Abstract
Despite a significant burden of neurobehavioral and psychiatric comorbidities in children with Down syndrome (DS), and the general increased risk for anxiety in individuals with intellectual disabilities (ID), children with DS have significantly lower odds of anxiety. Understanding the specific mechanisms of anxiety in DS could inform the development of new treatments. This study performed a comprehensive investigation of genomic variants that contribute to anxiety disorders in DS, as well as variants shared in other mental disorders. We employed deep learning algorithms using neural network models in combination with one of the largest whole-genome sequencing (WGS) cohorts of 1479 DS individuals and family members, including 255 DS probands diagnosed with at least one type of mental disorder, of whom 74 had confirmed anxiety disorders. We found that only a fraction (19%) of anxiety-specific corresponding gene variants previously reported overlap with those shared in anxiety in DS patients, suggesting distinct molecular mechanisms for anxiety in DS individuals. Functional overrepresentation analysis suggested that anxiety results from a complex interplay of genetic and environmental factors. Additionally, non-coding variants, particularly those proximal to splicing sites, play significant roles. Moreover, the variants associated with anxiety and other mental disorders are not uniquely distributed genome wide. Several loci, including 17q25, 16q23, 21q22, and 22q13, show greater weight in DS patients. Furthermore, 29 biomarkers containing recurrent anxiety-specific variants were identified to assist in the diagnosis of anxiety in the DS population. This pioneering study represents the first comprehensive exploration of anxiety disorders in DS utilizing WGS cohorts and advanced deep-learning AI models. The results indicate that anxiety disorder in DS patients has distinct molecular patterns from other mental disorders. The insights gained from our research offer valuable understanding of underlying mechanisms and hold promise for enhancing clinical diagnosis and potentially guiding more effective intervention strategies in this vulnerable population.
Similar content being viewed by others
Introduction
Anxiety disorders in children and adolescents have become a global healthcare concern because of the rapid growth in prevalence over the past decades. Based on Centers for Disease Control and Prevention (CDC) statistics, between 2016 and 2019, 9.4% of children aged 3–17 years were diagnosed with anxiety, which accounts for approximately 5.8 million individuals in the US. This rate has shown a notable upward trend in recent decades (https://www.cdc.gov/childrensmentalhealth/features/anxiety-depression-children.html). Although anxiety is often considered alongside other mental disorders and is one of the most common co-occurring conditions, it has distinct characteristics that differentiate it from other mental illnesses. For instance, anxiety is primarily driven by emotional dysregulation [1], whereas other behavioral disorders, such as attention deficit hyperactivity disorder (ADHD)—a neurodevelopmental disorder—exhibit fundamentally different core features [2]. Even among mood disorders, anxiety and depression, though commonly comorbid and influenced by stress, have distinct clinical presentations. Anxiety is characterized by excessive worry and fear, whereas depression is marked by persistent sadness and loss of interest in activities (https://www.webmd.com/depression/depression-or-anxiety). These differences also extend to their diagnosis and treatment, as outlined in the Diagnostic and Statistical Manual of Mental Disorders (DSM-5). For example, ADHD treatment typically involves stimulant or non-stimulant medications, while anxiety management often includes antidepressants or cognitive behavioral therapy (CBT). Even among mood disorders, treatment approaches vary—anxiety therapy focuses on managing triggers, panic attacks, and excessive worry, whereas depression therapy emphasizes addressing feelings of hopelessness, low mood, and disinterest in activities. These distinctions underscore the importance of recognizing anxiety as a separate clinical entity, necessitating tailored approaches for its diagnosis and treatment.
The prevalence of neurobehavioral and psychiatric comorbidity in children with Down syndrome (DS) ranges from 18–38% [3], underscoring a significant burden of mental health issues within this population. Moreover, individuals with intellectual disabilities (ID) are generally at increased risk for developing anxiety disorders [4]. While almost all individuals with DS experience some degree of ID, it is noteworthy that DS is associated with significantly lower odds of anxiety [4]. The extra copy of chromosome 21 (trisomy 21) in DS affects brain development and function; however, the genetic effects on the brain in DS extend well beyond chromosome 21 with genome-wide alterations in gene expression impacting a substantial number of genes across multiple brain regions [5]. Epigenetic factors also modulate gene expression in the human brain, which may be particularly relevant in DS patients [6]. What makes the study of anxiety in children with DS unique is the interplay between trisomy 21 and its genome-wide effects on brain development and function, which may lead to lower anxiety levels despite the higher prevalence of mental disorders typically associated with ID. Currently, more research is needed to fully understand the specific mechanisms. Understanding the unique ways anxiety manifests in the DS population despite the overall lower risk can lead to more accurate diagnoses and tailored interventions. Furthermore, identifying and addressing anxiety early can prevent the exacerbation of other behavioral and emotional challenges, promoting better overall mental health and well-being in children with ID. Understanding the protective factors related to anxiety in DS could also inform the development of new treatments for anxiety in other populations by identifying specific genetic, neurobiological, and environmental mechanisms that contribute to lower anxiety levels in individuals with DS.
