Abstract
Acute promyelocytic leukemia (APL) is driven by the oncoprotein PML/RARα, which destroys the architecture of PML nuclear bodies (NBs). PML NBs are critical to tumor suppression, and their disruption mediated by PML/RARα accelerates APL pathogenesis. However, the mechanisms of PML NB disruption remain elusive. Here, we reveal that the failure of NB assembly in APL results from neddylation-induced aberrant phase separation of PML/RARα. Mechanistically, PML/RARα is neddylated in the RARα moiety, and this neddylation enhances its DNA-binding ability and further impedes the phase separation of the PML moiety, consequently disrupting PML NB construction. Accordingly, deneddylation of PML/RARα restores its phase separation process to reconstruct functional NBs and activates RARα signaling, thereby suppressing PML/RARα-driven leukemogenesis. Pharmacological inhibition of neddylation by MLN4924 eradicates APL cells both in vitro and in vivo. Our work elucidates the neddylation-destroyed phase separation mechanism for PML/RARα-driven NB disruption and highlights targeting neddylation for APL eradication.

Similar content being viewed by others
Introduction
In more than 90% of acute promyelocytic leukemia (APL) cells, promyelocytic leukemia (PML) fuses to retinoic acid receptor α (RARα) because of t (15; 17) chromosomal translocation, which destroys the architecture of PML nuclear bodies (NBs) [1]. PML NB is a membraneless structure 0.2–1 μm in diameter that is critical to tumor suppression [2]. PML NBs recruit partners to promote cell apoptosis and/or senescence, regulate the cell cycle, inhibit cell migration, and hinder tumor-associated angiogenesis to finally inhibit carcinogenesis [3,4,5]. Thus, PML NB disruption by PML/RARα is crucial to leukemogenesis [6]. Although the phenomenon of NB disruption has been observed for a long time, its mechanism remains poorly understood. Elucidating the mechanism of PML/RARα-mediated NB disruption is of critical importance to understand the pathogenesis of APL.
To determine the mechanism of PML/RARα-mediated NB disruption, we screened ~300 signaling pathway inhibitors and identified two neddylation inhibitors that could induce PML/RARα to reform NBs. Neddylation is a posttranslational modification in which the ubiquitin-like (UBL) molecule NEDD8 conjugates to substrates by the E1/E2/E3 enzymatic cascade, which is involved in multiple physiological and pathological processes [7, 8]. Here, we discuss the role of neddylation in the PML/RARα-disrupted NB process.
Recently, mounting evidence has suggested that phase separation, a physical process, plays a pivotal role in cellular condensate assembly [9, 10], such as several membraneless organelles (nucleolus, Cajal body, and Notch intracellular ___domain (NICD) nuclear body) [11,12,13]. Considering that PML NB is a classical membraneless organelle, it may also show the same liquid-like features. Importantly, PML NBs appear to be dynamic in cellulo [14, 15]. Different quantities, sizes, and concentrated partners of PML NBs are frequently observed in the same cell type, and these dynamic characteristics are highly stress-sensitive. Additionally, the PML NB architecture is highly consistent with the scaffold-client model of phase separation [16]. Consistent with the dynamic characteristics and phase separation-like structural composition of PML NBs, we suspect that PML may induce phase separation to construct PML NBs. In contrast, the PML/RARα fusion protein cannot form an NB structure, which perhaps just results from aberrant phase separation. Moreover, posttranslational modification is recognized as a pivotal regulator of phase separation [17]. Therefore, the correlation among neddylation, phase separation and PML NB disruption warrants further investigation.
In this study, we report that PML/RARα is neddylated in the RARα region, and inhibition of the neddylation of PML/RARα markedly induces the reassembly of PML NBs. Mechanistically, the neddylation of PML/RARα enhances its DNA-binding ability and further disrupts the phase separation of the PML moiety, consequently destroying PML NB formation. Deneddylation of PML/RARα can restore the phase separation of fusion proteins, followed by PML NB reconstruction as well as RARα signaling reactivation, significantly blocking the self-renewal of APL cells and suppressing leukemogenesis. Herein, we offer new insights into the mechanism by which the neddylation of PML/RARα disrupts PML NB formation through weakening phase separation and suggest targeting neddylation as a potential therapeutic option against APL.
Results
High-content screening identifies neddylation inhibitors that induce PML/RARα to reassemble NB structures
In APL, PML NBs are known to be disrupted by PML/RARα, which contributes to leukemogenesis. Accordingly, we found that PML/RARα disrupted PML NBs in a dose-dependent manner (Fig. 1A). To explore the mechanism by which PML/RARα disrupts NBs, we established a cell-based high-content screening assay to screen for signals that would induce PML/RARα to reform NBs (Fig. 1B). ATO is the positive control that induces the reaggregation of PML/RARα [18, 19]. As displayed in Fig. 1C and Table S1, among the ~300 signaling pathway inhibitors, MLN4924 and TAS4464, two inhibitors of the NEDD8-activating enzyme E1, exerted the strongest upregulation effect on the large foci numbers of PML/RARα-GFP. Further immunofluorescence results showed that MLN4924 induced PML/RARα-HA to reassemble NB-like foci, whose number and diameter were similar to classic PML NBs rather than ATO-induced PML/RARα aggregates (Fig. 1D). Additionally, the MLN4924-restored PML/RARα NBs exhibited good solubility in detergent-containing buffers (RIPA buffer), similar to classic PML NBs but not to ATO-bonded aggregates (Fig. S1). These results indicate the similarity between reformed PML/RARα NBs and classic PML NBs.
A The effect of PML/RARα on the PML NBs formation. H1299 cells were infected with PML-Flag and different dosage of PML/RARα-HA lentivirus. B Schematic view of the cell-based PML/RARα-GFP high-content screening (HCS) system. H1299 cells were infected with PML/RARα-GFP lentivirus and then treated with a collection of ~300 signaling pathway inhibitors (1 μM). ATO is the positive control that induces the reaggregation of PML/RARα by directly binding to cysteine residues of PML/RARα. C The number of large foci (foci size≥200 nm) per 200 cells in each well after inhibitor treatment as shown in Fig. 1B. D Representative fluorescence images of PML and PML/RARα treated with MLN4924 or ATO. H1299 cells were infected with PML/RARα-HA or PML-HA lentivirus followed by treatment with MLN4924 (1 μM) for 48 h and ATO (1 μM) for 12 h. The average diameter of foci was calculated and the number of large foci (foci size ≥ 200 nm) per 100 cells was counted. E The PML NBs formation when co-expressed with different dosage of PML/RARα upon MLN4924 treatment. H1299 cells were infected with PML-Flag and different dosage of PML/RARα-HA lentivirus, followed by treatment with MLN4924 (1 μM) for 48 h. F Formation of PML NBs in NB4 cells triggered by MLN4924. NB4 cells were treated with MLN4924 (40 nM) for the indicated times and then immunostained with anti-PML antibody and DAPI staining. G The endogenous PML NBs in ATRA-resistant APL cell lines after exposed to MLN4924. NB4R1 and NB4R2 cells were treated with MLN4924 (40 nM) for 24 h and immunostained with anti-PML antibody and DAPI staining. (F, G) The average diameter of foci was calculated and the number of large foci (foci size≥200 nm) per 100 cells was counted. Data are represented as mean ± SD (n = 3). **, p < 0.01; ***, p < 0.001, vs. PML/RARα (D) or Control group (F, G). Statistical significance of differences between groups was determined with one-way ANOVA analysis (D and F) or two-tailed unpaired Student’s t-test (G).
In APL, PML/RARα fusion coexists with PML; thus, we used a system that accounts for the allelic balance to verify the effect of MLN4924. Similarly, PML NB structures destroyed by different dosages of PML/RARα were all strongly reassembled by MLN4924 (Fig. 1E). More supportively, MLN4924 promoted the reformation of endogenous PML NBs in PML/RARα-positive APL cells (NB4) (Fig. 1F). Similar results were also obtained in two ATRA-resistant APL cell lines (NB4R1 and NB4R2) (Fig. 1G). Thus, it appears that neddylation inhibitors block the disruption of PML/RARα on PML NBs to finally reassemble NB structures.
PML/RARα NBs reformed by neddylation inhibitors show functional similarity to classic PML NBs
Because of the high similarity between reformed PML/RARα NBs and classic PML NBs both in foci size and solubility, we next investigated whether the reformed PML/RARα NBs had the same recruitment function as classic PML NBs. The recruitment of partners into NBs is strongly associated with PML sumoylation [20]. Thus, we first tested the sumoylation of PML/RARα. As presented in Fig. 2A and S2, PML/RARα NBs reformed by MLN4924 were colocalized with SUMO-1 and SUMO-2/3. Similar results were also obtained for ATRA-resistant PML/RARα mutants (∆F286 and R276Q) and ATO-resistant mutants of PML/RARα (A216V and L218P) (Fig. S3) [21]. Moreover, a marked increase in SUMO-conjugated PML/RARα species was induced by MLN4924 (Fig. 2B). We further verified that DAXX, a well-studied sequestered partner in PML NBs [22], was highly colocalized with MLN4924-reformed PML/RARα NBs (Fig. 2C). These results suggest that MLN4924-reformed PML/RARα NBs may have the same recruitment function as PML NBs.
