Abstract
Solute carrier family 25 member 51 (SLC25A51) was recently identified as the mammalian mitochondrial NAD+ transporter essential for mitochondria functions. However, the role of SLC25A51 in human disease, such as cancer, remains undefined. Here, we report that SLC25A51 is upregulated in multiple cancers, which promotes cancer cells proliferation. Loss of SLC25A51 elevates the mitochondrial proteins acetylation levels due to SIRT3 dysfunctions, leading to the impairment of P5CS enzymatic activity, which is the key enzyme in proline biogenesis, and the reduction in proline contents. Notably, we find fludarabine phosphate, an FDA-approved drug, is able to bind with and inhibit SLC25A51 functions, causing mitochondrial NAD+ decrease and proteins hyperacetylation, which could further synergize with aspirin to reinforce the anti-tumor efficacy. Our study reveals that SLC25A51 is an attractive anti-cancer target, and provides a novel drug combination of fludarabine phosphate with aspirin as a potential cancer therapy strategy.

Similar content being viewed by others
Introduction
Nicotinamide adenine dinucleotide (NAD+) is a crucial cofactor that functions in various redox reactions, including glycolysis, citric acid cycle and oxidative phosphorylation [1]. In addition, NAD+ is also the substrate of numerous NAD+ consuming enzymes, such as poly (ADP-ribose) polymerases (PARPs) [2], Sirtuins [3], and CD38 [4], which plays vital roles in DNA damage repair, gene expression regulation, cell cycle progress and cellular metabolism. Cancer cells have been reported to sustain a higher concentration of NAD+ than normal cells, in order to support the fast generation of energy and macromolecules, as well as to counteract the excess ROS production [5]. Various enzymes involved in NAD+ biosynthesis, including nicotinamide phosphoribosyltransferase (NAMPT) and nicotinic acid phosphoribosyltransferase (NAPRT), are aberrantly up-regulated in multiple cancers and indicated to promote tumorigenesis and cancer progression [6,7,8]. Besides, the NAD+ lowering agents, such as FK866 and KPT-9274, both are selective inhibitors of NAMPT that have entered the early clinical trials, demonstrated a strong capacity to reduce the NAD+ levels and induce apoptosis in cancer cells, indicating the great potential of NAD+-depleting in cancer treatment [9, 10].
The distribution of cellular NAD+ is compartmentalized, and more than 40% NAD+ is present in the mitochondria, which is crucial for cell survival under stress [11]. Recently, Solute carrier family 25 member 51 (SLC25A51), also known as MCART1, was identified as a mammalian mitochondrial NAD+ transporter that is capable of mitochondria taking up NAD+ and maintaining the homeostasis of mitochondrial NAD+ pool [12,13,14]. It is reported that loss of SLC25A51 caused a significant decrease in mitochondrial NAD+ level, and impaired mitochondrial metabolism, respiratory capacity and mitochondrial ATP generation [13]. However, the pathological roles of SLC25A51 in human diseases, such as malignant tumors, remain unexplored.
Lysine acetylation is one of the most abundant and crucial protein post-translational modifications, reversibly controlled by histone acetyltransferases (HATs) and histone deacetylases (HDACs), which involves in the regulation of diverse cellular processes, including gene transcription, cell cycle, signaling transduction and metabolism [15]. Mitochondrial acetylation homeostasis has been shown critical for mitochondrial functions, since major metabolic enzymes in mitochondria that involve in energy production, amino acid metabolism, fatty acid metabolism and urea cycle can be regulated by acetylation [16]. SIRT3 is an NAD+ dependent and mitochondria located deacetylase that dominates the regulation of mitochondrial protein acetylation [17]. SIRT3 is found elevated in numerous cancers, and promotes cancer progression via sustaining mitochondrial energy homeostasis, reducing ROS level and preventing apoptosis [18].
In this study, we identified SLC25A51 as an oncoprotein which was upregulated in various cancers. Loss of SLC25A51 impaired SIRT3 enzymatic activity and mitochondrial acetylation homeostasis, leading to the inhibition of AKT/mTOR signaling pathway and defect in cell proliferation via proline synthesis limitation. In addition, we discovered that fludarabine phosphate was a potential inhibitor of SLC25A51, and could synergize with aspirin to break the mitochondrial acetylation homeostasis and inhibit tumor growth in vivo and in vitro.
Results
SLC25A51 is upregulated in cancers and an unfavorable prognostic factor
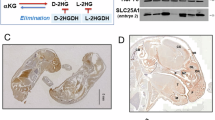
In order to investigate the role of SLC25A51 in cancers, we firstly examined the expression levels of SLC25A51 in the TCGA and GTEx database, and found SLC25A51 was elevated in the most of cancers compared with the normal tissues (Supplementary Fig. 1A), including colon adenocarcinoma (COAD) and rectum adenocarcinoma (READ) (Fig. 1A). The similar observation was obtained from a GEO database of colorectal adenocarcinoma (Fig. 1B). In addition, the immunohistochemical staining analysis of 69 pairs of colorectal cancer tissues and adjacent normal tissues from patients was performed to examine the protein levels of SLC25A51. The result showed that SLC25A51 was notably up-regulated in tumor samples (Fig. 1C, D), which was further confirmed by the immunoblotting analysis (Fig. 1E). Besides, through assessing the endogenous expression levels of SLC25A51 in 8 different colon cancer cell lines and one normal human colon mucosal epithelial cell line, NCM460, we found that SLC25A51 was overexpressed at both mRNA and protein levels in most cancer cells compared with NCM460 cells (Fig. 1F, G). To further explore the clinical features of SLC25A51, we analyzed the relationship between SLC25A51 expression levels and clinical stages based on TCGA database, finding that the expression level of SLC25A51 was positively associated with the progression of several cancers, including colorectal cancer, adrenal cancer, gastric cancer, cholangiocarcinoma and liver cancer (Supplementary Fig. 1B). Moreover, patients with higher SLC25A51 expression showed a lower overall survival time rate in comparison with the lower expression group, based on the results of IHC analysis (Fig. 1H) and the GEO database (GSE17536) (Fig. 1I), indicating that SLC25A51 expression level was correlated with poor prognosis in colorectal cancer. Additionally, the negative relationship between SLC25A51 expression level and prognosis was further confirmed in multiple cancers via examining the TCGA database (Supplementary Fig. 1C). In summary, these data concluded that SLC25A51 is commonly upregulated in cancers, and positively associated with cancer progression and poor prognosis.
A Scatter plots of SLC25A51 expression levels of normal tissues and tumor tissues in colon adenocarcinoma (COAD, left) and rectum adenocarcinoma (READ, right) from TCGA and GTEx database. B Scatter plots of SLC25A51 expression values of normal tissues and tumor tissues in colorectal adenocarcinoma from GEO database (GSE20916). C Representative images of the IHC analysis of colorectal cancer (CRC) and the according adjacent tissues with SLC25A51 antibodies. Scale bars, 5X, 500 μm and 40X, 50 μm. D Statistical analysis of relative SLC25A51 protein levels of tumor and adjacent tissues. E Western blot analysis of SLC25A51 protein levels of 10 pairs of CRC and adjacent tissues. F qPCR analysis of SLC25A51 mRNA levels comparing one normal colon epithelial cell line with eight CRC cell lines. G Western blot analysis of SLC25A51 protein levels comparing one normal colon epithelial cell line with eight CRC cell lines. H Kaplan-Meier plots of CRC patients stratified by SLC25A51 protein levels based on the IHC analysis in C. I Kaplan-Meier plots of colon cancer patients stratified by SLC25A51 mRNA levels from the GEO database (GSE17536). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Data are representative of three independent experiments with similar results. Bars, mean ± SD.
SLC25A51 deficiency inhibits cancer cell proliferation
As SLC25A51 is overexpressed in cancers, we next examined the potential role of SLC25A51 on cancer cell proliferation. We firstly generated SLC25A51 knockdown cancer cell lines, and found that SLC25A51 deficiency significantly impaired the colony formation ability (Fig. 2A and Supplementary Fig. 2A, B) and cell proliferation rates (Fig. 2B and Supplementary Fig. 2C) of multiple cancer cell lines including colon, lung and breast cancers. Besides, we also found that loss of SLC25A51 impaired the proliferative capacity of SW480 and H1299 cells through the EdU incorporation assay (Fig. 2C and Supplementary Fig. 2D). To further determine the influence of SLC25A51 on tumor growth in vivo, 4 weeks old BALB/c nude mice were subcutaneously injected with SLC25A51 knockdown or wild type HCT116 cells. The results showed that the tumor with SLC25A51 deficiency dramatically reduced in both tumor growth rate and tumor weight (Fig. 2D–F). The similar phenomenon was also observed in CT26 xenograft tumor model on BALB/c mice (Supplementary Fig. 2E–G). In addition, we examined the Ki67 fluorescence intensity of tumors as the proliferation marker, and the results revealed that loss of SLC25A51 obviously decreased the percentage of Ki67+ cancer cells (Fig. 2G, H). Taken together, the above data demonstrated that SLC25A51 promotes cancer cells proliferation in vivo and in vitro, which is common in a variety of tumors.