There are challenges and opportunities in anxiety research within the DS population. Diagnosing anxiety in children can be particularly challenging, as children often have difficulty articulating their symptoms of anxiety [7]. Anxiety in children can manifest through a wide range of behaviors that might be mistaken for other issues, such as behavioral problems, attention difficulties, or even physical ailments like stomachaches and headaches. Diagnosing anxiety in children with DS is even more challenging due to communication difficulties and cognitive impairments. The identification of risk genes and mutations related to anxiety has been limited potentially due to the interplay of relatively stable genetic influences [8] and significant environmental factors [9]. Furthermore, very few studies have been conducted to determine to what degree non-coding region variants’ may impact anxiety disorders despite evidence suggesting that non-coding DNA variants affecting important 5′ and 3′ regulatory as well as intronic sequences are associated with central nervous system disorders, including anxiety, via altering transcription factor or microRNA binding sites [10]. This lack of focused research makes it even more challenging to understand and address anxiety in the DS population.
In this study, we leverage a genome-wide, unbiased, data-driven analysis using one of the largest databases encompassing DS individuals with anxiety and other types of mental disorders, as part of the Gabriella Miller Kids First program project (https://kidsfirstdrc.org/), to elucidate the underlying mechanisms of anxiety disorders, with a specific focus on identifying biomarker genes and variants that distinguish anxiety from other mental disorders within the DS population. By applying deep learning models using neural networks, we aim to quantitatively assess the contributions of genomic variants, including both coding and non-coding regions, to the diagnosis of anxiety. The new insights gained could lead to the development of validated biomarkers that could aid in the diagnosis and treatment of anxiety in individuals with complex clinical phenotypes, such as DS.
Methods
Patient recruitment
Patients diagnosed with DS were recruited from the Center for Applied Genomics (CAG) at Children’s Hospital of Philadelphia (CHOP). Diagnoses of mental disorders, including anxiety disorders, were made using the International Classification of Diseases (ICD) codes ICD-9/ICD-10. All subjects were systematically documented in the electronic medical records (EMRs) of CHOP, a phenotype system established in 2003. CAG at CHOP maintains a de-identified abstraction of clinical data extracted from the CHOP EMR database of patients who have provided research consent. This database encompasses longitudinal information regarding patient visits, diagnoses, medical history, prescriptions, procedures, and laboratory tests, with all data meticulously coded and de-identified.
A total of 1479 whole-genome sequencing (WGS) datasets were generated from blood (>95%) or saliva samples of participating individuals (Fig. 1A). Of those, 709 were DS patients, 425 were healthy individuals who are family members of the probands, and an additional 345 healthy controls were from independent families without DS patients. Among the 709 cases, 255 DS patients were diagnosed with at least one type of mental disorders and 74 DS patients had confirmed anxiety disorders based on EMR records, including established therapeutic treatment plans. An additional 74 RNA sequencing (RNA-seq) samples from human peripheral blood mononuclear cells (PBMCs) were included in the analysis (a subset of 709 individuals), including 25 DS patients diagnosed with anxiety and 49 DS patients without anxiety but diagnosed with at least one type of other mental disorders. All patients were receiving health care at CHOP and were recruited during hospital visits, including emergency rooms, ambulatory settings, or surgical settings, through general pediatric clinics or CHOP’s pediatric specialty practices. Parental consent was obtained for individuals under 18 years of age, and assent was also obtained for subjects aged 7–17 years. The informed consent allowed samples to be obtained and analyzed using the genomic technologies in this study to address the proposed research questions.
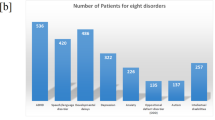
A Pie chart shows the portions of patients in this study, including Down Syndrome patients diagnosed with anxiety, diagnosed with other mental disorder, have no diagnosis of mental disorder, healthy controls from Down Syndrome patients’ family, and indepedent healthy controls; B Number of Down Syndrome patients for eight major mental disorder diagnosis.