A Co-localization of PML/RARα and SUMO1 in NBs following MLN4924 treatment. H1299 cells were infected with PML/RARα-HA lentivirus followed by treatment with MLN4924 (1 μM) for 48 h. B MLN4924 upregulates the sumoylation modification level of PML/RARα. COS7 cells were transfected with pCDNA3.0-PML/RARα-HA for 24 h, followed by MLN4924 treatment (1 μM) for 48 h. The input band at ~100 KDa detected by anti-NEDD8 antibody on input is Cullin-NEDD8 conjugates. C Co-localization of DAXX with reformed NBs induced by MLN4924. PML/RARα-HA expressing H1299 cells were treated with MLN4924 (1 μM) for 48 h. D Schematic view of the BioID method to identify the transient partner proteins in reconstructed PML/RARα NBs and classical PML NBs. H1299 cells were infected with pCDH-PML/RARα-BirA*, pCDH-PML-BirA* or pCDH-NLS-BirA* lentivirus for 5 days, and the PML/RARα-BirA* group was treated with MLN4924 (1 μM) for 48 h before harvest. The specific biotin-labeled proteins in cells were assessed by the peptide intensity ratio (vs. NLS) ≥ 2. E Overlay proteins both specifically identified in PML/RARα-BirA* cells treated with MLN4924 and in PML-BirA* cells. The proteins are arranged in order by their score. Red star represents the proteins previously reported as recruited partners in classic PML NBs. F Top 20 clusters from Metascape pathway enrichment analysis of the overlay proteins. The heatmap of enriched terms is colored based on p-values. G Effect of MLN4924 on PML/RARα NB reconstruction in two different PML KO subclones. PML KO H1299 subclones (#1 and #2) were infected with PML/RARα-HA lentivirus, followed by treatment with MLN4924 (1 μM) for 48 h. H The average diameter of foci was calculated and the number of large foci (foci size ≥ 200 nm) per 100 cells was counted in Fig. 2G. Data are represented as mean ± SD (n = 3). ***, p < 0.001, vs. Control group. Statistical significance of differences between groups was determined with two-tailed unpaired Student’s t-test.
To further confirm this hypothesis, we examined the partner proteins in the reformed PML/RARα NBs. Because of the remarkable dynamic activities of NBs, we identified the transient partner proteins in NBs utilizing the proximity-dependent biotin identification (BioID) method [23, 24] (Fig. 2D). In total, 159 proteins were identified to physically interact with both reformed PML/RARα NBs and classic PML NBs (Fig. 2E and Table S2). Notably, 25 proteins, as indicated in Fig. 2E, were previously reported as recruited proteins in classic PML NBs [25,26,27,28,29,30]. Gene Ontology (GO) analysis revealed that these 159 proteins were highly enriched in four pathways associated with classic PML NBs, including sumoylation of transcription cofactors, DNA repair, sumoylation of DNA replication proteins and transcriptional regulation by TP53 (Fig. 2F). These results suggest that MLN4924-reformed PML/RARα NBs recruit similar partners to classic PML NBs, indicating that they may show functional similarity.
A critical question is whether the reconstructed NBs simply result from PML/RARα and are independent of PML. In the four PML KO subclones generated by CRISPR/Cas9 (Fig. S4A, B), MLN4924-treated PML/RARα also reformed into NBs (Fig. 2G, H and S4C), revealing that this NB reconstruction is independent of PML.
PML/RARα is neddylated at the Lys227 and Lys360 sites in the RARα moiety
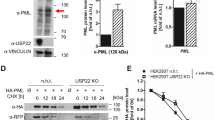
To dissect the mechanisms underlying neddylation inhibitor-induced NB reassembly, we first investigated whether their own deneddylation activities played key roles in this process. The results demonstrated that both shNEDD8 and shUbc12 as well as the deneddylation enzyme NEDP1 dramatically induced the reconstruction of PML/RARα NBs (Fig. S5), suggesting the involvement of neddylation in the process of PML/RARα NB reassembly. Further considering the fact that PML/RARα NB reformation is independent of PML (Fig. 2G), we hypothesized that PML/RARα might be directly modified by NEDD8. As expected, NEDD8 was covalently conjugated to PML/RARα, which was completely prevented by MLN4924 (Fig. 3A). Consistently, Ubc12 dramatically increased the level of neddylated PML/RARα, while the dominant-negative mutant of Ubc12 (C111S) did not (Fig. 3B). Moreover, PML/RARα neddylation was strongly attenuated by NEDP1 (Fig. 3C). Together, these results clearly indicate that PML/RARα is a target for neddylation.
A MLN4924 inhibits the neddylation of PML/RARα. COS7 cells were transfected with PML/RARα-HA for 24 h followed by MLN4924 treatment (1 μM) for 24 h. B The neddylation E2-conjugating enzyme Ubc12 promotes the neddylation of PML/RARα. COS7 cells were transfected with PML/RARα-HA combined with Ubc12-Flag or Ubc12 (C111S)-Flag. l.e., long exposure; s.e., short exposure. C Deneddylation enzyme NEDP1 attenuates the neddylation of PML/RARα. COS7 cells were transfected with PML/RARα-HA combined with NEDP1-Flag. D Neddylation levels of PML/RARα, PML, and RARα. COS7 cells were transfected with PML/RARα-HA, PML-HA, and RARα-HA for 48 h. E Representative tandem mass spectrometry spectra of peptides VSLDIDLWDKFSELSTK and VDMLQEPLLEALKVYVR. Lysine 227 and 360 were the potent neddylation sites. F Effect of potential neddylation site mutations on PML/RARα neddylation. COS7 cells were transfected with different PML/RARα-HA (WT, K227R, K360R, and 2KR (K227/360R)) for 48 h. The neddylation of RARα segment in the RRL cell-free system. G Recombinant GST-RARα (LBD) were incubated with or without RRL at 37 °C for 30 mins. H Recombinant GST-RARα (LBD) (WT) and GST-RARα (LBD) (2KR) were incubated with or without RRL.
Next, we sought to identify the potential neddylation sites of PML/RARα. First, NEDD8 was conjugated to both PML/RARα and RARα, but not the PML protein (Fig. 3D), indicating that PML/RARα was neddylated in the RARα moiety. Then, we generated two RARα segment mutants: (1) 61-183aa, containing the DNA-binding ___domain DBD (88-153aa); (2) 183-417aa: the ligand-binding ___domain LBD [31]. The LBD segment was neddylated, whereas the RARα segment (61-183aa) containing DBD was not (Fig. S6A). Next, we enriched the neddylated LBD segment for mass spectrometry analysis and found five lysines as candidate neddylation sites (Fig. S6B, C). Mutagenesis scanning revealed that Lys227 and Lys360 were critical sites (Fig. 3E), as K227R, K360R, and the double mutation 2KR (K227/360R) obviously reduced PML/RARα neddylation (Fig. 3F), while the other 3 lysine mutations did not (Fig. S6D). Additionally, because the LBD segment possesses 14 lysines, we also generated lysyl replacement mutants to further confirm the neddylation sites. K227R, K360R, and K399R of RARα showed significantly reduced neddylation levels (Fig. S6E). In PML/RARα, K227R, and K360R, but not K399R, the neddylation levels weakened (Fig. S6F). Furthermore, robust neddylation of the recombinant LBD segment was also observed in a cell-free system using rabbit reticulocyte lysate (RRL) (Fig. 3G). In contrast, the neddylation level was downregulated by the double mutation 2KR (K227/360R) (Fig. 3H). Overall, these findings demonstrate that the Lys227 and Lys360 sites in the RARα region are the major neddylation sites of PML/RARα.
Neddylation of PML/RARα impedes NB assembly by inhibiting the initial nucleation process
To further characterize the role of neddylation of PML/RARα in NB assembly, we first examined the subcellular localization of neddylation-deficient mutants of PML/RARα. The results showed that K227R, K360R, and 2KR all facilitated PML/RARα NB structures (Fig. 4A). Similarly, PML NBs disrupted by PML/RARα were also reconstructed by neddylation-deficient mutation (Fig. S7). Furthermore, K227R, K360R, and 2KR condensates were colocalized with SUMO (Fig. S8) and DAXX (Fig. 4B). Similar results were obtained in PML KO subclones (Fig. S9). Moreover, a marked increase in sumoylated PML/RARα was also observed in neddylation-deficient mutants (Fig. 4C). Taken together, these results establish that deneddylation of PML/RARα promotes functional NB reconstruction.
