Clin Psychol Sci. 2016;4:1028–46.
Meier MH. Cannabis use and psychosocial functioning: evidence from prospective longitudinal studies. Curr Opin Psychol. 2021;38:19–24.
Meier MH, Olive MF, Jenks OA, Wernik SR. Cannabis use and cognitive functioning across the lifespan. Curr Addict Rep. 2024;11:384–95.
Hall KS, Cohen HJ, Pieper CF, Fillenbaum GG, Kraus WE, Huffman KM, et al. Physical performance across the adult life span: correlates with age and physical activity. J Gerontol A Biol Sci Med Sci. 2017;72:572–8.
Lange-Maia BS, Karvonen-Gutierrez CA, Kazlauskaite R, Strotmeyer ES, Karavolos K, Appelhans BM, et al. Impact of chronic medical condition development on longitudinal physical function from mid-to early late-life: the study of women’s health across the nation. J Gerontol A Biol Sci Med Sci. 2020;75:1411–7.
Poulton R, Moffitt TE, Silva PA. The dunedin multidisciplinary health and development study: overview of the first 40 years, with an eye to the future. Soc Psychiatry Psychiatr Epidemiol. 2015;50:679–93.
Richmond-Rakerd LS, D’Souza S, Andersen SH, Hogan S, Houts RM, Poulton R, et al. Clustering of health, crime and social-welfare inequality in 4 million citizens from two nations. Nat Hum Behav. 2020;4:255–64.
Belsky DW, Caspi A, Corcoran DL, Sugden K, Poulton R, Arseneault L, et al. DunedinPACE, a DNA methylation biomarker of the pace of aging. eLife. 2022;11:e73420.
Bowtell DD. Rapid isolation of eukaryotic DNA. Anal Biochem. 1987;162:463–5.
Jeanpierre M. A rapid method for the purification of DNA from blood. Nucleic Acids Res. 1987;15:9611.
Pidsley R, Y Wong CC, Volta M, Lunnon K, Mill J, Schalkwyk LC. A data-driven approach to preprocessing Illumina 450K methylation array data. BMC Genomics. 2013;14:1–10.
Davis S, Du P, Bilke S, Triche J, Bootwalla M. methylumi: Handle Illumina methylation data. R package version 2.30.0. 2019. https://doi.org/10.18129/B9.bioc.methylumi.
Lehne B, Drong AW, Loh M, Zhang W, Scott WR, Tan ST, et al. A coherent approach for analysis of the Illumina HumanMethylation450 BeadChip improves data quality and performance in epigenome-wide association studies. Genome Biol. 2015;16:37.
Robins LN, Cottler L, Bucholz KK, Compton W. Diagnostic interview schedule for DSM-IV. St Louis, MO: Washington University School of Medicine; 1995.
Robins LN, Helzer JE, Croughan J, Ratcliff KS. National Institute of Mental Health Diagnostic Interview Schedule: its history, characteristics, and validity. Arch Gen Psychiatry. 1981;38:381–9.
Meier MH, Caspi A, Ambler A, Hariri AR, Harrington H, Hogan S, et al. Preparedness for healthy ageing and polysubstance use in long-term cannabis users: a population-representative longitudinal study. Lancet Healthy Longev. 2022;3:e703–e14.
Knodt AR, Meier MH, Ambler A, Gehred MZ, Harrington H, Ireland D, et al. Diminished structural brain integrity in long-term cannabis users reflects a history of polysubstance use. Biol Psychiatry. 2022;92:861–70.
Reuben A, Sugden K, Arseneault L, Corcoran DL, Danese A, Fisher HL, et al. Association of neighborhood disadvantage in childhood with DNA methylation in young adulthood. JAMA Netw Open. 2020;3:e206095.
Moffitt TE, Arseneault L, Belsky D, Dickson N, Hancox RJ, Harrington H, et al. A gradient of childhood self-control predicts health, wealth, and public safety. Proc Natl Acad Sci USA. 2011;108:2693–8.
Milne BJ, Caspi A, Harrington H, Poulton R, Rutter M, Moffitt TE. Predictive value of family history on severity of illness: the case for depression, anxiety, alcohol dependence, and drug dependence. Arch Gen Psychiatry. 2009;66:738–47.