Data processing and variant detection by WGS
WGS was conducted at 30X coverage, as part of the Gabriella Miller Kids First project. Variant call format (VCF) files for WGS were generated using the Illumina DRAGEN (Dynamic Read Analysis for GENomics) Bio-IT Platform (Illumina, San Diego, CA), aligned to the GRCh38/hg38 human genome assembly. Variant annotations were produced using the ANNOVAR software developed by our group with default parameters [11]. Variants were then classified into coding regions (encompassing nonsynonymous and synonymous variants), introns, 5′ untranslated regions (UTR), and non-coding RNA (ncRNA) regions. The distances of intronic variants to the closest exon sites were calculated based on the GRCh38/hg38 reference, and ncRNA targets were determined using LncTarD version 2.0 [12].
Identification of anxiety-specific genomics variants
A variant is considered recurrent if it resides at the same genomic locus with the same alternative allele and occurs in more than one individual. We searched for recurrent variants that occurred in at least three individuals in the DS patient group and were absent in healthy controls, including both family members and independent healthy individuals, to minimize the impact of gene dosage effects in DS patients. The variants’ loading matrix was constructed, where each row represents a patient, each column represents a recurrent variant, and each entry indicates the occurrences of the specific variant for the corresponding patient. Each column was treated as a feature vector, and variants with high correlation coefficients (Pearson correlation coefficient ≥0.7) were merged. Additionally, exonic variants in the same exon were merged. A matrix model was deployed to quantify the contributions of each variant to anxiety or other mental disorders, either positively or negatively. Specifically, the variants’ loading matrix and clinical phenotype matrix were used as inputs, and the Moore–Penrose inverse was calculated as the weight of the corresponding phenotypes (anxiety and other mental disorders) for each variant. To select the most representative signals, we required the weight of the variants to be at least 50-fold higher than expected weights. As a result, genomic variants with a positive weight for anxiety and a negative weight for other mental disorders were considered anxiety-specific variants. Meanwhile, variants with positive weights for both anxiety and other mental disorders were considered shared variants between anxiety and other mental disorders.
Ranking the variants with deep learning
A deep learning model with multi-layer perceptron (MLP) was applied to further rank anxiety-protective and anxiety-predisposing variants using the Scikit-learn package (version 0.21.3) [13] for DS patients with anxiety, DS patients with a mental disorder but without an anxiety diagnosis, and DS patients without any mental disorders. The optimization of parameters for the deep learning model, including maximum iterations, alpha value in L2 regularization, activation functions, solvers, learning rate, number of layers, and number of neurons per layer, was conducted using the ‘gp_minimize’ function from the scikit-optimize 0.7.2 Python library. To account for randomness, permutation importance for each variant in the MLP model was computed 20 times. The mean importance of each variant was calculated from the permutations, and variants with negative mean importance scores were removed. Subsequently, the selected variants were mapped to their corresponding genes based on genomic ___location, and functional overrepresentation analysis was performed using WebGestalt (WEB-based Gene SeT AnaLysis Toolkit) [14] and the DAVID Bioinformatics platform [15]. Differential expression tests for RNA-seq data were performed using the Illumina DRAGEN (Dynamic Read Analysis for GENomics) Bio-IT Platform and DESeq2 [16]. The cell-type-specific enrichment analysis for selected genes were performed by WebCSEA [17].
Results
Functional overrepresentation of anxiety-specific and shared variants in mental disorders
A total of 609 genes containing at least one anxiety-specific variant were identified. Functional analysis reveals overrepresentation of gene sets in the GO:0098862 cluster of actin-based cell projections (FDR = 0.039), GO:0045505 dynein intermediate chain binding (FDR = 0.0067), schizophrenia (FDR = 0.042), and mental disorders (FDR = 0.089) (Fig. 2A). A total of 1770 genes contain genomic variants that contribute to either anxiety or other mental disorders. Biological and/or clinical overrepresentations include Adhesion (FDR = 4.7E–10)), nervous system malformations (FDR = 6.7E–4), the Cadherin signaling pathway (FDR = 0.0078), and hsa04730 Long-term depression (FDR = 0.15). (Fig. 2B). The functional overrepresentations pertaining to genes associated with anxiety-specific variants, as well as those implicated in anxiety variants shared among other mental disorders, exhibit marked disparities. Moreover, the degree of overlap between these two gene sets is notably limited, constituting 19.7 and 6.8% respectively (Fig. 2C). These findings underscore the discernible patterns within the underlying molecular pathways and distinctive characteristics specific to anxiety disorders in contrast to other psychiatric conditions.
A Gene sets overrepresented with anxiety-specific genomic variants; B Gene sets overrepresented with anxiety variants shared in other mental disorders; C Venn diagram for anxiety-specific variant corresponding genes and genes containing variants shared in anxiety and other mental disorders; D Importance/weight for genomic variants in different loci.