A The effect of neddylation site mutations on PML/RARα NB formation. H1299 cells were infected with different PML/RARα-HA contrasts (WT, K227R, K360R, and 2KR) lentivirus for 5 days. The average diameter of foci was calculated and the number of large foci (foci size ≥ 200 nm) per 100 cells was counted. ***, p < 0.001, vs. PML/RARα(WT). Statistical significance of differences between groups was determined with one-way ANOVA analysis. 2KR means K227R and K360R in RARα moiety. B Localization of DAXX in reformed PML/RARα NBs when its neddylation sites were mutated. H1299 cells were infected with different PML/RARα-HA contrasts (WT, K227R, K360R, and 2KR) lentivirus for 5 days. C PML/RARα neddylation-defect mutants exhibit obvious sumoylation modification. COS7 cells were transfected with different PML/RARα-HA constructs (WT, K227R, K360R, and 2KR) for 48 h. D Time course of PML/RARα sumoylation modification during MLN4924 treatment. COS7 cells were transfected with PML/RARα-HA, followed by treatment with MLN4924 (1 μM) for the indicated times. E Time course of DAXX recruitment into reformed PML NBs during MLN4924 exposure. H1299 cells were infected with PML/RARα-HA lentivirus, and then were treated with MLN4924 (1 μM) for the indicated times. F Effect of classical sumoylation site mutations on NB reformation and DAXX recruitment induced by neddylation inhibition. H1299 cells were infected with different PML/RARα-HA constructs (WT, NEDD8-2KR, SUMO-3KR, or NEDD8 + SUMO-5KR) lentivirus for 5 days. NEDD8-2KR means K227R and K360R in RARα moiety, SUMO-3KR means K65R, K160R, and K499R in PML moiety, and NEDD8 + SUMO-5KR means above 5 lysine mutations both in PML and RARα.
The formation of PML NBs is a multistep process, that includes nucleation, PML sumoylation, and the recruitment of partners [20, 32, 33]. Thus, we wanted to deeply elucidate the direct step that is disrupted by the neddylation of PML/RARα. Time-course experiments showed that the neddylation of PML/RARα dropped immediately after exposure to MLN4924 for 2 h, while the sumoylation of PML/RARα was slightly elevated during the first 8–12 h and then sharply increased over 24 h (Fig. 4D). Meanwhile, cells began to form small NB structures at 4–6 h, and the structures became larger at 8–12 h (Fig. S10). Furthermore, colocalization of DAXX with reformed NBs was obviously observed until 24 h (Fig. 4E). Therefore, the process is initiated by the loss of PML/RARα-NEDD8 conjugation (2 h), followed by initial renucleation (4–12 h), hypersumoylation (8–12 h), and DAXX colocalization. These results indicated that deneddylation-induced PML/RARα nucleation occurred earlier than sumoylation and DAXX recruitment. We further examined the impact of neddylation on a sumoylation-deficient mutant of PML/RARα (SUMO-3KR), which is considered to hardly affect the formation of NBs but fails to recruit NB-related proteins [32]. As presented in Fig. 4F and S11, SUMO-3KR lost the ability to promote the colocalization of DAXX, as expected, but retained the renucleation ability induced by abolishing neddylation (NEDD8 + SUMO-5KR) or MLN4924 treatment without the accumulation of sumoylation modification. These results suggest that sumoylation modification is dispensable for PML/RARα nucleation, which is consistent with a previous study showing that PML sumoylation does not underlie NB formation [32]. Collectively, we infer that neddylated PML/RARα impedes NB assembly by inhibiting the initial nucleation process.
Neddylation of PML/RARα disrupts the nucleation process by weakening the phase separation ability of the PML moiety
To further gain insight into the mechanism underlying deneddylation-induced PML/RARα renucleation, we carefully analyzed the formation processes of PML/RARα NBs. Interestingly, the PML/RARα microspeckles underwent fusion processes to form larger PML/RARα foci upon MLN4924 treatment (Fig. 5A and Movie S1), which was very similar to the phase separation process. Furthermore, FRAP analysis revealed that the recovery ability of PML NBs was high as assessed by the mobile fraction (Mf = 0.71) and half-life (t1/2 = 3.38 min), and the recovery curve showed an exponential time dependence (Fig. 5B). In contrast, PML/RARα microspeckles recovered less efficiently than PML NBs. Then, MLN4924- or neddylation-deficient mutation-reconstructed NBs exhibited a higher recovery extent as assessed by the mobile fraction (Mf), which was similar to PML NBs but different from ATO-induced aggregates (Fig. 5C and Table S3). Thus, we suspect that PML may induce phase separation to construct PML NBs, while PML/RARα fails to form NBs due to aberrant phase separation, and deneddylation of PML/RARα may recover the phase separation process to reconstruct NBs.
A Time-lapse imaging of the fusion of NBs reformed by MLN4924. H1299 cells were infected with PML/RARα-GFP lentivirus for 3 days, followed by MLN4924 treatment (1 μM). Images were acquired using an ImageXpress Pico scanner (Molecular Devices). The fusion of two PML/RARα droplets is indicated by different circles. B The FRAP recovery curve of the PML-GFP fluorescent intensity. H1299 cells were infected with PML-GFP lentivirus for 3 days. Data are represented as mean ± SD (n = 3). C Recovery rates of reformed NBs after photobleaching. H1299 cells were infected with PML/RARα(WT)-GFP or PML/RARα(2KR)-GFP lentivirus for 3 days. Then PML/RARα(WT)-GFP infected cells were treated with MLN4924 (1 μM) for 24 h or ATO (1 μM) for 12 h. Data are represented as mean ± SD (n = 3). D The saturation concentration (Csat) of PML. Calculation of the apparent phase separated fraction was with (ICondensate)/(ITotal), where ICondensate is the condensate fluorescence intensity, ITotal the fluorescence intensity of the image. E Formation of PML droplets in buffers containing different concentrations of NaCl. F Decay time (τ) vs. length scale (l) for PML droplets prepared in 300 mM NaCl. The linear slope represents the inverse capillary velocity (η/γ). G Plot showing the time course of the recovery after photobleaching PML-GFP droplets. Data are represented as mean ± SD (n = 3). (H, I) In vitro phase separation of PML, PML/RARα and deneddylated-PML/RARα. GFP-His, PML-GFP-His, PML/RARα(WT)-GFP-His, and PML/RARα(2KR)-GFP-His were expressed in COS7 cells for 48 h. PML/RARα(WT)-GFP-His expressed cells were treated with MLN4924 (1 μM) for 48 h. Then all the proteins were purified using 6 × His-affinity beads from cell lysate. Droplet formation was performed under the crowding conditions. Representative fluorescence and DIC images of the droplets (H). Quantification of the size of droplets are shown (I). Data are represented as mean ± SD (n = 300). J The in vitro phase-separated fraction of PML/RARα and deneddylated-PML/RARα at different protein concentrations. Proteins were expressed in COS7 cells for 48 h and purified using 6×His-affinity beads from cell lysates. Droplet formation was performed at room temperature using the nonionic crowder dextran-70 (10%) and 300 mM NaCl. K ChIP analysis of wild-type or mutant PML/RARα binding to the endogenous RARB promoter. 293T cells were transfected with different PML/RARα-HA contrasts (WT, 2KR, ΔDBD, and C88G) for 48 h. Data are represented as mean ± SD (n = 3). L The effect of DNA-binding defective mutation on PML/RARα NB formation. H1299 cells were infected with different PML/RARα-HA contrasts (WT, 2KR, ΔDBD, and C88G) lentivirus for 48 h. The average diameter of foci was calculated and the number of large foci (foci size ≥ 200 nm) per 100 cells was counted. M The FRAP recovery curve of the PML/RARα(ΔDBD)-GFP fluorescent intensity. H1299 cells were infected with PML/RARα(ΔDBD)-GFP lentivirus for 3 days. Data are represented as mean ± SD (n = 3). N The effect of DNA on the in vitro phase separation of PML/RARα. Purified PML/RARα(WT)-GFP-His and PML/RARα(2KR)-GFP-His were used to analysis the droplet formation under the crowding conditions. PGL4.14-RARE plasmid was added to the in vitro system. Representative fluorescence images of the droplets (Left). Quantification of the size of droplets are shown (Right). Data are represented as mean ± SD (n = 300). ***, p < 0.001. vs. indicated (I, K, L, N). The significance analysis was conducted by one-way ANOVA analysis (I, K, L, and N).
Phase separation is presumed to play a pivotal role in the formation of PML NBs [3, 34], but vigorous experimental evidence is lacking. Here, we performed an in vitro phase separation assay. Purified PML-GFP rapidly formed microsized liquid droplets under the nonionic crowder dextran-70 in a protein concentration-dependent manner, and its saturation concentration (Csat) was 0.36 μM (Fig. 5D, S12A). Moreover, droplet formation was sensitive to salt concentration (Fig. 5E, S12B). Meanwhile, PML droplets underwent fusion with an inverse capillary velocity of η/γ ≈ 8.06 s/μm, which is a ratio of the viscosity to surface tension [16, 35], indicating that PML droplets are viscous liquids (Fig. 5F, S12C). In vitro FRAP analysis also indicated that PML-GFP molecules diffused within droplets and exchanged between droplets and the surrounding solution (Fig. 5G). Additionally, the in vitro-formed PML drops were dissolved in 10% 1,6-hexanediol (1,6-HD), an aliphatic alcohol that disrupts weak hydrophobic interactions in phase separation [24, 36, 37] (Fig. S12D). Meanwhile, recombinant DAXX (rDAXX), rather than the SUMO-interacting motif (SIM) mutant of DAXX (ΔSIM), colocalized with PML drops in vitro (Fig. S12E). Overall, these results indicate that PML possesses the ability to promote phase separation to assemble PML NBs.