Stephenson M, Bollepalli S, Cazaly E, Salvatore JE, Barr P, Rose RJ, et al. Associations of alcohol consumption with epigenome‐wide DNA methylation and epigenetic age acceleration: individual‐level and co‐twin comparison analyses. Alcohol Clin Exp Res. 2021;45:318–28.
Sun Y-Q, Richmond RC, Suderman M, Min JL, Battram T, Flatberg A, et al. Assessing the role of genome-wide DNA methylation between smoking and risk of lung cancer using repeated measurements: the HUNT study. Int J Epidemiol. 2021;50:1482–97.
Mansell G, Gorrie-Stone TJ, Bao Y, Kumari M, Schalkwyk LS, Mill J, et al. Guidance for DNA methylation studies: statistical insights from the Illumina EPIC array. BMC Genomics. 2019;20:1–15.
Cohen J. A power primer. Psychol Bull. 1992;112:155–9.
Sugden K, Hannon EJ, Arseneault L, Belsky DW, Corcoran DL, Fisher HL, et al. Patterns of reliability: assessing the reproducibility and integrity of DNA methylation measurement. Patterns. 2020;1:100014.
Dempster EL, Wong CC, Burrage J, Hannon E, Quattrone D, Trotta G, et al. Methylomic signature of current cannabis use in two first-episode psychosis cohorts. Mol Psychiatry. 2025;30:1277–86. https://doi.org/10.1038/s41380-024-02689-0
Allen JP, Danoff JS, Costello MA, Hunt GL, Hellwig AF, Krol KM, et al. Lifetime marijuana use and epigenetic age acceleration: a 17-year prospective examination. Drug Alcohol Depend. 2022;233:109363.
Cordero AIH, Li X, Yang CX, Ambalavanan A, MacIsaac JL, Kobor MS, et al. Cannabis smoking is associated with advanced epigenetic age. Eur Respir J. 2024;63:2400458.
Clark SL, McGinnis EW, Zhao M, Xie L, Marks GT, Aberg KA, et al. The impact of childhood mental health and substance use on methylation aging into adulthood. J Am Acad Child Adolesc Psychiatry. 2024;63:825–34.
Vogel CF, Van Winkle LS, Esser C, Haarmann-Stemmann T. The aryl hydrocarbon receptor as a target of environmental stressors–implications for pollution mediated stress and inflammatory responses. Redox Biol. 2020;34:101530.
Wu H, Dong X. Immunological role and clinical prognostic significance of P2RY6 in lung adenocarcinoma: a multi-omics studies and single-cell sequencing analysis. World J Surg Oncol. 2023;21:341.
Kõks S, Kõks G. Activation of GPR15 and its involvement in the biological effects of smoking. Exp Biol Med. 2017;242:1207–12.
Sun C, Wang L, Yang X-X, Jiang Y-H, Guo X-L. The aberrant expression or disruption of desmocollin2 in human diseases. Int J Biol Macromol. 2019;131:378–86.
Clarke RA, Lee S, Eapen V. Pathogenetic model for Tourette syndrome delineates overlap with related neurodevelopmental disorders including Autism. Transl Psychiatry. 2012;2:e158.
Vogel CF, Haarmann-Stemmann T. The aryl hydrocarbon receptor repressor–more than a simple feedback inhibitor of AhR signaling: clues for its role in inflammation and cancer. Curr Opin Toxicol. 2017;2:109–19.
Li Q, Lin N. 534P DSC2 promotes the proliferation, metastasis and drug resistance of lung cancer by activating the PI3K/AKT pathway. Ann Oncol. 2023;34:S1678.
Haase T, Müller C, Stoffers B, Kirn P, Waldenberger M, Kaiser FJ, et al. G protein-coupled receptor 15 expression is associated with myocardial infarction. Int J Mol Sci. 2022;24:180.
Brodehl A, Belke DD, Garnett L, Martens K, Abdelfatah N, Rodriguez M, et al. Transgenic mice overexpressing desmocollin-2 (DSC2) develop cardiomyopathy associated with myocardial inflammation and fibrotic remodeling. PLoS ONE. 2017;12:e0174019.
Ghosh M, Naderi S. Cannabis and cardiovascular disease. Curr Atheroscler Rep. 2019;21:1–6.