Unique molecular signatures for selected variants in different genomic loci
Variants in genomic loci can exert diverse effects on genetic functionality. For instance, nonsynonymous mutations can induce alterations in amino acid composition or even trigger frameshifts in protein sequences. Intronic variants may disrupt splicing mechanisms, while variants in the 5′ untranslated region (UTR) have the potential to influence upstream open reading frames (uORFs), internal ribosome entry sites (IRESs), microRNA binding sites, and structural elements implicated in mRNA stability regulation, pre-mRNA splicing, and translation initiation (Ryczek, Łyś, & Makałowska, 2023). Our investigation reveals that genes corresponding to different anxiety-specific variants exhibit distinct functional patterns compared to variants shared among anxiety and other mental disorders. Additionally, variants across different loci demonstrate diverse functional profiles (Fig. 3). Particularly noteworthy is the observation that non-coding variants, especially those situated within intronic regions and in close proximity to exons, exhibit significantly stronger overrepresentation scores (Fig. 3E, F). This underscores the potentially pivotal role of splicing mechanisms in mental disorders, a role that may be more significant than previously acknowledged.
A, B Nonsynonymous variants for anxiety-specific variant corresponding genes and genes with variants shared in anxiety and other mental disorders; C, D Synonymous variants for anxiety-specific variant corresponding genes and genes with variants shared in anxiety and other mental disorders; E, F Intronic variants for anxiety-specific variant corresponding genes and genes with variants shared in anxiety and other mental disorders; G, H 5′ UTR variants for anxiety-specific variant corresponding genes and genes with variants shared in anxiety and other mental disorders.
Discussion
Unique genomic patterns in anxiety disorders of DS patients
Rapidly developing AI algorithms, particularly deep learning models, have the potential to provide new insights into genome variants associated with anxiety disorders. Data-driven analytic strategies can mimic the impacts of various factors given sufficiently large datasets. In this study, WGS data from blood and saliva samples were generated for 709 DS individuals, including 255 probands diagnosed with at least one type of mental disorder, of whom 74 had a diagnosis of anxiety (Fig. 1B). For comparative analysis, WGS was conducted on 425 healthy individuals who were family members of the DS probands, with an additional 345 independent healthy controls included to mitigate false positive results. The Multi-layer Perceptron (MLP) deep learning model was deployed to rank the importance of genomic variants while fitting the clinical phenotype of DS children with anxiety, DS children with mental disorders other than anxiety, DS children without mental disorders, and healthy family members of DS patients. Unlike traditional machine learning models designed for predictive diagnostics, this study does not aim to predict anxiety diagnoses in individuals with Down syndrome (DS). Instead, our objective is to identify genomic variants specifically associated with anxiety, distinguishing them from those linked to other mental disorders within this unique population. The linear algebra calculations provided the contributing weights for each variant to specific diagnoses. We focused on identifying genomic variants that significantly contribute to anxiety, but not to other mental disorders or control groups. Meanwhile, we also explored variants associated with both anxiety and other mental disorders considering shared molecular pathways among psychiatry disorders [18].
Although anxiety is often categorized alongside other mental disorders such as autism, attention deficit hyperactivity disorder (ADHD), depression, and obsessive-compulsive disorder (OCD), our results indicate that genomic variant signatures for anxiety disorder exhibit distinct patterns compared to these other psychiatric conditions. The most direct evidence is the limited overlap of corresponding genes associated with anxiety-specific variants and those shared with other mental disorders (Fig. 2C). Meanwhile, the functional category analysis also reveals little similarity between these two groups of variants, as shown in Fig. 3. Additionally, the cell-type-specific analysis identified ciliated cells, endothelial cells, and neurons as the most significantly enriched cell types for anxiety-specific genes (Fig. 4A). Notably, ciliary dysfunction has been implicated in anxiety in both humans and mice, suggesting a potential role of cilia in anxiety regulation [19, 20]. Furthermore, studies have reported an association between increased circulating endothelial progenitor cell levels and anxiety severity in patients with depression [21]. Conversely, anxiety has also been shown to impair endothelial function, which serves as an early risk factor for cardiovascular diseases [22]. Notably, genes shared between anxiety and other mental disorders did not show a significant association with neuronal cell types (Fig. 4B), unlike anxiety-specific genes. This finding suggests that distinct cell-type associations underlie the genetic architecture of anxiety compared to other mental illnesses.