Next, we detected the phase separation ability of wild-type PML/RARα and deneddylated PML/RARα. Enriched PML/RARα from MLN4924-treated cells or PML/RARα(2KR)-transfected cells were found to be indeed deneddylated (Fig. S13), and deneddylated PML/RARα typically formed liquid droplets under dextran-70, whereas the phase separation of wild-type PML/RARα was weakened considerably at the same protein concentration (Fig. 5H, I). Moreover, under serial protein concentrations, the fraction of phase-separated PML/RARα in the deneddylated groups (P/R + MLN4924 and P/R(2KR)) was significantly higher than that in the P/R(WT) group (Fig. 5J, S14A). Additionally, rDAXX also colocalized with PML/RARα(2KR) lipid droplets (Fig. S14B). Taken together, these data confirm that inhibition of neddylation recovers the phase separation process of PML/RARα and leads to NB reconstruction.
Finally, considering that neddylation may strengthen the protein’s DNA-binding ability [38] and the DNA binding is involved in regulating phase separation [39], we wanted to investigate the role of DNA binding in recovered phase separation by deneddylation. First, in CHIP analysis, similar to DNA-binding defective mutants (PML/RARα(ΔDBD) and PML/RARα(C88G)) [40], neddylation-deficient PML/RARα (2KR) almost completely lost its ability to bind the RARB promoter region (Fig. 5K). Meanwhile, we overexpressed wild-type PML/RARα(WT) or neddylation-deficient PML/RARα(2KR) in mouse hematopoietic stem/progenitor cells (mHSPCs) (Fig. S15A). The results showed that the mRNA levels of RARα target genes (such as Tgm2 and Rarb) were actually diminished in the PML/RARα(WT)-transformed cells, whereas this was moderately reversed in PML/RARα(2KR)-transformed mHSPCs (Fig. S15B). Similar upregulation of RARα target genes could be also found upon MLN4924 treatment in NB4 cells (Fig. S15C). These results suggest that deneddylation interferes with the DNA-binding ability of PML/RARα and reverses the suppression of RARα signaling. Based on this, we detected localization of these two DNA-binding-defective PML/RARα mutants, PML/RARα(ΔDBD) and PML/RARα(C88G). As shown in Fig. 5L, both mutants showed NB-like structures, whose foci diameter and large foci number were similar to those of PML/RARα(2KR). Meanwhile, the FRAP analysis also indicated the high recovery ability of PML/RARα(ΔDBD), which was similar to that of PML/RARα(2KR) (Fig. 5M). More importantly, we found that compared to PML/RARα (2KR), RARE-containing DNA induced a stronger inhibitory effect on droplet formation of PML/RARα (WT) in vitro (Fig. 5N). Altogether, these results suggest that inhibition of DNA binding underlies the mechanism by which deneddylation promotes the phase separation of PML/RARα.
Neddylation is critical for PML/RARα-driven leukemogenesis
PML/RARα is a crucial driving factor of APL pathogenesis. To explore the role of PML/RARα neddylation during APL pathogenesis, we overexpressed wild-type PML/RARα (WT) or neddylation-deficient PML/RARα (2KR) in mHSPCs and evaluated their oncogenic ability both in vitro and in vivo (Fig. 6A). First, we confirmed the NB reconstruction of PML/RARα (2KR) in mHSPCs (Fig. 6B). Moreover, PML/RARα (WT)-expressing cells could be indefinitely replated in methylcellulose as expected, while PML/RARα (2KR)-expressing cells failed (Fig. 6C), demonstrating that inhibiting neddylation of PML/RARα destroyed its ability to immortalize primary hematopoietic stem/progenitor cells. Additionally, PML/RARα (WT)-transformed cells, rather than 2KR-expressing cells, grew faster than cells transduced with blank pMSCV in liquid culture (Fig. S16A). More differentiation-blocked promyelocytes were observed in cells expressing PML/RARα (WT), rather than PML/RARα (2KR) (Fig. S16B). Furthermore, an in vivo leukemogenesis assay showed that compared with the 2KR-overexpressing mHSPC group, apparent weight loss was only observed in mice injected with WT-overexpressing mHSPCs (Fig. 6D). Finally, overall survival was significantly extended in mice transplanted with 2KR-overexpressing mHSPCs compared with WT-overexpressing mHSPCs (Fig. 6E). Therefore, these data indicate that the deneddylation of PML/RARα abolishes its oncogenic transformation ability and APL leukemogenesis.
A Schematic generation of PML/RARα driven APL cells and evaluation of the self-renewal capacity of PML/RARα transformed mHSPCs both in vitro and in vivo. B Foci structure of PML/RARα in mHSPCs after transduction. Cells were immunostained with anti-HA antibody, followed by confocal imaging. The average diameter of foci was calculated and the number of large foci (foci size ≥ 200 nm) per 100 cells was counted. 2KR means K227R and K360R in RARα moiety. C Replating activity of mHSPCs in methylcellulose medium. mHSPCs were infected with pMSCV, PML/RARα(WT), or PML/RARα(2KR) lentivirus and then were cultured in methylcellulose medium. The colony number from the first to the fifth round of colonies is indicated. Data are represented as mean ± SD (n = 3). D, E Leukemogenesis of PML/RARα(WT) and neddylation-deficient PML/RARα (2KR) in vivo. mHSPCs transduced with PML/RARα (WT) or PML/RARα (2KR) lentivirus were injected via the tail vein into NSG mice. The body weight and survival times of NSG mice were recorded. Data are represented as mean ± SD (n = 6). F, G RNA-Seq analysis of mHSPCs infected with PML/RARα (WT) or PML/RARα (2KR) lentivirus. F Differentially expressed genes between PML/RARα (WT) and PML/RARα (2KR). G Top 15 clusters (including≥10 differentially expressed genes) from KEGG pathway enrichment analysis of the changed genes. H Heatmap display of 18 differentially expressed genes involved in pathways regulating the pluripotency of stem cells. I Expression levels of three stem cell markers, SCA-1, CD117, and CD34 in mHSPCs transduced with PML/RARα (WT) or PML/RARα (2KR). Data are represented as mean ± SD (n = 3). *, p < 0.05; **, p < 0.01; ***, p < 0.001, vs. PML/RARα (WT). Statistical significance of differences was determined with two-tailed unpaired Student’s t-test (B–D, I) or Log-rank test (E).
To genetically gain more genetic insight into the reduced clonogenicity caused by PML/RARα neddylation, we performed RNA-Seq analysis. In total, 1380 genes (558 upregulated and 822 downregulated) were markedly changed in the 2KR group compared with the WT group (Fig. 6F and Table S4). The top 15 enriched pathways were listed from Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis, and 18 genes involved in regulating the pluripotency of stem cells were dramatically enriched (Fig. 6G). Furthermore, 16 of the 18 genes (88.9%) were downregulated in the 2KR group (Fig. 6H), indicating that the stemness of 2KR-overexpressing mHSPCs might be lower than that of WT-overexpressing mHSPCs. Consistent with these results, the expression levels of early progenitors and stem cell markers (SCA-1, CD117, and CD34) were significantly downregulated in 2KR-transduced mHSPCs compared with WT-expressing mHSPCs (Fig. 6I). Thus, these results indicate that deneddylation of PML/RARα suppresses stemness pathways to reduce the self-renewal ability of APL cells. Together, these data strongly demonstrate that the neddylation of PML/RARα is critical to its transforming potential and that inhibiting the neddylation of PML/RARα obviously suppresses APL leukemogenesis.
Inhibiting the neddylation of PML/RARα leads to APL eradication
Finally, to assess whether the deneddylation of PML/RARα induced by MLN4924 was beneficial for APL treatment, we first detected the impact of MLN4924 on APL cell proliferation and cell viability. We found that MLN4924 effectively inhibited the proliferation and viability of NB4 cells (Fig. 7A) and induced cell apoptosis in a time-dependent manner (Fig. 7B). Notably, MLN4924 also obviously induced apoptosis in primary APL blasts (Fig. 7C, S17). Interestingly, cell apoptosis was selectively induced by MLN4924 in APL cells (NB4 and drug-resistant NB4R1 and NB4R2) but not in non-APL cells (HL60 and OCI-AML2) (Fig. 7D). Next, we further detected the effect of MLN4924 on the clone formation ability of APL cells. From the clonogenic activity in methylcellulose, we found that in contrast to normal mHSPCs, PML/RARα-expressing mHSPCs were more sensitive to MLN4924 than another oncogenic driver (AML1/ETO)-transduced cells (Fig. 7E). Meanwhile, MLN4924 also selectively destroyed the clone formation ability of APL cells (NB4, NB4R1, and NB4R2). In contrast, MLN4924 at the same concentration did not influence the clone formation activity of non-APL AML cells (HL60 and OCI-AML2) (Fig. 7F). Additionally, we determined the effect of the neddylation inhibitor on cell differentiation. MLN4924 partially induced the differentiation of NB4 cells, as assessed by increased expression of CD11b (Fig. 7G) and enhanced NBT reduction activity (Fig. S18). In conclusion, these results together indicate that neddylation inhibitors can selectively eradicate PML/RARα-positive APL cells in vitro.
