Abstract
With the development of public health, female diseases have become the focus of current concern. The unique reproductive anatomy of women leads to the development of gynecological diseases gradually become an important part of the socio-economic burden. Epigenetics plays an irreplaceable role in gynecologic diseases. As an important mRNA modification, m6A is involved in the maturation of ovum cells and maternal-fetal microenvironment. At present, researchers have found that m6A is involved in the regulation of gestational diabetes and other reproductive system diseases, but the specific mechanism is not clear. In this manuscript, we summarize the components of m6A, the biological function of m6A, the progression of m6A in the maternal-fetal microenvironment and a variety of gynecological diseases as well as the progression of targeted m6A treatment-related diseases, providing a new perspective for clinical treatment-related diseases.
Similar content being viewed by others
Facts
-
m6A Modification in Female Reproduction: The role of N6-methyladenosine (m6A) in female reproduction is an emerging field, with evidence suggesting its involvement in oocyte maturation, embryonic development, and placental function, which are crucial for maternal-fetal health.
-
Epigenetic Regulation in Gynecological Diseases: m6A modification has been implicated in various gynecological diseases, including cervical cancer, ovarian cancer, endometrial cancer, and choriocarcinoma, potentially affecting their progression and treatment outcomes.
-
Technological Advances in m6A Detection: Recent developments in high-throughput sequencing and other detection technologies have expanded our ability to study m6A modifications at single-base resolution, offering new opportunities for understanding the mechanisms of gene regulation.
-
m6A’s Impact on Immune Cells: The influence of m6A on immune cells within the maternal-fetal microenvironment, such as dendritic cells, natural killer cells, and macrophages, suggests a significant role in modulating immune tolerance and responses during pregnancy.
-
Clinical Implications and Challenges: The potential of m6A as a biomarker and therapeutic target and biomarker in gynecological diseases is evident, but the complexity of its regulatory mechanisms and the need for a comprehensive understanding of its role in different pathological contexts present challenges for clinical application.
Open Questions
-
How does m6A modification specifically regulate the balance between immune tolerance and response in the maternal-fetal microenvironment, and what are the long-term implications for both maternal and fetal health?
-
What are the precise molecular mechanisms by which m6A modification influences the progression of various gynecological cancers, and how can this knowledge be leveraged to develop targeted therapies?
-
Given the recent advancements in m6A detection technologies, what new insights can be gained about the role of m6A in non-malignant gynecological conditions such as endometriosis and polycystic ovary syndrome?
-
Can m6A modification patterns serve as reliable biomarkers for the diagnosis, prognosis, and treatment response monitoring in gynecological diseases, and what validation is needed to implement them in clinical practice?
-
How do environmental factors and lifestyle choices impact m6A methylation patterns in the context of female reproduction and gynecological health, and what preventive strategies can be developed based on this knowledge?
Introduction
Epitranscriptomics refers to the dynamic and reversible process of chemically changing several kinds of RNA molecules, including mRNAs and non-coding RNAs. These modifications play a critical role in governing RNA functionality and determining its fate, as well as influencing the gene expression process [1, 2]. The discovery of over 170 RNA alterations in various organisms has led to an increasing number of studies being conducted to elucidate their precise pathways in the development of disease [2]. The primary epitranscriptomic modifications consist of N6-methyladenosine (m6A) [3], N1-methyladenosine (m1A) [4], 5-methylcytidine (m5C) [5], and pseudouridine (Ψ) [6]. Out of all the chemical modifications in eukaryotic messenger RNAs, m6A is regarded as the most prevalent [3]. The particular mechanism of m6A modification in RNA involves the insertion of a methyl group (CH3) at the N6 position of adenine A in the RNA molecule. As an essential element of the epigenetic, m6A modification dynamically regulates multiple aspects of gene expression, containing RNA splicing, stability, translation, and degradation. Furthermore, m6A modification interacts closely with other epigenetic mechanisms, including DNA methylation and histone alterations, collectively contributing to the precise regulation of gene expression. DNA methylation is typically correlated to gene silencing, while m6A modification and histone modifications primarily center on post-transcriptional regulation. The coordinated interplay of these three modifications plays an essential role in biological processes, regulating cellular fate, immune responses, and the development of various diseases [7, 8].