Machado AS, Bragança M, Vieira-Coelho M. Epigenetic effects of cannabis: A systematic scoping review of behavioral and emotional symptoms associated with cannabis use and exocannabinoid exposure. Drug Alcohol Depend. 2024;263:111401.
Buzzi B, Koseli E, Moncayo L, Shoaib M, Damaj MI. Role of neuronal nicotinic acetylcholine receptors in cannabinoid dependence. Pharmacol Res. 2023;191:106746.
Mohammad GS, Joca S, Starnawska A. The cannabis-induced epigenetic regulation of genes associated with major depressive disorder. Genes. 2022;13:1435.
Gurillo P, Jauhar S, Murray RM, MacCabe JH. Does tobacco use cause psychosis? Systematic review and meta-analysis. Lancet Psychiatry. 2015;2:718–25.
Waisman Campos M, Serebrisky D, Castaldelli-Maia JM. Smoking and cognition. Curr Drug Abuse Rev. 2016;9:76–9.
Thomson WM, Poulton R, Broadbent JM, Moffitt TE, Caspi A, Beck JD, et al. Cannabis smoking and periodontal disease among young adults. JAMA. 2008;299:525–31.
Christiansen C, Castillo-Fernandez J, Domingo-Relloso A, Zhao W, El-Sayed Moustafa J, Tsai P-C, et al. Novel DNA methylation signatures of tobacco smoking with trans-ethnic effects. Clin Epigenetics. 2021;13:1–13.
Tsaprouni LG, Yang T-P, Bell J, Dick KJ, Kanoni S, Nisbet J, et al. Cigarette smoking reduces DNA methylation levels at multiple genomic loci but the effect is partially reversible upon cessation. Epigenetics. 2014;9:1382–96.
Le TT, Mendez D, Warner KE. The benefits of quitting smoking at different ages. Am J Prev Med. 2024;67:684–8.
ElSohly MA, Chandra S, Radwan M, Gon C, Church JC. A comprehensive review of cannabis potency in the USA in the last decade. Biol Psychiatry Cogn Neurosci Neuroimaging. 2021;6:603–6.
Freeman TP, Craft S, Wilson J, Stylianou S, ElSohly M, Di Forti M, et al. Changes in delta‐9‐tetrahydrocannabinol (THC) and cannabidiol (CBD) concentrations in cannabis over time: systematic review and meta‐analysis. Addiction. 2021;116:1000–10.
Rychert M, Wilkins C. Implementation of the medicinal cannabis scheme in New Zealand: six emerging trends. N Z Med J. 2024;137:73–86.
Institute for Health Metrics and Evaluation (IHME). GBD Compare Data Visualization. Seattle, WA: IHME, University of Washington, 2024. Available from: http://vizhub.healthdata.org/gbd-compare (Accessed July 3 2024).
Acknowledgements
We thank the Dunedin Study members, their families, and friends for their long-term involvement. The Dunedin Multidisciplinary Health and Development Research Unit is based at University of Otago within the Ngāi Tahu tribal area who we acknowledge as first peoples, tangata whenua (transl. people of this land). We thank Dunedin Study Unit research staff and previous Study Director, Emeritus Distinguished Professor, the late Richie Poulton, for his leadership during the Study’s research transition from young adulthood to aging (2000–2023) and Study founder, Dr Phil A. Silva. We would like to acknowledge the assistance of the Molecular Genomics Core at the Duke Molecular Physiology Institute, Duke University School of Medicine, for the generation of data for the manuscript. The Dunedin Multidisciplinary Health and Development Research Unit is supported by the New Zealand Health Research Council (programme Grant 16-604), and has also received funding from the New Zealand Ministry of Business, Innovation and Employment. This research received support from the US National Institute on Aging (grants R01AG069939, R01AG032282, R01AG073207) and the UK Medical Research Council (grant MR/X021149/1).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The premise and analysis plan for this project were pre-registered at https://sites.duke.edu/moffittcaspiprojects/files/2024/07/Meier_2024_CannabisDNAm.pdf.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Caspi, A., Sugden, K., Moffitt, T.E. et al. DNA methylation profiles of long-term cannabis users in midlife: a comprehensive evaluation of published cannabis-associated methylation markers in a representative cohort. Mol Psychiatry (2025). https://doi.org/10.1038/s41380-025-03042-9
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41380-025-03042-9