A The effect of MLN4924 on the proliferation and viability of NB4 cells. NB4 cells were treated with MLN4924 for indicated times, followed by trypan blue exclusion test. Data are presented as mean ± SD (n = 3). B The effect of MLN4924 on the apoptosis of NB4 cells. NB4 cells were treated with MLN4924 (40 nM) for indicated times, and the apoptosis-inducing ability of MLN4924 was confirmed by PI/Annexin V assay. C The effect of MLN4924 on the apoptosis of primary APL blast cells. Primary APL blast cells were directly separated from the bone marrow of patients using lymphocyte monocytes separation medium. Then the apoptosis rate of cells was detected after treated with MLN4924 (2 μM) for 5 days. D The apoptosis rate induced by MLN4924 in APL and non-APL cells. NB4, NB4R1, NB4R2, HL60, and OCI-AML2 cells were treated with MLN4924 (40 nM) for 5 days. E The effect of MLN4924 on the clonogenic activity of PML/RARα in methylcellulose. mHSPCs were infected with pMSCV, PML/RARα or AML1/ETO, and then were treated with MLN4924 (0, 40, 80, 160 and 320 nM) for 24 h. Treated cells were finally cultured in methylcellulose medium for 2 weeks. F The effect of MLN4924 on clone formation of APL and non-APL cells. NB4, NB4R1, NB4R2, HL60, and OCI-AML2 cells were treated with MLN4924 (40 nM) for 2 weeks. The clone inhibition rate was determined by calculating as 100× (clone numbers of the treated group)/ (clone numbers of the control group). G The differentiation-inducing ability of MLN4924 in NB4 cells. NB4 cells were treated with MLN4924 (40 nM) for indicated times or ATRA (40 nM) for 1 day, and then CD11b expression was evaluated. (H–L) The effect of MLN4924 on the tumor burden of NOD/SCID mice. NB4 cells were intravenously transplanted into NOD/SCID mice and then mice were treated with MLN4924 (30 mg/kg and 60 mg/kg) (i.p.). H The schematic diagram of animal experiments. I, J The APL burden detection in bone marrow of the sacrificed mice. When the control leukemic mice were moribund, we sacrificed three mice in each group to detect the APL burden in bone marrow. Cell morphological analysis of bone marrow cells were obtained from NOD/SCID mice and subjected to Wright–Giemsa staining (I). The population of human CD45-positive and mouse CD45-negative (hCD45+mCD45−) leukemia cells in the bone marrow of NOD/SCID mice were detected (J). The effect of MLN4924 on the body weight of APL-burden NOD/SCID mice. Data are presented as mean ± SE (n = 7). (L) The effect of MLN4924 on the survival times of APL-burden NOD/SCID mice. n.s., p > 0.05; *, p < 0.05; **, p < 0.01; ***, p < 0.001. vs. Control (A–D, F, G, and K, L) or vs. pMSCV (E). The significance analysis was conducted by one-way ANOVA analysis (A, E, K), two-tailed unpaired Student’s t-test (B-D, F-G), or Log-rank test (L).
Thus, we evaluated the in vivo antitumor activity of MLN4924 in an APL xenograft mouse model (Fig. 7H). First, the reduction in leukemic blasts in bone marrow after MLN4924 treatment was confirmed by Wright-Giemsa staining (Fig. 7I). In addition, the proportion of hCD45+mCD45- cells, which reflected the overall leukemic burden, was significantly increased in the control group but not in the MLN4924 group (Fig. 7J). Furthermore, compared with control leukemic mice, MLN4924 completely reversed the APL burden-induced body weight decline (Fig. 7K) and significantly prolonged the survival of APL leukemic mice (Fig. 7L). Collectively, these results demonstrate that MLN4924 represses the invasion of leukemic blasts in APL mice, further supporting the therapeutic potential of neddylation inhibitors in APL patients.
Discussion
PML/RARα is recognized as a major oncogenic driver of APL [41]. Here, we reveal that the carcinogenicity of PML/RARα is dependent on its neddylation. A subsequent study indicated that PML/RARα is neddylated in the RARα region. Interestingly, we provide the experimental evidence for the first time to confirm that PML NB assembly is a process of phase separation. In contrast, neddylation of PML/RARα in the RARα region disrupts the phase separation of its PML moiety by enhancing its DNA-binding ability, which further destroys the construction of PML NBs. Finally, deneddylation of PML/RARα reconstructs functional PML NBs, reactivates RARα signaling, and leads to APL eradication, suggesting deneddylation of PML/RARα as a novel therapeutic strategy against APL. Overall, the identification of PML/RARα neddylation provides a better understanding of the fundamental mechanism responsible for the disruption of the phase separation of PML NBs by PML/RARα in APL pathogenesis, but also potentially provides a rationale for targeting neddylation for APL treatment.
PML/RARα has been reported to be modified by phosphorylation [42], sumoylation [43], ubiquitination [44, 45], and ISG15ylation [46], but this is the first time that neddylation has been found. In addition to Cullin-RING ubiquitin ligases, a small number of non-Cullin targets of NEDD8 have been identified, including several transcription factors (such as p53, TAp73, E2F1, SREBP1c, IRF7, and JunB). Neddylation regulates their transcriptional activity by altering protein subcellular localization, protein-protein, and protein-DNA interactions, and protein stability [7, 47,48,49]. Here, we find that neddylation controls the transcriptional suppressive activity of PML/RARα by enhancing its DNA-binding ability. For the mechanisms underlying the DNA-binding regulation of PML/RARα by neddylation, given that neddylation of Cullin-RING ligases can induce conformational rearrangement [50], we propose that neddylation may influence the protein conformation of PML/RARα to further regulate its DNA binding ability.
Phase separation increasingly appears to be a fundamental mechanism for organizing intracellular space [51]. We provide experimental evidence for the first time to confirm phase separation as an important mechanism to organize PML NBs as an intracellular space to suppress cancer pathogenesis. PML/RARα also dramatically loses its phase separation ability and fails to assemble NBs, followed by APL initiation. Our work provides a distinct example of phase separation being necessary for normal cellular function, with the loss of phase separation leading to disease phenotypes. Additionally, despite increasing interest in the study of phase separation, the development of phase separation modulators remains in its infancy [52]. Here, we show the feasibility of finding phase separation modulators using a high-throughput screening model in vivo and provide an example for phase separation reconstruction by altering the posttranslational modification of scaffold proteins.
Phosphorylation [53], acetylation [54], methylation [55, 56] and ubiquitination [57, 58] have been found to regulate phase separation. And this study is the first to connect neddylation with phase separation. Moreover, loss of DNA-binding ability was found to be involved in the deneddylation-enhanced phase separation of PML/RARα in this study. This is similar to the recent observation that DNA binding regulates the formation of phase-separated droplets, such as HP1α [59], cGAS [37], and VRN1 [60]. Obviously, it is worth further dissecting the role and mechanism of DNA-binding ability in the neddylation-regulated phase separation of PML/RARα.
Both transcriptional repression of RARα signaling and PML NB disruption by PML/RARα are important for APL leukemogenesis [1]. Here, the reactivation of RARα target genes upon neddylation inhibition suggests that deneddylation of PML/RARα reverses the transcriptional repression of RARα signaling. In addition to canonical RAREs, PML/RARα is also found to bind noncanonical RAREs to regulate the expression of genes that are essential for APL pathogenesis [61]; thus, loss of the DNA-binding ability of PML/RARα by deneddylation may be beneficial for APL suppression. The disruption of PML NBs by PML/RARα, which blunts p53 signaling [5] and impairs DNA damage repair [6], is believed to contribute to APL pathogenesis. In MLN4924-reformed PML/RARα NBs, we also enriched posttranslational modification regulators that activate p53 and DNA damage repair-related proteins. Meanwhile, new partners were also identified in our study. Additional studies are needed to further clarify their contributions to APL pathogenesis and therapy response.
MLN4924, as a NEDD8-activating enzyme (NAE) inhibitor, has been reported to exhibit promising therapeutic activity in various malignancies [62,63,64]. Considering the possible effects of MLN4924 on other neddylation substrates, such as Cullin-RING ubiquitin ligases (CRLs), we evaluated the role of PML/RARα in the inhibitory effect of MLN4924 on APL cells. Our results showed that MLN4924 selectively eradicated PML/RARα-positive APL cells rather than PML/RARα-negative cells in vitro (Fig. 7D–F), indicating that this eradication may mainly result from the neddylation inhibition of PML/RARα. However, it is undeniable that other neddylation substrates are possibly influenced when MLN4924 is exposed to APL cells, which may partly contribute to the relatively imperfect survival benefit in vivo (Fig. 7L). Therefore, further investigation of the specific E3 ligases involved in the neddylation of PML/RARα by applying systems, such as the yeast 2-hybrid screen, may facilitate the design of a more specific inhibitor.