A Wild type and SLC25A51 knockdown cells as indicated were seeded into 6 cm dishes at 3 ×103 per dish, and the cell colonies were stained with crystal violet after 7 days. B Wild type and SLC25A51 knockdown cells were seeded into 6-well plates at 105 cells per well and the cell numbers were counted every day. C The representative image of EdU incorporation assay in wild type and SLC25A51 knockdown SW480 cells (left). The percentage of cells positively stained with EdU was quantified (right). Scale bars (bottom right), 50μm. D BALB/c Nude mice were inoculated with SLC25A51 knockdown or wild type HCT116 cells. Tumor volume was measured every other day. E The representative image of wild type and SLC25A51 knockdown tumors that dissected at the end of experiments. F The HCT116 tumor weight was determined at the final day. G The representative image of Ki67 staining (green) in wild type and SLC25A51 knockdown CT26 tumors. Scale bars (bottom right), 50μm. H The percentage of cells positively stained with Ki67 antibodies was quantified. **P < 0.01, ***P < 0.001, ****P < 0.0001. Data are representative of three independent experiments with similar results. Bars, mean ± SD.
SLC25A51 affects mitochondrial acetylation homeostasis
To further explore the mechanisms of SLC25A51 in the regulation of cancer cell proliferation, we firstly validated that SLC25A51 knockdown indeed decreased the mitochondrial NAD+ levels in LoVo and H1299 cells (Supplementary Fig. 3A). As SIRT3, SIRT4 and SIRT5 are the major mitochondria located NAD+ consuming enzymes [19], and the fluctuation of NAD+ levels was reported to influence the enzymatic activity of SIRT3, leading to the unbalanced mitochondrial acetylation homeostasis [20], we speculated that SLC25A51 knockdown could affect their activities. Therefore, we extracted mitochondria from SLC25A51 knockdown LoVo, HCT116, DLD1, SW480 and CT26 cells, finding that the mitochondrial acetylation levels were dramatically elevated after loss of SLC25A51, whereas the cytoplasm acetylation levels remained almost unchanged (Fig. 3A and Supplementary Fig. 3B), which was consistent with the recent discoveries that SLC25A51 had little influence on the whole cell NAD+ levels [12, 13]. On the contrary, the succinylation levels of mitochondrial proteins, which is controlled by SIRT5 [21], stayed unaltered after SLC25A51 knockdown (Fig. 3B and Supplementary Fig. 3C). To explore whether the hyper-acetylation of mitochondria is associated with the dysfunction of SIRT3, we overexpressed SIRT3 in both wild-type and SLC25A51 silenced cells, and found that loss of SLC25A51 notably impaired SIRT3-mediated deacetylation of the global mitochondrial proteins as well as SOD2, one of the most canonical substrates of SIRT3 [22, 23], while the cytoplasm-located protein, α-Tubulin, was unaffected (Fig. 3C, D, and Supplementary Fig. 3D). Additionally, further knockout of SIRT3 in SLC25A51 silenced cells showed little promotion effect on the mitochondrial acetylation levels (Supplementary Fig. 3E), indicating that the lack of NAD+ indeed impaired the function of SIRT3. Since there were a few researches indicating that SIRT4 also possessed a weak deacetylation activity [24, 25], we knocked down SIRT4 in wild-type and SLC25A51 deficient cells, which revealed that loss of SIRT4 rarely influenced the mitochondrial acetylation levels (Supplementary Fig. 3F). Besides, knockdown of SIRT5 in SLC25A51 silenced cells could further elevate the succinylation levels of mitochondrial proteins (Supplementary Fig. 3G), suggesting that the enzymatic activity of SIRT5 was barely influenced by SLC25A51. Moreover, the colony formation and proliferation assays showed that, knockout of SIRT3 significantly impaired the proliferative capability of wild-type cells compared to those SLC25A51 knockdown cells (Supplementary Fig. 3H, I), suggesting that SIRT3 was involved in the SLC25A51-mediated promotion of cancer cells proliferation. In collection, these data demonstrated that SLC25A51 deficiency mainly impaired SIRT3 functions, leading to the hyperacetylation of mitochondrial proteins.
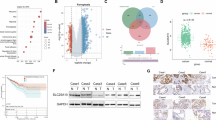
A Western blot analysis of the acetylation levels of mitochondrial and cytoplasmic proteins of wild type and SLC25A51 knockdown LoVo (left) and HCT116 (right) cells. The relative acetylation ratio is normalized to CS and α-Tubulin respectively. B Western blot analysis of the mitochondrial proteins acetylation levels and succinylation levels of wild type and SLC25A51 knockdown HCT116 cells. The relative acetylation ratio is normalized to CS. C SLC25A51 knockdown LoVo cells were transfected with SIRT3 expressing plasmids, and the acetylation levels of mitochondrial proteins were detected by western blot. D Wild-type and SLC25A51 knockdown LoVo cells were transfected with SIRT3 expression plasmids, and the acetylation levels of SOD2 and α-Tubulin were analyzed via western blotting. E Schematic of pan-acetyllysine antibody based immunoprecipitated proteomics analysis of mitochondrial proteins extracted from wild type and SLC25A51 knockdown LoVo cells. F The silver staining of acetylated mitochondrial proteins immunoprecipitating with anti-acetyllysine agaroses in LoVo cells. G Overlapped proteins of annoted mitochondrial gene sets and acetylation elevated proteins after SLC25A51 deficiency (fold change > 1.5). H The bar graph shows the enrichment analysis of hyper-acetylated mitochondrial proteins after SLC25A51 knockdown in the terms of GO biological process. I Western blot analysis of the acetylation levels of representative proteins from the MS results in LoVo and H1299 cells.
To investigate the most influenced mitochondrial proteins after SLC25A51 loss, we purified the mitochondria from wild type and SLC25A51 knockdown LoVo cells and performed the pan-acetyllysine-antibody based immunoprecipitation for LC-MS/MS analysis to catch maximum hyper-acetylated mitochondrial proteins (Fig. 3E, F). We defined 46 upregulated acetylated mitochondrial proteins after SLC25A51 deficiency (Fig. 3G). To determine the role of these proteins, we performed a cluster analysis, which indicated that these proteins were highly related to amino acids metabolism and proline biogenesis (Fig. 3H). We further confirmed the MS results via analyzing the acetylation levels of several representative proteins. Significantly, the acetylation levels of Pyrroline-5-carboxylate reductase 1 (PYCR1) and Delta-1-pyrroline-5-carboxylate synthase (P5CS), which are tandem enzymes that catalyze the production of proline from glutamate [26, 27], were notably up-regulated in SLC25A51 deficiency LoVo and H1299 (Fig. 3I). In summary, we find that loss of SLC25A51 causes mitochondrial hyper-acetylation due to SIRT3 dysfunction, and identified the most influenced mitochondrial proteins, including PYCR1 and P5CS.