A Top enriched cell types of anxiety-specific genes; B top enriched cell types of genes shared among anxiety and other types of mental disorders; C environmental factors, including injuries, poisoning, surgeries and medications treatments for DS patients with anxiety, other types of mental disorders, and no reported mental issues; D Environmental factor differences between anxiety-specific patients versus patients with other types of mental disorders.
Distinct clinical comorbid factors
One potential explanation for the distinct patterns of anxiety disorders in DS patients is the significant influence of clinical contributing factors on anxiety disorders. Among the selected intronic variants close to splicing sites, we identified a number of contributing factors that may increase the propensity for anxiety. Previous studies of myasthenia gravis (MG) genes (FDR = 0.2) found that anxiety is one of the major concerns among MG individuals, and having anxiety symptoms is notably more common than, for example, having autoimmune diseases [23]. Insulin resistant diabetes and pediatric renal diseases have a higher prevalence in the DS population, while insulin resistance in the brain induces mitochondrial and dopaminergic dysfunction, leading to anxiety and depressive-like behaviors [24]. Children with renal diseases have increased odds, approximately 5–6 times, of also carrying a diagnosis of anxiety compared to the general population [25]. The comorbid factors could have a bidirectional impact, where they may not only contribute to anxiety but also be a result of the process. For example, people with anxiety may perform poorly in perceptual-motor performance (e.g., picking up a cup from a table). A list of selected genes with anxiety-specific variants related to motor activity was found to be significantly overrepresented (FDR = 0.026).
To assess the environmental factors such as the impact of treatment, including medications and surgical procedures received by patients with anxiety, we extracted comprehensive treatment data from the electronic health records (EHRs) of 709 DS patients. The proportion of drug treatments (regardless of indication), surgical procedures performed, and daily problems reported were calculated among individuals diagnosed with anxiety, those with other mental disorders, and the overall DS patient population. As shown in Fig. 4C, no significant differences were observed in the medications or surgeries received across these groups. However, we found that the incidence of injury and poisoning was notably higher among DS patients with anxiety compared to those with other mental disorders (Fig. 4D). A potential explanation is that individuals with anxiety may be more susceptible to accidents due to heightened worry and preoccupation, which can impair focus and reaction time. This reduced awareness of potential hazards may increase their risk of injury. Prior research in sports science has demonstrated that anxiety affects athletic performance and is closely linked to the risk of sports-related injuries [26]. Additionally, mental illnesses, including anxiety and depression, have been associated with a higher likelihood of self-poisoning [27].
Genetic marker identification and importance of non-coding variants
With anxiety disorders exhibiting unique genomic patterns compared to other mental disorders in the DS population, identifying anxiety-specific genetic markers may assist with accurate diagnosis and intervention in this special patient group. In this study, mutation detection was expanded to the whole genome instead of focusing solely on the coding region. Variants that do not cause amino acid alterations, such as synonymous mutations, have been shown to have implications in psychiatric research on the human brain [28]. Several lines of compelling evidence suggest that non-coding DNA variants affecting important 5′ and 3′ regulatory as well as intronic sequences are associated with central nervous system disorders such as anxiety by altering transcription factor or microRNA binding sites [10]. As shown in Fig. 2D, importance measurements are higher in non-coding variants compared to coding regions, suggesting regulatory mechanisms for anxiety disorders compared to other types of mental disorders in the DS population. Meanwhile, biological pathways associated with anxiety and brain functions have been revealed in corresponding genes without amino acid alterations. Axon guidance mediated by the Slit/Robo pathway could be a novel target for precision medicine and the treatment of depression [29], and in animal studies, Slit2 transgenic mice showed anxiety-like behaviors [30]. Synonymous variant corresponding genes were found to be enriched in idiopathic pulmonary fibrosis (FDR = 0.2), which causes anxiety as a common consequence [31]. Variants in intronic regions, for example, have been identified as targets of MiR-125B, which is imprinted only in the human brain and mediates learning, memory, and anxiety in response to external factors like stress [32]. For the 43 selected variants in ncRNA exon regions (Table 1), their target genes include FGFR1, which is involved in multiple psychiatric disorders [33], SMAD4, whose gene expression is altered in the brain and blood of schizophrenia patients [34], NFE2L2, which has a pathophysiological role and potential as a target for psychiatric disorders [35], ILF3, which is significantly associated with the risk of bipolar disorder [36], miR-940, a microRNA upregulated in major depression disorders [37], and CTNNB1, whose mutation leads to dysfunction of the Wnt signaling pathway that regulates gene transcription and further disturbs synaptic plasticity, neuronal apoptosis, and neurogenesis [38].