In summary, we demonstrate for the first time that failed assembly of PML NBs in APL results from aberrant phase separation of neddylated PML/RARα, representing a conceptual leap towards understanding how PML NBs are disrupted in APL. Furthermore, we discover a therapeutic strategy for APL by targeting neddylation to reform functional PML NBs and reactivate RARα signaling. Collectively, this study offers important insight into PML/RARα-driven APL pathogenesis and treatment.
Materials and methods
Human patient blasts, cells, and cell culture
Primary patient blasts (APL-1~10) from bone marrow of patients (The First Affiliated Hospital or Children’s Hospital of Zhejiang University) were isolated using lymphocyte monocyte separation medium (GE Healthcare).
H1299, NB4, NB4R1, and NB4R2 cells were cultured in RPMI 1640 medium, and 293FT, 293T, and COS7 cells were cultured in DMEM, supplemented with 10% fetal bovine serum (Gibco BRL) and 0.1% penicillin/streptomycin. All the cell lines were routinely tested for mycoplasma using a Mycoplasma Detection Kit (Bimake, Houston, TX, USA) and were authenticated utilizing short tandem repeat (STR) profiling every 6 months. The source of all cell lines is supplied in Table S5.
Plasmids and reagents
The coding sequences were subcloned into the pCDNA3.0, pCDH, pCDH-GFP and/or pCDH-MSCV-GFP (pMSCV) plasmids. The packaging plasmid pRΔ8.9 and envelope plasmid pMD.G-VSVG were kindly provided by Dr. D.B. Kohn (University of Southern California). The full-length coding sequences of PML and RARα were amplified from the cDNA of the cell lines, the sequence for PML/RARα was synthesized, the cDNAs of Ubc12 and NEDP1 were purchased from Vigene Biosciences (Jinan, China). The shRNA oligonucleotides were cloned into pLKO.1. The sequences of shRNA are supplied in Table S6. The specific antibodies used for western blotting, immunofluorescence, and flow cytometry are listed in Table S7. The information of all used buffers is supplied in Table S8.
Target selective inhibitor library (299 compounds) was purchased from Selleck (Houston, USA). MLN4924 was supplied by Cayman Chemical Company (Ann Arbor, Michigan). ATO, 1,6-hexanediol, and dextran-70 were purchased from Sigma-Aldrich (St. Louis, MO).
Lentivirus production, concentration and transduction
Virus production, titration and transduction were performed as described previously [65].
High-content screening
H1299 cells were infected with PML/RARα-GFP lentivirus and seeded into 96-well microplates (Corning). Cells were automatically treated with target selective inhibitor library using a Tecan Fluent 780 liquid handling workstation (Tecan, Männedorf, Switzerland). After culturing for 12 h, cells were fixed with 4% paraformaldehyde followed by DAPI staining. Next, high-throughput acquisition of GFP and DAPI signals from each cell was performed using an ImageXpress Pico scanner (Molecular Devices). Finally, the number of large foci (foci size ≥ 200 nm) was counted using the ImageXpress Pico, and the average diameter of foci was calculated using ImageJ.
Solubility analysis of NBs
The solubility of NBs was determined according to the literature [18]. H1299 cells were infected with pCDH-PML/RARα-GFP lentivirus for 5 days and treated with MLN4924 or ATO for 12 h. The cells were subsequently lysed in 1% NP40 buffer or RIPA buffer. Cell lysates were smeared onto glass slides, and directly observed under fluorescence microscope. Additionally, RIPA cell lysates were centrifuged to separate the soluble and insoluble fractions, followed by western blot analysis.
Identification of recruited proteins in NBs
BioID technique was adopted to identify the recruited proteins by fusing PML/RARα or PML with humanized BirA (R118G), known as BirA* [24]. The infected cells were incubated with biotin before harvest and lysed in lysis buffer B to remove cytoplasmic proteins. Next, the pellets were lysed in 4% SDS buffer and incubated with anti-streptavidin beads. Finally, the beads were washed with 1% Briji buffer. Then, the samples were analyzed by mass spectrometry at Shanghai Biotechnology Corporation. The mass spectrometry data were analyzed using MaxQuant software version 1.5.3.17 (Max Planck Institute of Biochemistry in Martinsried, Germany), and two indicators were obtained in the raw data, Peptide intensity, and Score. Then we calculated the peptide intensity ratio (vs. NLS) of each group and selected the proteins whose peptide intensity ratio (vs. NLS) ≥ 2. Finally, we ordered the overlay proteins based on Score.
Generation of PML KO cells by CRISPR/Cas9
The generation assay of PML KO cells mediated by CRISPR/Cas9 was performed as previously described [24]. The sgRNA oligonucleotide sequences and primers for PML fragment’s PCR are supplied in Tables S9 and S10.
Immunoprecipitation
To detect the neddylation and sumoylation of PML/RARα, the cells were lysed in 4% SDS buffer. Next, the cell lysates were diluted 10 times with RIPA lysis buffer to the SDS concentration of 0.4%, and incubated with anti-HA beads. Finally, the beads were washed with 1% NP40 buffer before being boiled with loading buffer.
Fluorescence recovery after photobleaching (FRAP)
FRAP experiments were performed using an Olympus FV3000 confocal with microscope a 100× oil objective. A 488 nm laser was used to photobleach. Time-lapse images were recorded with a 2.4 s interval for a total of 250 images. The mobile fraction (Mf) and half-life (t1/2) were measured according to the literature [66].
Protein expression and purification
For eukaryotic purification, COS7 cells were transfected with indicated plasmids. The cells were lysed in RIPA lysis buffer. Protein was purified using 6×His-affinity beads. Next, the protein was exchanged into exchange buffer using Zeba™ Spin Desalting Columns.
For prokaryotic purification, plasmids were transformed into E. coli cells. The bacteria were lysed using a high-pressure bacteria breaker (Union Biotech). Protein was purified using a Ni-NTA or GST column (GE Healthcare). The eluted protein was dialysed in dialysed buffer.
In vitro phase separation assays
The purified protein and the crowding agent were mixed and deposited in coverslip-bottom, non-binding 384-well plates (Cellvis) at room temperature. Images of the phase separation of protein were acquired using a Leica TCS SP8 confocal microscope. The determination of saturation concentration (Csat) was according to literature [67].
Fusion kinetics
Droplet formation was recorded on an Olympus FV3000 confocal with microscope a 100× oil objective. Fusion events were manually spotted, cropped out, and analyzed in ImageJ. The inverse capillary velocity η/γ was measured according to the literature [35, 68].
ChIP assay
Cells were cross-linked with 1% formaldehyde and stopped by adding glycine. Harvested cells were resuspended in lysis buffer B and centrifuged to remove cytoplasmic proteins. Then the nuclei pellet was resuspended in 1% SDS buffer, and sonicated to shear DNA using the Bioruptor (Diagenode). The supernatant was incubated with HA beads. Then the beads were washed with low salt buffer, high salt buffer, LiCl buffer, and TE buffer, and finally eluted with elution buffer. Cross-linking was reversed with the addition of NaCl (0.2 M) and Proteinase K for 2 h at 42 °C and 8–10 h at 65 °C. DNA was extracted and analyzed by RT-PCR for genomic sequence from the RARB promoter region containing RARE using PCR primers in Table S10.
Mouse hematopoietic stem/progenitor cell (mHSPC) separation, infection, and clonogenic assay
Bone marrow cells were first obtained from the femurs of 5-fluorouracil-treated C57BL/6 mice, and then mHSPCs were selected using the Easysep Mouse Hematopoietic Progenitor Cell Isolation Kit (Stemcell Technologies). The detailed culture condition is supplied in supplemental Methods. Infection was obtained by centrifugation of the cells in viral supernatant at 1800 rpm for 40 min. The transduced mHSPCs were plated in methylcellulose medium (M3434; StemCell Technologies) for the clonogenic assay.
mHSPCs were cultured in IMDM medium supplemented with recombinant mouse SCF (50 ng/ml; Peprotech), recombinant mouse IL-3 (10 ng/ml; Peprotech), recombinant mouse IL-6 (10 ng/ml; Peprotech), 1% bovine serum albumin (Yeasen Biotech Co., Ltd.), hydrocortisone (1 μM; Sigma-Aldrich), 2-mercaptoethanol (100 μM; Sigma-Aldrich), L-glutamine (2 mM), 20% fetal bovine serum (Gibco BRL) and 1% penicillin/streptomycin.
RNA-seq analysis
RNA-seq analysis was performed at Shanghai Biotechnology Corporation as described previously [69]. Raw data for RNA-sequencing are deposited at the NCBI GEO (GSE165046).