Hyperacetylation impairs P5CS enzymatic activity
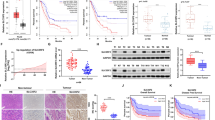
Our recent study has discovered that PYCR1 is a substrate of SIRT3, and hyper-acetylation could impair the assembling of PYCR1 holoenzyme, leading to the reduction in PYCR1 enzymatic activity, and thus proliferation blockage in cancer cells [28]. Therefore, we firstly validated that SLC25A51 knockdown impaired PYCR1 oligomerization in LoVo cells (Supplementary Fig. 4A). However, the role of P5CS acetylation has not been investigated yet. Through analyzing the TCGA and GTEx database, we firstly verified that P5CS (encoded by ALDH18A1) was upregulated in tumor tissues (Supplementary Fig. 4B), and showed a negative relationship with the patient prognosis in several cancers, including colon, lung and breast cancers (Supplementary Fig. 4C). In order to identify the HAT for P5CS, we co-transfected P5CS with different HATs, including CBP, p300, PCAF, Tip60 and hMOF, and found that CBP dominantly augmented the acetylation level of P5CS (Fig. 4A). Besides, we purified hyper-acetylated P5CS via ectopically expressing CBP for mass spectrometry analysis, which identified K347 and K432 as the main acetylated lysines of P5CS (Supplementary Fig. 4D). As SIRT3 is the primary deacetylase in mitochondria, we co-transfected P5CS with SIRT3, and found that SIRT3 was capable to decrease P5CS acetylation levels in a dose-dependent manner (Fig. 4B). The in vitro deacetylation assay revealed that SIRT3 was able to deacetylate P5CS only when the NAD+ was added (Supplementary Fig. 4E). In addition, we detected a notably higher acetylation level of P5CS in SIRT3 knockout cells (Fig. 4C). Moreover, we confirmed that SIRT3 directly interacted with P5CS in mitochondria via endogenous co-immunoprecipitation assay, mitochondrial fractionation and immunofluorescence staining (Fig. 4D, E, and Supplementary Fig. 4F). Besides, we also found that the dissociation constant (Kd) of P5CS and SIRT3 was about 30 nM via microscale thermophoresis (MST) assay (Fig. 4F). Since the formation of tetramer is essential for P5CS functions [29], we examined whether acetylation could regulate the oligomerization of P5CS. We treated cell lysates with 0.025% glutaraldehyde, and the result showed that both SIRT3 knockout and CBP overexpression impeded the dimer and tetramer formation of P5CS (Fig. 4G and Supplementary Fig. 4G). Additionally, SLC25A51 knockdown inhibited the assembly of P5CS holoenzyme (Fig. 4H and Supplementary Fig. 4H), and the enzymatic activity of P5CS (Fig. 4I) in LoVo and H1299 cells. Furthermore, we detected the amino acids contents in SLC25A51 deficient LoVo cells via the amino acid analyzer, and found that proline was the only amino acid that decreased significantly, while glutamate, the primary precursor for proline biogenesis [30] increased (Fig. 4J). Besides, we also observed a notable increase in cystine, which is kept in balance with glutamate due to the cystine/glutamate antiporter SLC7A11 [31], thus one possible explanation is that elevated glutamate induced an increase in cystine input, which caused the accumulation of cystine. Besides, proline has been shown essential for eliminating the oxidative stress [32]. Thus, SLC25A51 knockdown cells might require an elevated cystine level for glutathione production to protect from the oxidative stress. Furthermore, the reduction in proline content was validated in SLC25A51 knockdown LoVo and H1299 cells via the proline quantification (Fig. 4K), which was consistent with loss of SIRT3 (Supplementary Fig. 4I). Taken together, these results demonstrated that hyper-acetylation of P5CS, as the consequence of SLC25 A51 deficiency, inhibits the enzymatic activity of P5CS, leading to the diminish in cellular proline content.
A HEK293T cells were transfected with plasmids as indicated, and P5CS acetylation was analyzed by immunoblot. B P5CS-Flag plasmid was co-transfected with different amounts of HA-SIRT3 plasmids into HEK293T cells, and the acetylation level of P5CS was measured by western blot. C The cell lysates of SIRT3 knockout U2OS cells were immunoprecipitated with anti-acetyllysine agaroses, and SLC25A51 acetylation level was detected. D HEK293T cell lysates were immunoprecipitated with control IgG or anti-SIRT3 antibody, and then detected by anti-SIRT3 and anti-P5CS antibodies. E Representative immunofluorescence staining of endogenous SIRT3 (red), P5CS (green) and TOM20 (grey) in LoVo cells. Scale bars (bottom right), 10μm. F Microscale thermophoresis assay of SIRT3-His and P5CS-Flag proteins. G Cell lysates of SIRT3 knockout HCT116 cells expressing P5CS-Flag were treated with 0.025% glutaraldehyde and analyzed by Western blot analysis. H Cell lysates of SLC25A51 knockdown LoVo cells expressing P5CS-Flag were treated with 0.025% glutaraldehyde and analyzed by Western blot analysis. I The enzymatic activity of P5CS in wild type and SLC25A51 knockdown LoVo (left) and H1299 (right) cells. J Cellular amino acids contends of wild type and SLC25A51 knockdown LoVo cells were measured and exhibited as the heat map. K The cellular proline content of wild type and SLC25A51 knockdown LoVo (left) and H1299 (right) cells. *P < 0.05, **P < 0.01. Data are representative of three independent experiments with similar results. Bars, mean ± SD.
Proline replenishment rescues proliferation defects in SLC25 A51 knockdown cancer cells
Since a number of researches have pointed out that proline was indispensable for cancer cells proliferation [33,34,35], we therefore investigated that whether the inhibition effect on SLC25A51 knockdown cancer cells was related to proline deficiency. We firstly added L-proline into the cell culture medium, which verified that proline supplement was capable to elevate the cellular proline level (Supplementary Fig. 5A). In addition, the replenishment of proline was able to partially rescue the colony formation ability that was inhibited by SLC25A51 knockdown in LoVo, H1299 and MCF-7 cells (Fig. 5A and Supplementary Fig. 5B). Besides, proline supplement could also promote the proliferation rates of SLC25A51 deficient LoVo and H1299 cells, while the effect was unobvious in wild type cells (Fig. 5B). The EdU incorporation assay consistently revealed that the addition of proline significantly rescued the defect in proliferation ability caused by SLC25A51 knockdown (Fig. 5C, D and Supplementary Fig. 5C). Since proline starvation was reported to suppress AKT/mTOR signaling pathway, which are notable regulators of cancer cell survival and proliferation via mediating cell cycle progress and metabolism [36, 37], we speculated that knockdown of SLC25A51 impaired cell proliferation was connected to AKT/mTOR pathway inactivation due to proline starvation. To confirm this hypothesis, we firstly performed an RNA-sequencing analysis, and found a large fraction of down-regulating genes after SLC25A51 knockdown was enriched in AKT/mTOR signaling pathway (Fig. 5E, F). Next, we found that loss of SLC25A51 significantly decreased the phosphorylation levels of AKT, mTOR and S6K in vivo and in vitro (Fig. 5G, H and Supplementary Fig. 5D). Moreover, replenishment of proline counteracted the inhibitory effect on AKT/mTOR pathway due to the loss of SLC25A51 (Fig. 5I and Supplementary Fig. 5E). Collectively, we demonstrated that the deficiency in proline biosynthesis causes the proliferative arrest of SLC25A51 knockdown cancer cells, which is related to the inactivation of AKT/mTOR pathway.
A Wild type and SLC25A51 knockdown LoVo (top) and HCT116 (bottom) cells were seeded into 6 cm dishes at 3 ×103 per dish, after 7 days growing with or without proline addition (5 mM), the cell colonies were stained with crystal violet. B Wild type and SLC25A51 knockdown LoVo (left) and H1299 (right) cells were seeded into 6-well plates at 105 cells per well with or without proline (5 mM), and the cell numbers were counted every day. C The percentage of cells positively stained with EdU was quantified. D The representative image of EdU incorporation assay in wild type and SLC25A51 knockdown SW480 cells grew with or without proline addition (5 mM). Scale bars (bottom right), 50 μm. E The bar graph shows the enrichment analysis of down-regulated genes from RNA-sequencing analysis after SLC25A51 knockdown in the terms of KEGG pathway. F Heatmap of down-regulated genes after SLC25A51 knockdown that were involved in AKT/mTOR pathway. G Western blot analysis of the phosphorylation levels of AKT, mTOR and S6K in SLC25A51 knockdown HCT116 cells. H Western blot analysis of the phosphorylation levels of AKT and S6K in SLC25A51 knockdown CT26 tumors. I Western blot analysis of the phosphorylation levels of AKT, mTOR and S6K in SLC25A51 knockdown LoVo cells that grew with or without proline supplement (5 mM). ns, not significant, *P < 0.05, **P < 0.01, ***P < 0.001. Data are representative of three independent experiments with similar results. Bars, mean ± SD.