Several chromosome loci show overrepresentations of functional corresponding genes for both anxiety-specific variants and variants shared in anxiety and other psychiatric disorders, including chr17q25.3 (13 genes with anxiety-specific variants, FDR = 0.0014; 28 genes with shared variants FDR = 3.1E–5), and chr16p13.3 (18 genes with anxiety-specific variants, FDR = 0.0013; 40 genes with shared variants, FDR = 3.1E–5) (Table 2). A previous clinical trial reported that the chr17q25 region is significantly associated with white matter lesions [39]; meanwhile, altered white matter substance is found to be a vulnerability marker in individuals at high risk of clinical anxiety [40]. It should be noted that copy number abnormality in chr21q22.3 has been reported in autism spectrum disorder, anxiety, and severe depression [41]. Considering all the probands are DS subjects with trisomy 21, the chr21q22.3 region could be especially important as DS anxiety disorder target region.
The list of 29 genetic markers for anxiety-specific diagnosis in DS populations was generated based on the combined impact of contributions to the integrated clinical phenotype from deep learning models, along with the contributions to anxiety and other mental disorders from the linear algebra model with functional overrepresentation results. In other words, these biomarkers have the highest weight in fitting the clinical diagnosis for each patient, with the highest contributions to anxiety but no contributions or even negative contributions to other mental disorders (Table 3). Examples include variations in the 5′ UTR region of CCK, which is known to trigger anxiety and panic attacks in humans [42], a nonsynonymous mutation in exon 9 of KRT7, a gene participating in apoptosis and synaptic transmission in methylation profiling of schizophrenia [43], and an intronic variation close to exon 11 of STK11IP, a diagnostic marker for major depression [44]. To further validate the genetic markers identified, we selected an additional 25 blood samples from DS anxiety patients and 49 samples from DS patients without anxiety but with other mental illnesses. The differential expression tests of RNA sequencing between the two groups showed that five genetic markers demonstrated at least a 1.5-fold change in expression levels between DS patients with anxiety and those with other mental disorders. Among them, three markers were upregulated in the anxiety group (PBXIP1, TNN, KIR2DL1), while two markers were downregulated (KRT7, C16orf86). Notably, PBXIP1, which showed a 2.3-fold increase in expression in the anxiety group, and KRT7, which exhibited only 0.4-fold expression in this group, both contain selected nonsynonymous variations. As a result, the list could be a valuable resource for assisting in the diagnosis of anxiety, not other types of mental disorders in the DS group.
In summary, this pioneering study represents the first comprehensive evaluation of anxiety disorders in DS utilizing WGS cohorts and advanced deep learning AI models. The results indicate that anxiety disorder in DS patients has distinct molecular patterns from other mental disorders. The new insights gained from our research offer a valuable understanding of underlying mechanisms. The genetic markers identified in our study hold promise for enhancing the clinical diagnosis of anxiety and guiding more effective intervention strategies in this vulnerable population.
Availability of data and materials
The KidFirst data could be accessed at the Kids First Data Resource Portal (DRC, https://portal.kidsfirstdrc.org/login).
References
Penninx BW, Pine DS, Holmes EA, Reif A. Anxiety disorders. Lancet. 2021;397:914–27.
Austerman J. ADHD and behavioral disorders: assessment, management, and an update from DSM-5. Cleve Clin J Med. 2015;82:S2–7.
Capone G, Goyal P, Ares W, Lannigan E. Neurobehavioral disorders in children, adolescents, and young adults with down syndrome. Am J Med Genet C Semin Med Genet. 2006;142C:158–72.
Whitney DG, Shapiro DN, Peterson MD, Warschausky SA. Factors associated with depression and anxiety in children with intellectual disabilities. J Intellect Disabil Res. 2019;63:408–17.
Olmos-Serrano JL, Kang HJ, Tyler WA, Silbereis JC, Cheng F, Zhu Y, et al. Down syndrome developmental brain transcriptome reveals defective oligodendrocyte differentiation and myelination. Neuron. 2016;89:1208–22.
El Hajj N, Dittrich M, Böck J, Kraus TF, Nanda I, Müller T, et al. Epigenetic dysregulation in the developing down syndrome cortex. Epigenetics. 2016;11:563–78.
Chiu A, Falk A, Walkup JT. Anxiety disorders among children and adolescents. Focus (Am Psychiatr Publ). 2016;14:26–33.
Waszczuk MA, Zavos HM, Eley TC. Genetic and environmental influences on relationship between anxiety sensitivity and anxiety subscales in children. J Anxiety Disord. 2013;27:475–84.