Animal studies
1 × 107 transduced mHSPCs were injected into 4~5-week-old female NSG mice (Shanghai Model Organisms Center, Inc.) via the tail vein. There were 6 mice in each group. 1 × 107 NB4 cells were injected into 6–7-week-old female NOD/SCID mice (SLRC Laboratory Animal Inc.) and then mice were treated with MLN4924 (30 mg/kg and 60 mg/kg) (i.p.). There were 7 mice in each group.
Statistical analysis
Cell-based experiments as well as in vitro phase separation experiments were performed at least three biological replicates with three technical replicates. For animal study, the number of mice used was 6-7. The number of each experiment used for analyses is indicated in each figure legend. For all the parameters measured, the values for all samples under different experimental conditions were averaged, and the SD or SE was calculated. Statistical significance of differences between groups was determined by unpaired two-tailed Student’s t-test analysis, Tukey tests as part of one-way ANOVA, or Log-rank test. Statistical test used for a particular experiment is mentioned in figure legends.
Data availability
Raw data for RNA-sequencing are deposited at the NCBI Gene Expression Omnibus (GSE165046). Other data is available in the main text or the supplementary materials. For more additional data related to this subject, please contact the corresponding author.
References
de The H, Pandolfi PP, Chen Z. Acute promyelocytic leukemia: a paradigm for oncoprotein-targeted cure. Cancer Cell. 2017;32:552–60.
Lallemand-Breitenbach V, de The H. PML nuclear bodies: from architecture to function. Curr Opin Cell Biol. 2018;52:154–61.
Li Y, Ma X, Wu W, Chen Z, Meng G. PML nuclear body biogenesis, carcinogenesis, and targeted therapy. Trends Cancer. 2020;6:889–906.
Niwa-Kawakita M, Ferhi O, Soilihi H, Le Bras M, Lallemand-Breitenbach V, de The H. PML is a ROS sensor activating p53 upon oxidative stress. J Exp Med. 2017;214:3197–206.
Ablain J, Rice K, Soilihi H, de Reynies A, Minucci S, de The H. Activation of a promyelocytic leukemia-tumor protein 53 axis underlies acute promyelocytic leukemia cure. Nat Med. 2014;20:167–74.
Voisset E, Moravcsik E, Stratford EW, Jaye A, Palgrave CJ, Hills RK, et al. Pml nuclear body disruption cooperates in APL pathogenesis and impairs DNA damage repair pathways in mice. Blood. 2018;131:636–48.
Enchev RI, Schulman BA, Peter M. Protein neddylation: beyond cullin-RING ligases. Nat Rev Mol Cell Biol. 2015;16:30–44.
Zhou L, Zhang W, Sun Y, Jia L. Protein neddylation and its alterations in human cancers for targeted therapy. Cell Signal. 2018;44:92–102.
Boeynaems S, Alberti S, Fawzi NL, Mittag T, Polymenidou M, Rousseau F, et al. Protein phase separation: a new phase in cell biology. Trends Cell Biol. 2018;28:420–35.
Shin Y, Brangwynne CP. Liquid phase condensation in cell physiology and disease. Science 2017;357:aaf4382.
Feric M, Vaidya N, Harmon TS, Mitrea DM, Zhu L, Richardson TM, et al. Coexisting liquid phases underlie nucleolar subcompartments. Cell. 2016;165:1686–97.
Riback JA, Zhu L, Ferrolino MC, Tolbert M, Mitrea DM, Sanders DW, et al. Composition-dependent thermodynamics of intracellular phase separation. Nature. 2020;581:209–14.
Pak CW, Kosno M, Holehouse AS, Padrick SB, Mittal A, Ali R, et al. Sequence determinants of intracellular phase separation by complex coacervation of a disordered protein. Mol Cell. 2016;63:72–85.
Bernardi R, Pandolfi PP. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol. 2007;8:1006–16.
Weidtkamp-Peters S, Lenser T, Negorev D, Gerstner N, Hofmann TG, Schwanitz G, et al. Dynamics of component exchange at PML nuclear bodies. J Cell Sci. 2008;121:2731–43.
Alberti S, Gladfelter A, Mittag T. Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell. 2019;176:419–34.
Soding J, Zwicker D, Sohrabi-Jahromi S, Boehning M, Kirschbaum J. Mechanisms for active regulation of biomolecular condensates. Trends Cell Biol. 2020;30:4–14.
Zhang XW, Yan XJ, Zhou ZR, Yang FF, Wu ZY, Sun HB, et al. Arsenic trioxide controls the fate of the PML-RARalpha oncoprotein by directly binding PML. Science. 2010;328:240–3.
Jeanne M, Lallemand-Breitenbach V, Ferhi O, Koken M, Le Bras M, Duffort S, et al. PML/RARA oxidation and arsenic binding initiate the antileukemia response of As2O3. Cancer cell. 2010;18:88–98.
Lallemand-Breitenbach V, de The H. PML nuclear bodies. Cold Spring Harb Perspect Biol. 2010;2:a000661.
Noguera NI, Catalano G, Banella C, Divona M, Faraoni I, Ottone T, et al. Acute promyelocytic leukemia: update on the mechanisms of leukemogenesis, resistance and on innovative treatment strategies. Cancers 2019;11:1591.
Ishov AM, Sotnikov AG, Negorev D, Vladimirova OV, Neff N, Kamitani T, et al. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J Cell Biol. 1999;147:221–34.
Roux KJ, Kim DI, Raida M, Burke B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol. 2012;196:801–10.
Cao J, Chen X, Jiang L, Lu B, Yuan M, Zhu D, et al. DJ-1 suppresses ferroptosis through preserving the activity of S-adenosyl homocysteine hydrolase. Nat Commun. 2020;11:1251.
Guan D, Kao HY. The function, regulation and therapeutic implications of the tumor suppressor protein, PML. Cell Biosci. 2015;5:60.
Mimura Y, Takahashi K, Kawata K, Akazawa T, Inoue N. Two-step colocalization of MORC3 with PML nuclear bodies. J Cell Sci. 2010;123:2014–24.
Hsu KS, Kao HY. PML: Regulation and multifaceted function beyond tumor suppression. Cell Biosci. 2018;8:18.
McManus FP, Bourdeau V, Acevedo M, Lopes-Paciencia S, Mignacca L, Lamoliatte F, et al. Quantitative SUMO proteomics reveals the modulation of several PML nuclear body associated proteins and an anti-senescence function of UBC9. Sci Rep. 2018;8:7754.
Xu GL, Pan YK, Wang BY, Huang L, Tian L, Xue JL, et al. TTRAP is a novel PML nuclear bodies-associated protein. Biochem Biophys Res Commun. 2008;375:395–8.
Rasheed ZA, Saleem A, Ravee Y, Pandolfi PP, Rubin EH. The topoisomerase I-binding RING protein, topors, is associated with promyelocytic leukemia nuclear bodies. Exp Cell Res. 2002;277:152–60.
Xu A, Zhang N, Cao J, Zhu H, Yang B, He Q, et al. Post-translational modification of retinoic acid receptor alpha and its roles in tumor cell differentiation. Biochem Pharm. 2020;171:113696.
Sahin U, Ferhi O, Jeanne M, Benhenda S, Berthier C, Jollivet F, et al. Oxidative stress-induced assembly of PML nuclear bodies controls sumoylation of partner proteins. J Cell Biol. 2014;204:931–45.
Rabellino A, Carter B, Konstantinidou G, Wu SY, Rimessi A, Byers LA, et al. The SUMO E3-ligase PIAS1 regulates the tumor suppressor PML and its oncogenic counterpart PML-RARA. Cancer Res. 2012;72:2275–84.
Corpet A, Kleijwegt C, Roubille S, Juillard F, Jacquet K, Texier P, et al. PML nuclear bodies and chromatin dynamics: catch me if you can! Nucleic Acids Res. 2020;48:11890–912.
Zhang H, Elbaum-Garfinkle S, Langdon EM, Taylor N, Occhipinti P, Bridges AA, et al. RNA controls PolyQ protein phase transitions. Mol Cell. 2015;60:220–30.
Chong S, Dugast-Darzacq C, Liu Z, Dong P, Dailey GM, Cattoglio C, et al. Imaging dynamic and selective low-complexity ___domain interactions that control gene transcription. Science 2018;361:eaar2555.
Wegmann S, Eftekharzadeh B, Tepper K, Zoltowska KM, Bennett RE, Dujardin S, et al. Tau protein liquid-liquid phase separation can initiate tau aggregation. EMBO J 2018;37:e98049.
Li C, Zhang L, Qian D, Cheng M, Hu H, Hong Z, et al. RNF111-facilitated neddylation potentiates cGAS-mediated antiviral innate immune response. PLoS Pathog. 2021;17:e1009401.
Feric M, Misteli T. Phase separation in genome organization across evolution. Trends Cell Biol. 2021;31:671–85.
Durand B, Saunders M, Gaudon C, Roy B, Losson R, Chambon P. Activation function 2 (AF-2) of retinoic acid receptor and 9-cis retinoic acid receptor: presence of a conserved autonomous constitutive activating ___domain and influence of the nature of the response element on AF-2 activity. EMBO J. 1994;13:5370–82.