Fludarabine phosphate is a novel SLC25A51 inhibitor
As SLC25A51 was proved essential for tumor growth, we next investigated the potential inhibitor against SLC25A51. Due to the lack of the crystal structure of SLC25A51 or closely related homologue for homology modeling, we obtained the predicted structure of SLC25A51 from AlphaFold2 (Identifier: AF-Q9H1U9-F1) (Fig. 6A), which was used for the subsequent molecular docking [38, 39]. To discover the potential inhibitors of SLC25A51, we performed the ligand-based virtual screening of more than 2934 U.S. Food and Drug Administration (FDA)-approved drugs from Zinc database, and the top ten screening hits were displayed (Supplementary Table 1). Among these, fludarabine phosphate which is a purine antimetabolite approved for the treatment of adult patients with chronic lymphocytic leukemia (CLL) [40] was selected for further analysis due to its structural similarity to NAD+ (Fig. 6B) and anti-cancer capacity. The lowest-energy binding conformation of fludarabine phosphate within SLC25A51 showed that, fludarabine phosphate bound to the active sites of SLC25A51, including Lys91, Arg278 and Arg182 (Fig. 6C). We further validated the binding affinity of fludarabine phosphate and SLC25A51 through the cellular thermal shift assay (CETSA) [41], which indicated a shift in the denaturation temperature of SLC25A51 that incubated with fludarabine phosphate compared with DMSO (Fig. 6D). In addition, the MST assay revealed that the dissociation constant (Kd) between SLC25A51 and fludarabine phosphate was around 0.3 μM (Fig. 6E). Besides, fludarabine phosphate was capable to decrease the mitochondrial NAD+ levels in a dose- and time-dependent manner (Fig. 6F, G, Supplementary Fig. 6A, B). Consistently, fludarabine phosphate could also elevate the acetylation levels of mitochondrial proteins (Fig. 6H and Supplementary Fig. 6C), leading to the decrease in cellular proline content (Fig. 6I and Supplementary Fig. 6D), and AKT/mTOR pathway inhibition (Supplementary Fig. 6E). However, the suppressive effect of fludarabine phosphate on cancer cells was notably dampened by silencing SLC25A51 (Fig. 6J and Supplementary Fig. 6F), indicating that SLC25A51 is indeed a target of fludarabine phosphate. Taken together, we identified that fludarabine phosphate, an FDA-approved CLL drug, could bind with and inhibit the activity of SLC25A51, and cause the NAD+ content reduction in mitochondria of cancer cells.
A The predicted structure of human SLC25A51 from the AlphaFold2 database (identifier: AF-Q9H1U9-F1) and visualized in PyMOL. B Structure comparison of NAD+ and fludarabine phosphate. C Docking model of SLC25A51 and fludarabine phosphate. Fludarabine phosphate was represented by green sticks, while important residues engaged in the interactions with SLC25A51 were depicted as slate sticks. D Cellular thermal shift assay of LoVo lysates incubated with fludarabine phosphate or DMSO. E Microscale thermophoresis assay of fludarabine phosphate and SLC25A51-EGFP purified from HEK293T. F Measurement of mitochondrial NAD+ levels of LoVo cells treated with fludarabine phosphate for 24 hour at indicated concentrations. G Measurement of mitochondrial NAD+ levels of LoVo cells treated with fludarabine phosphate (40 μM) for indicated time. H Western blot analysis of mitochondrial proteins acetylation levels of LoVo cells treated with fludarabine phosphate for 24 hours at indicated concentrations. I The cellular proline content of HCT116 cells treated with fludarabine phosphate for 24 hours at indicated concentrations. J Cell viability analysis of wild type and SLC25A51 knockdown LoVo cells treated with fludarabine phosphate for 48 hours at indicated concentrations via CCK-8 kits. **P < 0.01, ***P < 0.001, ****P < 0.0001. Data are representative of three independent experiments with similar results. Bars, mean ± SD.
Aspirin boosts anti-cancer effects of SLC25A51 inhibition
Aspirin, a commonly used non-steroidal anti-inflammatory drug, has been reported able to acetylate a variety of proteins, such as cyclooxygenase (COX) [42] and p53 [43]. Therefore, we assumed that aspirin could augment the mitochondrial hyper-acetylation followed by SLC25A51 knockdown, and might synergize with fludarabine phosphate. Since there has no connection between aspirin and mitochondrial protein acetylation been reported yet, we firstly verified that aspirin was capable to acetylate mitochondrial proteins in a dose-dependent manner (Fig. 7A and Supplementary Fig. 7A), including PYCR1 and P5CS (Supplementary Fig. 7B). Furthermore, we found that aspirin could amplify the mitochondrial hyper-acetylation induced by both SLC25A51 deficiency and fludarabine phosphate treatment (Fig. 7B, C, and Supplementary Fig. 7C, D). Besides, the results also showed that aspirin significantly intensified the inhibitory effects on cancer cells colony formation ability (Fig. 7D and Supplementary Fig. 7E, F), proliferation rates (Fig. 7E, F, and Supplementary Fig. 7G, H) and cell viability (Supplementary Fig. 7I, J) taken by SLC25A51 knockdown as well as fludarabine phosphate treatment. The EdU incorporation assay further confirmed that the combination of aspirin and fludarabine phosphate most notably impaired the proliferative capacity of cancer cells (Fig. 7G, H, and Supplementary Fig. 7K). We next examined the combinational effect of aspirin and fludarabine phosphate in vivo. Through the xenograft tumor model, we found that the administration of aspirin showed a significant synergic effect with fludarabine phosphate on restricting HCT116 and CT26 cells growth in mice (Fig. 7I–K and Supplementary Fig. 8A–C), without causing the extra reduction in mice body weight compared with mice receiving a single use of fludarabine phosphate (Supplementary Fig. 8D). Additionally, the Ki67 staining confirmed such synergic effect on tumor proliferation capacity (Supplementary Fig. 8E). Mechanically, we found that the combination of aspirin and fludarabine phosphate most significantly impaired the phosphorylation levels of AKT/mTOR signaling pathway in vivo (Fig. 7L). Taken together, our data revealed that aspirin was capable to synergy with fludarabine phosphate to inhibit tumor growth through aggravating the mitochondrial acetylation imbalance.
A Representative immunofluorescence staining of pan-acetyllysine (green) and CS (mitochondrial marker, red) in aspirin treated (5 mM, 24 hours) H1299 cells. Scale bars (bottom right), 10 μm. B Western blot analysis of mitochondrial proteins levels of wild type and SLC25A51 knockdown HCT116 cells treated with aspirin (1 mM) or DMSO for 24 hours. The relative acetylation ratio is normalized to CS. C Western blot analysis of LoVo cells treated with fludarabine phosphate (40 μM) or aspirin (1 mM) or the combination of these two for 24 hours. D LoVo cells were seeded in the 6-well plates at 103 per well, after cells grew with aspirin or fludarabine phosphate or the combination of these two at indicated concentrations for 7 days, the colonies were fixed and stained with crystal violet. E Wild type and SLC25A51 knockdown LoVo cells were seeded into 6-well plates at 105 per well and grew with DMSO or aspirin (5 mM). The cell numbers were counted every day. F SW480 cells were seeded into 6-well plates at 105 per well and grew with fludarabine phosphate alone or the combination of aspirin and fludarabine phosphate at indicated concentrations. The cell numbers were counted every day. G The representative image of EdU incorporation assay in LoVo cells treated with fludarabine phosphate (10 μM) alone or the combination of fludarabine phosphate (10 μM) and aspirin (2 mM) for 24 hours. Scale bars (bottom right), 50 μm. H The percentage of cells positively stained with EdU was quantified. I BALB/c Nude mice were inoculated with HCT116 cells and inoculated intraperitoneally with vehicle, fludarabine phosphate alone, aspirin alone or the combination of fludarabine phosphate and aspirin as indicated. Tumor volume was measured every other day. J The representative image of HCT116 tumors that dissected at the end of experiments. K The tumor weight was determined at the final day. L Western blot analysis of the phosphorylation levels of AKT, mTOR and S6K in HCT116 tumors treated as indicated. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Data are representative of three independent experiments with similar results. Bars, mean ± SD.
Discussion
NAD+ metabolism has been verified to be crucial for the metabolic reprograming of cancer cells. The significantly elevated NAD+ level is observed in cancer cells to rapidly produce the mass of energy and macromolecules, which is known as the Warburg effect [44]. Besides, numerous enzymes involved in NAD+ metabolism, such as NAMPT, NMNAT and NAPRT, have been demonstrated to function in tumor progression, and targeting NAD+ metabolism becomes a potential cancer treatment [45]. In this study, we uncovered that SLC25A51, a recently discovered mitochondrial NAD+ transporter, is overexpressed in multiple cancers, and negatively associated with the patient prognosis. Loss of SLC25A51 significantly inhibits tumor growth in vivo and in vitro, through impairing the function of SIRT3, elevating the acetylation levels of mitochondrial proteins, and blocking the proline production.
Through virtual screening of FDA-approved drugs based on SLC25A51 predicted structure from AlphaFold2, we discovered that fludarabine phosphate could bind to the active sites of SLC25A51, and thus inhibit the function of SLC25A51, leading to the NAD+ decrease and mitochondrial proteins hyper-acetylation. Since the safety of fludarabine phosphate has been validated [46], our study could be instantly put into clinical use for those patients with SLC25A51 overexpression. As the lead compound, fludarabine phosphate could be forward modified to obtain the more specific targeted molecules of SLC25A51. For example, the mitochondrial inner membrane has a highly negative membrane potential due to the proton gradient for ATP synthesis, therefore, the compound with high lipophilicity and positive charges might have a stronger affinity for SLC25A51, which is located on mitochondrial inner membrane [13, 47]. Besides, our study also verifies the feasibility of occupying the activity center of SLC25A51 to hinder its functions, which is helpful for the further screening of SLC25A51 efficient inhibitors from commercial libraries, or de novo design the chemicals or peptides for SLC25A51 active sites according to the results of molecular docking. However, due to the technical limitation, the crystal structures of SLC25A51 alone and the complex with fludarabine phosphate have not been resolved.