Seok BJ, Jeon S, Lee J, Cho SJ, Lee YJ, Kim SJ. Effects of early trauma and recent stressors on depression, anxiety, and anger. Front Psychiatry. 2020;11:744.
Keszler G, Molnár Z, Rónai Z, Sasvári-Székely M, Székely A, Kótyuk E. Association between anxiety and non-coding genetic variants of the galanin neuropeptide. PLoS ONE. 2019;14:e0226228.
Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164.
Zhao H, Yin X, Xu H, Liu K, Liu W, Wang L, et al. LncTarD 2.0: an updated comprehensive database for experimentally-supported functional lncRNA–target regulations in human diseases. Nucleic Acids Res. 2023;51:D199–D207.
Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, et al. Scikit-learn: machine learning in python. J Mach Learn Res. 2011;12:2825–30.
Liao Y, Wang J, Jaehnig EJ, Shi Z, Zhang B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019;47:W199–W205.
Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550.
Dai Y, Hu R, Liu A, Cho KS, Manuel AM, Li X, et al. WebCSEA: web-based cell-type-specific enrichment analysis of genes. Nucleic Acids Res. 2022;50:W782–w790.
Morris G, Puri BK, Walker AJ, Maes M, Carvalho AF, Bortolasci CC, et al. Shared pathways for neuroprogression and somatoprogression in neuropsychiatric disorders. Neurosci Biobehav Rev. 2019;107:862–82.
Verkleij M, Appelman I, Altenburg J, Twisk J, Quittner AL, Haarman E. Anxiety and depression in dutch patients with primary ciliary dyskinesia and their caregivers: associations with health-related quality of life. ERJ Open Res. 2021;7:00274–2021.
Rödig N, Sellmann K, Dos Santos Guilherme M, Nguyen VTT, Cleppien D, Stroh A, et al. Behavioral phenotyping of Bbs6 and Bbs8 knockout mice reveals major alterations in communication and anxiety. Int J Mol Sci. 2022;23:14506.
Liou YJ, Chen MH, Hsu JW, Huang KL, Huang PH, Bai YM. Associations between increased circulating endothelial progenitor cell levels and anxiety/depressive severity, cognitive deficit and function disability among patients with major depressive disorder. Sci Rep. 2021;11:18221.
Sara JDS, Ahmad A, Toya T, Suarez Pardo L, Lerman LO, Lerman A. Anxiety disorders are associated with coronary endothelial dysfunction in women with chest pain and nonobstructive coronary artery disease. J Am Heart Assoc. 2021;10:e021722.
Nadali J, Ghavampour N, Beiranvand F, Maleki Takhtegahi M, Heidari ME, Salarvand S, et al. Prevalence of depression and anxiety among myasthenia gravis (MG) patients: a systematic review and meta-analysis. Brain Behav. 2023;13:e2840.
Kleinridders A, Cai W, Cappellucci L, Ghazarian A, Collins WR, Vienberg SG, et al. Insulin resistance in brain alters dopamine turnover and causes behavioral disorders. Proc Natl Acad Sci USA. 2015;112:3463–8.
Le P, Nguyen M, Vu T, Dao DP, Olson D, Zhang AY. Anxiety and depression in patients with retinitis pigmentosa. J Vitreoretin Dis. 2021;5:114–20.
Rogers DL, Tanaka MJ, Cosgarea AJ, Ginsburg RD, Dreher GM. How mental health affects injury risk and outcomes in athletes. Sports Health. 2024;16:222–9.
Zheng W, Han L, Fan Y, Yi M, Lu X, Yang J, et al. Association of mental health status between self-poisoning suicide patients and their family members: a matched-pair analysis. BMC Psychiatry. 2023;23:294.
Nishioka M, Bundo M, Iwamoto K, Kato T. Somatic mutations in the human brain: implications for psychiatric research. Mol Psychiatry. 2019;24:839–56.
Mahmud A, Avramescu RG, Niu Z, Flores C. Awakening the dormant: role of axonal guidance cues in stress-induced reorganization of the adult prefrontal cortex leading to depression-like behavior. Front Neural Circuits. 2023;17:1113023.
Huang G, Wang S, Yan J, Li C, Feng J, Chen Q, et al. Depression-/anxiety-like behavior alterations in adult Slit2 transgenic mice. Front Behav Neurosci. 2020;14:622257.
Lee YJ, Choi SM, Lee YJ, Cho YJ, Yoon HI, Lee JH, et al. Clinical impact of depression and anxiety in patients with idiopathic pulmonary fibrosis. PLoS ONE. 2017;12:e0184300.