Nasr R, Lallemand-Breitenbach V, Zhu J, Guillemin MC, de The H. Therapy-induced PML/RARA proteolysis and acute promyelocytic leukemia cure. Clin Cancer Res: Off J Am Assoc Cancer Res. 2009;15:6321–6.
Shima Y, Honma Y, Kitabayashi I. PML-RARalpha and its phosphorylation regulate PML oligomerization and HIPK2 stability. Cancer Res. 2013;73:4278–88.
Zhu J, Zhou J, Peres L, Riaucoux F, Honore N, Kogan S, et al. A sumoylation site in PML/RARA is essential for leukemic transformation. Cancer Cell. 2005;7:143–53.
Lallemand-Breitenbach V, Jeanne M, Benhenda S, Nasr R, Lei M, Peres L, et al. Arsenic degrades PML or PML-RARalpha through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat Cell Biol. 2008;10:547–55.
Tatham MH, Geoffroy MC, Shen L, Plechanovova A, Hattersley N, Jaffray EG, et al. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat Cell Biol. 2008;10:538–46.
Shah SJ, Blumen S, Pitha-Rowe I, Kitareewan S, Freemantle SJ, Feng Q, et al. UBE1L represses PML/RAR{alpha} by targeting the PML ___domain for ISG15ylation. Mol Cancer Ther. 2008;7:905–14.
Ju UI, Jeong DW, Seo J, Park JB, Park JW, Suh KS, et al. Neddylation of sterol regulatory element-binding protein 1c is a potential therapeutic target for nonalcoholic fatty liver treatment. Cell Death Dis. 2020;11:283.
Zhao M, Zhang Y, Yang X, Jin J, Shen Z, Feng X, et al. Myeloid neddylation targets IRF7 and promotes host innate immunity against RNA viruses. PLoS Pathog. 2021;17:e1009901.
Li H, Zhu H, Liu Y, He F, Xie P, Zhang L. Itch promotes the neddylation of JunB and regulates JunB-dependent transcription. Cell Signal. 2016;28:1186–95.
Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, Schulman BA. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell. 2008;134:995–1006.
Bergeron-Sandoval LP, Safaee N, Michnick SW. Mechanisms and consequences of macromolecular phase separation. Cell. 2016;165:1067–79.
Mullard A. Biomolecular condensates pique drug discovery curiosity. Nat Rev Drug Discov. 2019;18:324–6.
Kim TH, Tsang B, Vernon RM, Sonenberg N, Kay LE, Forman-Kay JD. Phospho-dependent phase separation of FMRP and CAPRIN1 recapitulates regulation of translation and deadenylation. Science. 2019;365:825–9.
Saito M, Hess D, Eglinger J, Fritsch AW, Kreysing M, Weinert BT, et al. Acetylation of intrinsically disordered regions regulates phase separation. Nat Chem Biol. 2019;15:51–61.
Hofweber M, Hutten S, Bourgeois B, Spreitzer E, Niedner-Boblenz A, Schifferer M, et al. Phase separation of FUS Is suppressed by its nuclear import receptor and arginine methylation. Cell. 2018;173:706–19. e713
Qamar S, Wang G, Randle SJ, Ruggeri FS, Varela JA, Lin JQ, et al. FUS phase separation is modulated by a molecular chaperone and methylation of arginine cation-pi interactions. Cell. 2018;173:720–34. e715
Dao TP, Kolaitis RM, Kim HJ, O’Donovan K, Martyniak B, Colicino E, et al. Ubiquitin modulates liquid-liquid phase separation of UBQLN2 via disruption of multivalent interactions. Mol Cell. 2018;69:965–78. e966
Sun D, Wu R, Zheng J, Li P, Yu L. Polyubiquitin chain-induced p62 phase separation drives autophagic cargo segregation. Cell Res. 2018;28:405–15.
Larson AG, Elnatan D, Keenen MM, Trnka MJ, Johnston JB, Burlingame AL, et al. Liquid droplet formation by HP1alpha suggests a role for phase separation in heterochromatin. Nature. 2017;547:236–40.
Zhou H, Song Z, Zhong S, Zuo L, Qi Z, Qu LJ, et al. Mechanism of DNA-induced phase separation for transcriptional repressor VRN1. Angew Chem. 2019;58:4858–62.
Tan Y, Wang X, Song H, Zhang Y, Zhang R, Li S, et al. A PML/RARalpha direct target atlas redefines transcriptional deregulation in acute promyelocytic leukemia. Blood. 2021;137:1503–16.
Liu C, Nie D, Li J, Du X, Lu Y, Li Y, et al. Antitumor effects of blocking protein neddylation in T315I-BCR-ABL Leukemia Cells and leukemia stem cells. Cancer Res. 2018;78:1522–36.
Milhollen MA, Traore T, Adams-Duffy J, Thomas MP, Berger AJ, Dang L, et al. MLN4924, a NEDD8-activating enzyme inhibitor, is active in diffuse large B-cell lymphoma models: rationale for treatment of NF-{kappa}B-dependent lymphoma. Blood. 2010;116:1515–23.
Kuo KL, Ho IL, Shi CS, Wu JT, Lin WC, Tsai YC, et al. MLN4924, a novel protein neddylation inhibitor, suppresses proliferation and migration of human urothelial carcinoma: In vitro and in vivo studies. Cancer Lett. 2015;363:127–36.
Ying M, Shao X, Jing H, Liu Y, Qi X, Cao J, et al. Ubiquitin-dependent degradation of CDK2 drives the therapeutic differentiation of AML by targeting PRDX2. Blood. 2018;131:2698–711.
Mitrea DM, Chandra B, Ferrolino MC, Gibbs EB, Tolbert M, White MR, et al. Methods for physical characterization of phase-separated bodies and membrane-less organelles. J Mol Biol. 2018;430:4773–805.
Guillen-Boixet J, Kopach A, Holehouse AS, Wittmann S, Jahnel M, Schlussler R, et al. RNA-induced conformational switching and clustering of G3BP drive stress granule assembly by condensation. Cell. 2020;181:346–61. e317
Elbaum-Garfinkle S, Kim Y, Szczepaniak K, Chen CC, Eckmann CR, Myong S, et al. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc Natl Acad Sci USA. 2015;112:7189–94.
Shao X, Xiang S, Fu H, Chen Y, Xu A, Liu Y, et al. CDK2 suppression synergizes with all-trans-retinoic acid to overcome the myeloid differentiation blockade of AML cells. Pharm Res. 2019;151:104545.
Acknowledgements
Dr. Lingtao Wu (University of Southern California) kindly donated NB4 cell line. NB4R1 cells were kindly gifted from Dr. He Huang (Zhejiang University) and NB4R2 cells were kindly provided by Dr. Jian Zhang (Shanghai Jiao Tong University). OCI-AML2 cells were kindly gifted from Dr. Yu Rao (Tsinghua Univesity).
Funding
This work was supported by grants from the National Natural Science Foundation of China (Nos. 81973354 to MY, No. 81803552 to XS); Leading Talent of “Ten Thousand Plan”—National High-Level Talents Special Support Plan and the Fundamental Research Funds for the Central Universities.
Author information
Authors and Affiliations
Contributions
MY and XS conceived the study and analyzed data; XS performed high-content screening; YC performed phase separation and BioID assay; XS and AX performed the mHSPC-related experiment; XS, YC, AX, DX, WD and YH performed the immunofluorescence and western blotting procedure; AX, WW, MC and ZX detected the therapeutic effect of MLN4924; WW generated the PML KO cells; WW and XZ quantified the immunofluorescence data; JC, QH, BY, YW and YZ conceived the experiments and helped organize the paper; MY, XS and YC wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics declarations
Written informed consents from patients and approval from the Institutional Research Ethics Committee of the hospital were obtained before the use of these clinical materials for research purposes. The Animal Research Committee at Zhejiang University approved all the animal studies, and animal care was provided in accordance with the institutional guidelines.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by K. Engeland
Rights and permissions
About this article
Cite this article
Shao, X., Chen, Y., Xu, A. et al. Deneddylation of PML/RARα reconstructs functional PML nuclear bodies via orchestrating phase separation to eradicate APL. Cell Death Differ 29, 1654–1668 (2022). https://doi.org/10.1038/s41418-022-00955-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-022-00955-8
This article is cited by
-
CPSF6-RARγ interacts with histone deacetylase 3 to promote myeloid transformation in RARG-fusion acute myeloid leukemia
Nature Communications (2025)
-
Protein neddylation and its role in health and diseases
Signal Transduction and Targeted Therapy (2024)
-
Phase separations in oncogenesis, tumor progressions and metastasis: a glance from hallmarks of cancer
Journal of Hematology & Oncology (2023)
-
VAPB-mediated ER-targeting stabilizes IRS-1 signalosomes to regulate insulin/IGF signaling
Cell Discovery (2023)
-
Defining the condensate landscape of fusion oncoproteins
Nature Communications (2023)