Mitochondria acetylation homeostasis is crucial for maintaining the metabolic functions of mitochondrial enzymes in cancer cells [16], and inhibition of SIRT3 has been shown as a potential strategy for cancer treatment in various cancers [48,49,50]. In this study, we find that disrupting mitochondria acetylation balance could impair the proline biogenesis pathway, causing the inactivation of proliferative signaling, such as AKT/mTOR pathway, in cancer cells. Significantly, we demonstrated that aspirin, one of the most commonly used chemical drugs for suppressing inflammation and preventing thrombus, is capable to acetylate mitochondrial proteins. Thus, we further find that aspirin is able to synergize with SLC25A51 inhibition to cause the irreversible excessive acetylation levels in mitochondria, and arrest tumor growth in vivo. This discovery gives a novel application for aspirin as the adjuvant of SLC25A51 or SIRT3 inhibitors, and also at least partially explains the mechanisms of aspirin on cancer prevention [51], since cancer cells are addicted to various mitochondrial metabolic processes.
However, there are some limitations in this study. Although SIRT3 is the important NAD+ consumer in mitochondria, there are still some other enzymes that utilize NAD+ but not investigated in this study, such as pyruvate dehydrogenase and isocitrate dehydrogenase that are connected to the energy production [52]. Besides, a recent study demonstrated that loss of SLC25A51 impaired the functions of electron transport chain (ETC) and reduced TCA cycle flux [13], indicating that SLC25A51-mediated NAD+ transport was also important for the activities of various NAD+-dependent proteins in mitochondria besides Sirtuin family. Moreover, although we demonstrate that proline addition could obviously rescue the proliferation defects caused by SLC25A51 knockdown, we yet cannot exclude the other metabolic pathways that could be influenced by SLC25A51, especially there are some other mitochondrial proteins hyper-acetylated after SLC25A51 deficiency, such as MTHFD1L and MTHFD2, indicating that one-carbon metabolism might be also altered [53]. Besides, the mechanisms of SLC25A51 up-regulated in cancer cells remains unelucidated. A recent CHIP sequencing result demonstrated that the oncoprotein Myc was enriched in the promoter regions of SLC25A51 [54], indicating that the hyper-activation of Myc might contribute to the SLC25A51 up-regulation, but still needs further confirmation.
Here, our study reveals a pro-tumor role of SLC25A51, promoting cancer cells proliferation via activating SIRT3 and sustaining the acetylation homeostasis of mitochondria. We also show that fludarabine phosphate is a potential inhibitor of SLC25A51, and could synergize with aspirin to suppress tumor growth. In conclusion, our study provides a novel target for cancer treatment, and discovers a potent drug combination that could be put into clinical use as soon as possible.
Materials and methods
Mice
All the experiments with mice were approved by the Institutional Animal Care and Use Committee of Peking University Health Science Center (permit number 2019PHE073). Male BALB/c Nude (4-6 weeks old), male BALB/c (4-6 weeks old) were obtained from the Department of Laboratory Animal Science of Peking University Health Science Center, Beijing. All the mice were housed according to standards, which including constant room temperature and humidity and 12 h: 12 h light cycle. The mice were randomly assigned to each group to minimize the effects of little difference between them.
Cell lines
HEK293T, HCT116, H1299, MCF-7, MDA-MB-231, 4T1 were obtained from American Type Culture Collection (ATCC). NCM460, DLD-1, HCT-8, RKO, SW480, SW620, HT-29 and LoVo were kindly provided by Zhanlong Shen (Peking University People’s Hospital). PC9 was a gift from Xiaoyan Qiu (Peking university). CT26 wild type was a kind gift from Xinshan Ye (Peking University). 4T1 and PC9 were cultured in RPMI-1640 supplemented with 10% Fetal Bovine Serum (FBS), 100 U/mL penicillin and 100 μg/mL streptomycin. The other cell lines were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% FBS, 100 U/mL penicillin and 100 μg/mL streptomycin. All cells were free from mycoplasma contamination.
Reagents and antibodies
Fludarabine Phosphate (HY-B0028) and Aspirin (HY-14654) were from MedChemExpress (MCE). L-proline (PHR1332), anti-HA agarose beads (E6779) and anti-Flag (M2 clone) agarose beads (A2220) were from Sigma-Aldrich. Protein A/G agarose beads (20241) were from Thermo Fisher Scientific. Anti-acetyl lysine agarose beads (ICP0388) were from Immunechem. SIRT3-6xhis (APE913Hu01) was from Cloud-Clone. Flag peptide (P9801) was from Beyotime.
The antibodies were commercially obtained: anti-SIRT3 (sc-365175), anti-β-actin (sc-8432), anti-α-tubulin (sc-8035), anti-CS (sc-390693), anti-SOD2 (sc-137254) and anti-Vinculin (sc-73614) were from Santa Cruz Biotechnology. Anti-phospho-AKT (Ser473, 4060), anti-phospho-mTOR (Ser2448, 5536), anti-mTOR (2983), anti-phospho-S6K (Thr389,9205), S6K (9202), SIRT3 (5490), anti-GAPDH (5174), anti-MTHFD1L (14998) and anti-acetylated-lysine (9441) were from Cell Signaling Technology. Anti-Ki67 (ab16667), anti-TOMM20 (ab289670), anti-acetyl-α-Tubulin (K40, ab179484) and anti-acetyl-SOD2 (K68, ab137037) were from Abcam. Anti-HA (26183) and Alexa Fluor 488 and 594 conjugated secondary antibodies (A21206 and A11032) were from Thermo Fisher Scientific. Anti-PYCR1 (13108-1-AP), anti-MTHFD2 (12270-1-AP) and anti-P5CS (17719-1-AP) were from Proteintech. Anti-SLC25A51 (CSB-PA875649LA01HU) was from CUSABIO. Anti-Succinyllysine (PTM-419) was from PTM BIO. Alexa Fluor 647 conjugated anti-Rabbit IgG (bs-0295G-AF647) was from Bioss.
Plasmid construction
For transient expression in eukaryotic cells, cDNA of P5CS, HATs and SIRT3 were amplified by PCR and cloned into pcDNA3.1 vectors (Invitrogen) with HA or Flag tag, and cDNA of SLC25A51 was amplified by PCR and cloned into pHBLV-EFFP vectors (HanBio).
shRNA and siRNA mediated knockdown
To generate SLC25A51 knockdown cell lines, 21 bp short hairpin RNA (shRNA) sequences were ligated into pLKO.1 (Sigma-Aldrich) plasmid and then co-transfected with viral packaging plasmids (psPAX2 and pMD2.G) into HEK293T cells. 48 h after transfection, the culture medium was collected and filtered through a 0.22 μm strainer. The targeted cells were infected with the viral supernatant for 48 hours and then selected with 1 μg/mL puromycin for one week. The oligonucleotides of shRNA: Human SLC25A51 shRNA-1: 5’-CCGGGCACTGAAATGTCATGGAATTCTCGAGAATTCCATGACATTTCAGTGCTTTTTG-3’; Human SLC25A51 shRNA-2: 5’-CCGGGCACTTATGTTTGGTCTGTATCTCGAGATACAGACCAAACATAAGTGCTTTTTG-3’; Mouse SLC25A51 shRNA-1: 5’-CCGGGCTCGAATACAGTCTCAGATTCTCGAGAATCTGAGACTGTATTCGAGCTTTTTG-3’; Mouse SLC25A51 shRNA-2: 5’-CCGGCGATAAGTTCACAAACACTTACTCGAGTAAGTGTTTGTGAACTTATCGTTTTTG-3’.
To transiently silence SIRT4 or SIRT5, the siRNA was transfected into cells using Lipofectamine RNAiMAX (Thermo Fisher Scientific), and cells were harvested after 48 h of transfection. The oligonucleotides of siRNA: Human SIRT4: CGAUUGCAAUACUGAACAUTT. Human SIRT5: CGUCCACACGAAACCAGAUTT.
Immunoprecipitation and co-immunoprecipitation
Harvested cells were lysed in Flag lysis buffer [50 mmol/L Tris-HCl (pH 7.9), 137 mmol/L NaCl, 1% Triton X-100, 0.2% Sarkosyl, 1 mmol/L NaF, 1 mmol/L Na3VO4, and 10% glycerol] containing protease inhibitor cocktail (Selleck), 1 mmol/L dithiothreitol and 1 mmol/L phenylmethyl sulfonyl fluoride. The supernatants were incubated with anti-Flag M2 agarose beads or anti-acetyllysine antibody agarose beads overnight at 4 °C. After washed with lysis buffer for five times, proteins were eluted via boiling with 1 × loading buffer and then analyzed by western blotting.