Chou MY, Cao X, Hou KC, Tsai MH, Lee CY, Kuo MF, et al. Mir125b-2 imprinted in human but not mouse brain regulates hippocampal function and circuit in mice. Commun Biol. 2023;6:267.
Deng Z, Deng S, Zhang MR, Tang MM. Fibroblast growth factors in depression. Front Pharmacol. 2019;10:60.
Wolf A, Yitzhaky A, Hertzberg L. SMAD genes are up-regulated in brain and blood samples of individuals with schizophrenia. J Neurosci Res. 2023;101:1224–35.
Morris G, Walker AJ, Walder K, Berk M, Marx W, Carvalho AF, et al. Increasing Nrf2 activity as a treatment approach in neuropsychiatry. Mol Neurobiol. 2021;58:2158–82.
Park CY, Zhou J, Wong AK, Chen KM, Theesfeld CL, Darnell RB, et al. Genome-wide landscape of RNA-binding protein target site dysregulation reveals a major impact on psychiatric disorder risk. Nat Genet. 2021;53:166–73.
Wu Z, Cai Z, Shi H, Huang X, Cai M, Yuan K, et al. Effective biomarkers and therapeutic targets of nerve-immunity interaction in the treatment of depression: an integrated investigation of the miRNA-mRNA regulatory networks. Aging. 2022;14:3569–96.
Zhuang W, Ye T, Wang W, Song W, Tan T. CTNNB1 in neurodevelopmental disorders. Front Psychiatry. 2023;14:1143328.
Verhaaren BF, de Boer R, Vernooij MW, Rivadeneira F, Uitterlinden AG, Hofman A, et al. Replication study of chr17q25 with cerebral white matter lesion volume. Stroke. 2011;42:3297–9.
Lu M, Yang C, Chu T, Wu S. Cerebral white matter changes in young healthy individuals with high trait anxiety: a tract-based spatial statistics study. Front Neurol. 2018;9:704.
Orru S, Papoulidis I, Siomou E, Papadimitriou DT, Sotiriou S, Nikolaidis P, et al. Autism spectrum disorder, anxiety and severe depression in a male patient with deletion and duplication in the 21q22.3 region: a case report. Biomed Rep. 2019;1:1–5.
Rehfeld JF. Cholecystokinin and panic disorder: reflections on the history and some unsolved questions. Molecules. 2021;26:5657.
Rukova B, Staneva R, Hadjidekova S, Stamenov G, Milanova T, Toncheva D. Genome-wide methylation profiling of schizophrenia. Balkan J Med Genet. 2014;17:15–23.
Jiang H, Fu CY. Identification of shared potential diagnostic markers in asthma and depression through bioinformatics analysis and machine learning. Int Immunopharmacol. 2024;133:112064.
Acknowledgements
Sample collection and biobanking for this study was supported by the Institutional Development Funds from the Children’s Hospital of Philadelphia (CHOP) to the Center for Applied Genomics (CAG) and CHOP´s Endowed Chair in Genomic Research (CAG). The sequencing data was provided through the Gabriella Miller Kids First Pediatric Research Program consortium (Kids First), supported by the Common Fund of the Office of the Director of the National Institutes of Health (www.commonfund.nih.gov/KidsFirst), awarded to CAG. We express our gratitude to the CAG staff and the patients and families who generously contributed biosamples to CAG/CHOP.
Funding
The study was supported by the Institutional Development Funds from the Children’s Hospital of Philadelphia to the Center for Applied Genomics, and The Children’s Hospital of Philadelphia Endowed Chair in Genomic Research to HH.
Author information
Authors and Affiliations
Contributions
Conceptualization and supervision, YL and HH; literature search, YL; data preparation & analysis, YL, HQQ, CX, FDM, HQ, ST, KN, KO, and TW; data interpretation, YL, HQQ, CX, JG, and HH; original draft writing, YL; review and revision led by YL, HQQ and HH, with all authors contributing and approving the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The author declares no competing interests.
Ethics approval and consent to participate
We confirm that all methods were carried out in accordance with relevant regulatory guidelines and regulations. All experimental protocols were approved by the Institutional Review Board (IRB) of the Children’s Hospital of Philadelphia (CHOP) with the IRB number: IRB 16-013278. Informed consent was obtained from all subjects. If subjects are under 18, consent was obtained from a parent and/or legal guardian with assent from the child if 7 years or older.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Liu, Y., Qu, HQ., Chang, X. et al. Deep learning algorithms reveal genomic markers for anxiety disorder in a large cohort of children with down syndrome. Mol Psychiatry (2025). https://doi.org/10.1038/s41380-025-03065-2
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41380-025-03065-2