For co-immunoprecipitation, cells were lysed in BC100 buffer [20 mmol/L Tris-HCl (pH 7.9), 100 mmol/L NaCl, 0.2% NP-40, and 20% glycerol] containing protease inhibitor cocktail (Selleck), 1 mmol/L dithiothreitol and 1 mmol/L phenylmethyl sulfonyl fluoride). The whole cell lysates were incubated with 2 μg of anti-SIRT3 antibody at 4 °C overnight, and further with Protein A/G agarose beads for another 2 hours at 4 °C. After washed with lysis buffer for five times, the proteins were eluted via boiling with 1 × loading buffer and then analyzed by western blotting.
Pan-Acetyllysine antibody immunoprecipitated proteomics analysis
Wild type or SLC25A51 knockdown LoVo cells from 10 cm dishes were harvested and purified mitochondria via the Cell Mitochondria Isolation Kit (C3601, Beyotime). The mitochondria pellets were then lysed in Flag lysis buffer. Anti-acetyllysine antibody agarose (ICP0388, Immunechem) was mixed with equal amount of proteins (around 400 μg) overnight at 4 °C. Eluted acetylated proteins were detected by semiquantitative LC-MS/MS (Q-Exactive HF, Peking university medical and health analysis center).
In vitro deacetylation assay
HA-SIRT3 was purified from HEK293T cells and the hyper-acetylated P5CS-Flag was purified from HEK293T cells co-transfected with CBP. The reaction mixture contained 10 × Ac-K Buffer (200 mM pH8.0 HEPES,10 mM DTT, 10 mM PMSF and 1 mg/ml BSA), 2 × De-Ac-K Buffer (8 mM MgCl2, 100 mM NaCl and 20% glycerol), and hyper-acetylated P5CS. SIRT3 and NAD+ (1 mM) were present or absent as indicated. The reaction mixture was incubated at 37 °C for 2 hours and then boiled in the SDS sample buffer followed by analysis via western blotting.
Colony formation and cell proliferation assay
For colony formation assay, 2 × 103 cells were seeded into 6 cm dishes and cultured for one week. Then, cells were fixed by 4% paraformaldehyde for 15 minutes and stained with 0.2% crystal violet solution for 10 minutes.
For cell proliferation assay, 1 × 105 cells were seeded into six-well plates. Cells were digested with trypsin and counted at 1-, 2-, 3- and 4-days post seeding via Countstar IC1000 (Inno-Alliance Biotech) stained with Trypan Blue.
RNA isolation and Real-time quantitative PCR
TRI Reagent (Sigma-Aldrich)) was used to extract total RNA from cells or tumors. Then, 2 μg total RNA was reversely transcribed into cDNA using the cDNA Synthesis Kit (Yeasen). The cDNA was diluted for 5-fold and mixed with qPCR SYBR Green Master Mix (Yeasen) and primer pairs. The Real-time quantitative PCR was performed on 7500 Real-time PCR System (Applied Biosystems). The primers used for qPCR: human SLC25A51 Fwd: TACCAACACTTACCAGGCTTTCA, Rev: CAAGACATTGCTGAGTCCATTCC. Human GAPDH Fwd: GTCTCCTCTGACTTCAACAGCG, Rev: ACCACCCTGTTGCTGTAGCCAA.
Measurement of mitochondrial NAD+ level
Cells in 10 cm dishes were harvested and the mitochondria was purified with the Cell Mitochondria Isolation Kit (C3601, Beyotime) according to the manufacturer’s instructions. The mitochondrial NAD+ content was measured by the CheKine™ NAD(H) Assay Kit (KTB1020, Abbkine) according to the manufacturer’s instructions.
Amino acid analysis
Wild type and SLC25A51 knockdown LoVo cells in 10 cm dishes were harvested, and 1 × 107 cells for each group were pelleted via centrifuged at 1000 g for 5 minutes at 4 °C. The cells were resuspended in 300 μL distilled water, and free thawed twice with −80 °C freezer. The cellular amino acids contents were analyzed by an automated amino acid analyzer (L-8900, Hitachi) with the specific ion-exchange resin.
Measurement of proline content
Cells in 10 cm dishes were digested with trypsin and 5 × 106 cells were collected for the measurement of proline content with the CheKine™ Micro Proline (PRO) Assay Kit (KTB1430) according to the manufacturer’s instructions.
Molecular docking
The docking was conducted using Schrödinger software package. The predicted 3D structure of SLC25A51 was retrieved from the Alphafold2 (Identifier: AF-Q9H1U9-F1) and prepared by Protein Preparation Wizard and all settings were kept default. The active sites of SLC25A51 were identified and characterized by SiteMap module from Schrödinger package, which were consistent with the recent report [13]. The FDA-approved drugs were prepared using LigPrep. The ligand docking was performed by Glide in the standard precision (SP) and extra precision (XP) modes. After docking, the top 10 hits with favorable conformations were chosen for further validation and visualized by PyMOL.
Microscale thermophoresis analysis
The binding affinity of fludarabine phosphate with SLC25A51, and SIRT3 with P5CS was measured by NanoTemper Monolith NT.115 instrument (NanoTemper Technologies, Germany). SLC25A51-EGFP and labeled SIRT3 were diluted with binding buffer (50 mM tri-HCl, pH 7.5, 10 mM KCl, 5 mM MgCl2 and 0.005% Tween 20) to ensure that the fluorescence intensity during the analysis was about 500 RU. After pre-test, the final concentration of SLC25A51 and SIRT3 after mixing with the compound was chosen at 100 nM.
SLC25A51-EGFP was purified from HEK293T cells. Fludarabine phosphate was serially diluted in the binding buffer (16 points, 1:2 dilutions, started at 200 μM), and then mixed and incubated with an equal volume of the diluted SLC25A51 at room temperature for 5 minutes.
SIRT3-6xhis was obtained from Cloud-Clone (APE913Hu01). SIRT3-6xhis was labeled using His-Tag Labeling Kit at room temperature for 30 minutes. P5CS-Flag was purified from HEK293T cells and serially diluted in the binding buffer (16 points, 1:2 dilutions, started at 500 nM), and then mixed and incubated with an equal volume of the diluted SIRT3-6xhis at room temperature for 5 minutes.
After incubation, the mixture was loaded into the premium treated capillaries and measured in NanoTemper Monolith NT.115 instrument (NanoTemper Technologies, Germany). The KD values were fitted by NanoTemper Monolith affinity software (NanoTemper Technologies, Germany) using 1:1 binding mode.
Cellular thermal shift assay
The assay was performed as described previously [55]. LoVo cells in 10 cm dishes were collected and lysed with RIPA lysis buffer with protease inhibitor cocktail. The cell lysates were centrifuged at 15,000 g for 15 minutes at 4 °C, and the supernatants were aliquoted into two, one was incubated with 40 μM Fludarabine phosphate while the other one was incubated with DMSO for 1 hour at room temperature. The respective lysates were divided into the smaller (50 μL) aliquots and heated individually at different temperatures (°C) for 5 minutes followed by cooling for 3 minutes at room temperature. The heated lysates were centrifuged at 15,000 g for 20 minutes at 4 °C, and the supernatants were analyzed by western blotting analysis.
Immunochemistry
59 pairs of colorectal tumor tissues and normal adjacent tissues were obtained from Department of Gastroenterological Surgery, Peking University People’s Hospital. Tissue samples were sectioned at the Servicebio, Wuhan. The sections were processed for deparaffinization and rehydration followed by antigen retrieval. To inactivate endogenous peroxidase, the whole section was covered in 3% H2O2 for 15 minutes. Next, slides were incubated with the SLC25A51 antibody (CUSABIO, 1:100), followed by secondary antibodies reaction (Servicebio). Subsequently, the slides were developed with 3, 3’-diaminobenzidine staining (ZSGB-Biotech) and counterstained with hematoxylin. The protein level SLC25A51 was classified into five groups according to the staining intensity. Grade I: low staining marked as 1; Grade II: faint staining as 2; Grade III: moderate staining as 3, Grade IV: strong staining as 4, Grade V: stronger staining as 5.
Immunofluorescent staining
For imaging of fixed cells, cells were seeded into cover glasses in the 6-well plate. After treatment, the cells were fixed in 4% Paraformaldehyde (PFA) and permeabilized by 0.3% Triton X-100 in PBS. The cells were then blocked with PBS containing 3% goat serum and 5% BSA, followed by primary antibodies incubation overnight at 4 °C. After washing with PBS with 0.1% Triton X-100, cells were incubated with secondary antibodies for 1 hour at 37 °C. Finally, cells were incubated with DAPI and mounted to slides.
For immunofluorescent staining of tumor tissue sections, tumors were fixed in 4% Paraformaldehyde (PFA), dehydrated in 30% sucrose and then embedded in optimal cutting temperature compound (O.C.T., Sakura) which were immediately frozen at −80 °C freezer. The tumor blocks were cryo-sectioned into 8 μm sections via Leica Cryostat (Leica CM1950). Tumor sections were washed twice with TBS, followed by antigen retrieval solution treatment (Solarbio). Then the tissue sections were blocked in the TBS containing 1% BSA, 10% goat serum for 1 hour at 37 °C and incubated with anti-Ki67 antibody (Abcam) overnight at 4 °C. Then, the sections were incubated with secondary antibodies for 1 hour at 37 °C, followed by DAPI incubation for 15 minutes, and finally mounted to slides. All immunofluorescent imaging was performed on ZEISS LSM 880.
In vivo mice experiments
Mice were inoculated with ten million HCT116 cells or two million CT26 cells in 100 μL PBS at fat pad of mammary gland. The tumor volume was measured manually with vernier caliper every other day until day 20 or so and the tumor volume was calculated as length (mm) × width (mm2) × 0.5. At the last day, the tumors were dissected and weighed after mice were sacrificed.
For drug administration, tumor bearing mice were treated intraperitoneally with aspirin (50 mg/kg) alone, fludarabine phosphate (100 mg/kg) alone, or the combination of fludarabine phosphate (100 mg/kg) and aspirin (50 mg/kg) at indicated days after tumor inoculation. For safety monitor, aspirin alone, fludarabine phosphate alone or the combination of fludarabine phosphate and aspirin were intraperitoneally given every day, and the body weight of mice was recorded.
EdU incorporation assay
Wild type and SLC25A51 knockdown cells were seeded into 12-well plates and cells were incubated with EdU for 2 hours when reaching 80% confluence, followed by fixed with 4% paraformaldehyde for 15 minutes, and permeated with 0.3% Triton X-100 for 15 minutes. The EdU staining was performed via BeyoClick™ EdU Cell Proliferation Kit with Alexa Fluor 488 (C0071S, Beyotime), followed by DAPI incubation for 10 minutes. The immunofluorescent imaging was performed on ZEISS LSM 880.
Cell viability assay
Wild type or SLC25A51 knockdown cells were seeded in 96-well plates at 1 × 104 cells per well, and grew overnight. After incubating with fludarabine phosphate or aspirin or the combination for 24 hours, cell viability assay was performed using Cell Counting Kit-8 (CCK8) (Dojindo), according to the manufacturer’s instruction.
Bioinformatics analysis
For expression analysis based on TCGA database, the mRNA level of SLC25A51 in COAD, READ and other types of cancer and normal tissue by RNA-sequencing were downloaded from UCSC Xena (https://xenabrowser.net/datapages/). Data of tumor tissue were from TCGA, while data of normal tissue were from TCGA and GTEx. The RNA-seq data in TPM (transcripts per million reads) format were log2 normalized before comparing. The two groups were performed by independent samples t-test. P values less than 0.05 were considered statistically significant (ns, p ≥ 0.05; *, p < 0.05; **, p < 0.01; ***, p < 0.001). The ggplot2 package in R was used to identify the relationship between SLC25A51 expression and clinical features based on TCGA database. The expression of SLC25A51 was used as independent variable types, while the dependent variable was clinical characteristics. The Kaplan–Meier curve was used to compare the differences in overall survival between patients with high expression and low expression of SLC25A51 using the Log rank test with “survival” and “survminer” packages in R.
For expression analysis based on GEO database, the expression values of SLC25A51 in normal and tumor tissues from GSE20916 were retrieved via the online tool GEO2R (NCBI). For survival analysis, patients from GSE17536 were divided into high or low risk group according to the SLC25A51 expression values, and the overall survival curve was analyzed by Log rank test.
Statistical analyses
The experiments were performed three times independently and the repeated experiments showed similar results. Statistical analyses were performed with a two-tailed unpaired Student t test by Graphpad prism 8.0. Data were presented as mean ± standard deviation (S.D.). A difference was considered statistically significant at a value of p < 0.05. p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001 p < 0.0001, which were considered statistically significant.
Data availability
The RNA-seq data was submitted to NCBI SRA database (BioProject ID: PRJNA953609). The full and uncropped western blots were uploaded as the Supplemental Material. Other datasets used and/or analyzed during the study are available from the corresponding author on reasonable request.
References
Berger F, Ramirez-Hernandez MH, Ziegler M. The new life of a centenarian: signalling functions of NAD(P). Trends Biochem Sci. 2004;29:111–8.
Kim MY, Zhang T, Kraus WL. Poly(ADP-ribosyl)ation by PARP-1: ‘PAR-laying’ NAD+ into a nuclear signal. Genes Dev. 2005;19:1951–67.
North BJ, Verdin E. Sirtuins: Sir2-related NAD-dependent protein deacetylases. Genome Biol. 2004;5:224.
Chini EN, Chini CCS, Espindola Netto JM, de Oliveira GC, van Schooten W. The Pharmacology of CD38/NADase: An Emerging Target in Cancer and Diseases of Aging. Trends Pharm Sci. 2018;39:424–36.
Chiarugi A, Dolle C, Felici R, Ziegler M. The NAD metabolome-a key determinant of cancer cell biology. Nat Rev Cancer. 2012;12:741–52.
Magni G, Di Stefano M, Orsomando G, Raffaelli N, Ruggieri S. NAD(P) biosynthesis enzymes as potential targets for selective drug design. Curr Med Chem. 2009;16:1372–90.
Pramono AA, Rather GM, Herman H, Lestari K, Bertino JR. NAD- and NADPH-Contributing Enzymes as Therapeutic Targets in Cancer: An Overview. Biomolecules. 2020;10:358.
Chowdhry S, Zanca C, Rajkumar U, Koga T, Diao Y, Raviram R, et al. NAD metabolic dependency in cancer is shaped by gene amplification and enhancer remodelling. Nature. 2019;569:570–5.
Ghanem MS, Monacelli F, Nencioni A. Advances in NAD-Lowering Agents for Cancer Treatment. Nutrients. 2021;13:1665.
Mitchell SR, Larkin K, Grieselhuber NR, Lai TH, Cannon M, Orwick S, et al. Selective targeting of NAMPT by KPT-9274 in acute myeloid leukemia. Blood Adv. 2019;3:242–55.
Yang H, Yang T, Baur JA, Perez E, Matsui T, Carmona JJ, et al. Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell. 2007;130:1095–107.
Luongo TS, Eller JM, Lu MJ, Niere M, Raith F, Perry C, et al. SLC25A51 is a mammalian mitochondrial NAD(+) transporter. Nature. 2020;588:174–9.
Kory N, Uit de Bos J, van der Rijt S, Jankovic N, Güra M, Arp N, et al. MCART1/SLC25A51 is required for mitochondrial NAD transport. Sci Adv. 2020;6:eabe5310.
Girardi E, Agrimi G, Goldmann U, Fiume G, Lindinger S, Sedlyarov V, et al. Epistasis-driven identification of SLC25A51 as a regulator of human mitochondrial NAD import. Nature Commun. 2020;11:6145.
Shvedunova M, Akhtar A. Modulation of cellular processes by histone and non-histone protein acetylation. Nat Rev Mol Cell Biol. 2022;23:329–49.
Guan KL, Xiong Y. Regulation of intermediary metabolism by protein acetylation. Trends Biochem Sci. 2011;36:108–16.
Ahn B-H, Kim H-S, Song S, Lee IH, Liu J, Vassilopoulos A, et al. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc Natl Acad Sci. 2008;105:14447–52.
Alhazzazi TY, Kamarajan P, Verdin E, Kapila YL. SIRT3 and cancer: Tumor promoter or suppressor? Biochimica et Biophysica Acta. 2011;1816:80–8.
Verdin E, Hirschey MD, Finley LW, Haigis MC. Sirtuin regulation of mitochondria: energy production, apoptosis, and signaling. Trends Biochem Sci. 2010;35:669–75.
Camacho-Pereira J, Tarrago MG, Chini CCS, Nin V, Escande C, Warner GM, et al. CD38 Dictates Age-Related NAD Decline and Mitochondrial Dysfunction through an SIRT3-Dependent Mechanism. Cell Metab. 2016;23:1127–39.
Rardin MJ, He W, Nishida Y, Newman JC, Carrico C, Danielson SR, et al. SIRT5 regulates the mitochondrial lysine succinylome and metabolic networks. Cell Metab. 2013;18:920–33.
Chen Y, Zhang J, Lin Y, Lei Q, Guan KL, Zhao S, et al. Tumour suppressor SIRT3 deacetylates and activates manganese superoxide dismutase to scavenge ROS. EMBO Rep. 2011;12:534–41.
Dikalova AE, Itani HA, Nazarewicz RR, McMaster WG, Flynn CR, Uzhachenko R, et al. Sirt3 Impairment and SOD2 Hyperacetylation in Vascular Oxidative Stress and Hypertension. Circ Res. 2017;121:564–74.
Lei MZ, Li XX, Zhang Y, Li JT, Zhang F, Wang YP, et al. Acetylation promotes BCAT2 degradation to suppress BCAA catabolism and pancreatic cancer growth. Signal Transduct Target Ther. 2020;5:70.
Zhang F, Wang D, Li J, Su Y, Liu S, Lei QY, et al. Deacetylation of MTHFD2 by SIRT4 senses stress signal to inhibit cancer cell growth by remodeling folate metabolism. J Mol Cell Biol. 2022;14:1–12.
Li Y, Bie J, Song C, Liu M, Luo J. PYCR, a key enzyme in proline metabolism, functions in tumorigenesis. Amino Acids. 2021;53:1841–50.
Hu CA, Khalil S, Zhaorigetu S, Liu Z, Tyler M, Wan G, et al. Human Delta1-pyrroline-5-carboxylate synthase: function and regulation. Amino Acids. 2008;35:665–72.
Chen S, Yang X, Yu M, Wang Z, Liu B, Liu M, et al. SIRT3 regulates cancer cell proliferation through deacetylation of PYCR1 in proline metabolism. Neoplasia. 2019;21:665–75.
Zhong J, Guo CJ, Zhou X, Chang CC, Yin B, Zhang T, et al. Structural basis of dynamic P5CS filaments. Elife 2022;11:e76107.
Murphy JM, Murch SJ, Ball RO. Proline is synthesized from glutamate during intragastric infusion but not during intravenous infusion in neonatal piglets. J Nutr. 1996;126:878–86.
Koppula P, Zhuang L, Gan B. Cystine transporter SLC7A11/xCT in cancer: ferroptosis, nutrient dependency, and cancer therapy. Protein Cell. 2021;12:599–620.
Krishnan N, Dickman MB, Becker DF. Proline modulates the intracellular redox environment and protects mammalian cells against oxidative stress. Free Radic Biol Med. 2008;44:671–81.
Westbrook RL, Bridges E, Roberts J, Escribano-Gonzalez C, Eales KL, Vettore LA, et al. Proline synthesis through PYCR1 is required to support cancer cell proliferation and survival in oxygen-limiting conditions. Cell Rep. 2022;38:110320.
Ding Z, Ericksen RE, Escande-Beillard N, Lee QY, Loh A, Denil S, et al. Metabolic pathway analyses identify proline biosynthesis pathway as a promoter of liver tumorigenesis. J Hepatol. 2020;72:725–35.
Alaqbi SS, Burke L, Guterman I, Green C, West K, Palacios-Gallego R, et al. Increased mitochondrial proline metabolism sustains proliferation and survival of colorectal cancer cells. PLoS One. 2022;17:e0262364.
Luo J, Manning BD, Cantley LC. Targeting the PI3K-Akt pathway in human cancer: rationale and promise. Cancer Cell. 2003;4:257–62.
Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22.
Varadi M, Anyango S, Deshpande M, Nair S, Natassia C, Yordanova G, et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022;50:D439–44.
Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–9.
Chun HG, Leyland-Jones B, Cheson BD. Fludarabine phosphate: a synthetic purine antimetabolite with significant activity against lymphoid malignancies. J Clin Oncol. 1991;9:175–88.
Molina DM, Jafari R, Ignatushchenko M, Seki T, Larsson EA, Dan C, et al. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science. 2013;341:84–7.
Lecomte M, Laneuville O, Ji C, DeWitt DL, Smith WL. Acetylation of human prostaglandin endoperoxide synthase-2 (cyclooxygenase-2) by aspirin. J Biol Chem. 1994;269:13207–15.
Marimuthu S, Chivukula RS, Alfonso LF, Moridani M, Hagen FK, Bhat GJ. Aspirin acetylates multiple cellular proteins in HCT-116 colon cancer cells: Identification of novel targets. Int J Oncol. 2011;39:1273–83.
Moreira JD, Hamraz M, Abolhassani M, Bigan E, Peres S, Pauleve L, et al. The Redox Status of Cancer Cells Supports Mechanisms behind the Warburg Effect. Metabolites 2016;6:33.
Demarest TG, Babbar M, Okur MN, Dan X, Croteau DL, Fakouri NB, et al. NAD+ metabolism in aging and cancer. Annu Rev Cancer Biol. 2019;3:105–30.
Grever M, Leiby J, Kraut E, Metz E, Neidhart J, Balcerzak S, et al. A comprehensive phase I and II clinical investigation of fludarabine phosphate. Semin Oncol. 1990;17:39–48.
Cho H, Cho Y-Y, Shim MS, Lee JY, Lee HS, Kang HC. Mitochondria-targeted drug delivery in cancers. Biochimica et Biophysica Acta. 2020;1866:165808.
Zuo Z, He L, Duan X, Peng Z, Han J. Glycyrrhizic acid exhibits strong anticancer activity in colorectal cancer cells via SIRT3 inhibition. Bioengineered. 2022;13:2720–31.
Sun S, Shi J, Wang X, Huang C, Huang Y, Xu J, et al. Atractylon inhibits the tumorigenesis of glioblastoma through SIRT3 signaling. Am J Cancer Res. 2022;12:2310–22.
Li M, Chiang Y-L, Lyssiotis CA, Teater MR, Hong JY, Shen H, et al. Non-oncogene Addiction to SIRT3 Plays a Critical Role in Lymphomagenesis. Cancer Cell. 2019;35:916–31.e919.
Thun MJ, Jacobs EJ, Patrono C. The role of aspirin in cancer prevention. Nat Rev Clin Oncol. 2012;9:259–67.
Li W, Sauve AA. NAD+ content and its role in mitochondria. Methods Mol. Biol. 2015;1241:39–48.
Ducker GS, Rabinowitz JD. One-carbon metabolism in health and disease. Cell Metab. 2017;25:27–42.
Walz S, Lorenzin F, Morton J, Wiese KE, von Eyss B, Herold S, et al. Activation and repression by oncogenic MYC shape tumour-specific gene expression profiles. Nature. 2014;511:483–7.
Peng Y, Zhang X, Zhang T, Grace PM, Li H, Wang Y, et al. Lovastatin inhibits Toll-like receptor 4 signaling in microglia by targeting its co-receptor myeloid differentiation protein 2 and attenuates neuropathic pain. Brain Behav Immun. 2019;82:432–44.
Acknowledgements
We thank Zhanlong Shen (Peking University People’s Hospital), Xinshan Ye (Peking university), Xiaoyan Qiu (Peking University) and Jiadong Wang (Peking University) for providing us cell lines. We thank Yaxin Lou (Peking University) for the assistance of mass spectrometry analysis; Lihua An (Peking university) for the analysis of amino acids contents; Liping Liu (Shanghai Institute of Materia Medica) for the assistance of molecular docking; Qian Wang (Peking University) for the assistance of MST assay. This work was supported by grants from National Natural Science Foundation of China (82172959, 81874147 and 81671389).
Author information
Authors and Affiliations
Contributions
YL, JB and JL designed the research. YL, JB and CS performed most of the experiments. TZ and ML assisted with the cell culture. LZ and CY collected the tumor samples.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics
This study was approved by the Ethics Committee of the Peking University Health Science Center.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, Y., Bie, J., Zhao, L. et al. SLC25A51 promotes tumor growth through sustaining mitochondria acetylation homeostasis and proline biogenesis. Cell Death Differ 30, 1916–1930 (2023). https://doi.org/10.1038/s41418-023-01185-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-023-01185-2
This article is cited by
-
Single-cell profiling of SLC family transporters: uncovering the role of SLC7A1 in osteosarcoma
Journal of Translational Medicine (2025)
-
BACH1 deficiency improves placental angiogenesis via SLC25A51-mediated mitochondrial NAD+ transport in intrahepatic cholestasis of pregnancy
Molecular Medicine (2025)
-
Mitochondrial NAD+ transporter SLC25A51 linked to human aortic disease
Nature Cardiovascular Research (2025)
-
Histidine triad nucleotide-binding protein 2 attenuates metabolic dysfunction-associated steatotic liver disease through NAD+-dependent sirtuin-3 activation
Experimental & Molecular Medicine (2025)
-
P5CS deacetylation mediated by SIRT2 facilitates tumor growth by enhancing mitochondrial respiration in hepatocellular carcinoma
Oncogene (2025)