Women’s reproductive health is a crucial component of societal progress [9]. Currently, due to the fast-paced socio-economic progress, there is a heightened global focus on reproductive health. However, a significant number of women continue to experience various reproductive health issues, including gynecological cancer, pregnancy-related ailments, infertility, gynecological chronic diseases, and more. These conditions can have detrimental effects on women’s physical health, family life, and economic well-being, and in severe cases, they can even be life-threatening [10, 11]. Hence, comprehending the mechanism and target of associated diseases is crucial for resolving issues pertaining to clinical treatment.
The study of epigenomics has steadily confirmed the connection between m6A modification and the physiological and pathological processes in the female reproductive system [12] (Fig. 1). As an illustration, it has been elucidated that m6A modification has a substantial influence on follicular development and oocyte growth. Based on RNA-seq and Me RIP-seq results, researchers discovered that Itsn2 is targeted for m6A modification by METTL3, thereby enhancing its stability and then affecting the oocyte meiotic process [12, 13]. Itsn2 is a critical adapter protein that has a significant role in regulating oocyte meiotic resumption [14]. The presence of m6A modification in ovarian tissue of individuals diagnosed with polycystic ovary syndrome (PCOS) is dysregulated, which disrupts follicular development and growth, ultimately leading to ovarian dysfunction and the pathogenesis of PCOS. This process involves the alteration of key signaling pathways, including PI3K/Akt and mTOR, which play critical roles in regulating ovarian function and folliculogenesis [15]. m6A regulators appear to be involved in reproductive aging. Sun et al. evaluated the levels of expression of FTO and m6A methylation levels within various germ cells and found that FTO expression gradually decreased but m6A showed an increasing trend during the aging process [16]. In order to ascertain the precise mechanism at play, additional research is required. Furthermore, pertinent investigations have suggested that m6A additionally contributes to mechanisms associated with gynecological cancer [17,18,19]. m6A modification regulates tumor growth and dissemination by regulating the expression levels of oncogenes and tumor suppressor genes. Consequently, clinical research may leverage m6A modification as a diagnostic and prognostic biomarker for various diseases affecting the female reproductive system, enabling personalized diagnosis and treatment through the identification of potential therapeutic targets. m6A modification is increasingly recognized for its significant role in female reproductive health. However, further investigation is essential to more comprehensively understand the correlation between m6A modification and female reproductive system function, as well as the deeper mechanisms involved.
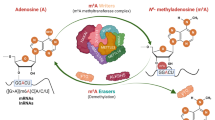
m6A writers (METTL3, METTL14, WTAP, RBM15/15B, VIRMA), m6A erasers (ALKBH5 and FTO), and m6A readers (YTH family protein and hnRNPs protein) are the primary regulators of the m6A modification. m6A methylation modification is involved in the regulation of alternative splicing, nuclear export, degradation, translation, and RNA stability. In the process of female reproduction, m6A RNA modification is also essential for oogenesis, ovulation, and embryonic development.
This manuscript presents an exhaustive review of m6A regulators and their biological function-related mechanisms, as well as the progress made in detecting m6A. Our emphasis is on the pivotal role of m6A modification in shaping and regulating the maternal-fetal immune microenvironment, as well as influencing the treatment and prognosis of obstetrics and gynecology-related diseases. We provide obstetricians and gynecologists with a new perspective to understand associated diseases more thoroughly, which will improve their future comprehension of these conditions.
m6A modification mechanism and detection method
m6A modification
m6A writer
The m6A writers refer to a group of enzymes or complexes that are mainly tasked with the methylation of the target. METTL3, METTL14, and Wilms’ tumor-associated protein (WTAP) will be the subject of this section [8]. For instance, the initial two are regulated by WTAP. Moreover, ancillary subunits, including RNA-binding motif protein 15 (RBM15), zinc finger CCCH ___domain protein 13 (ZC3H13), and vir-like m6A methyltransferase-associated subunit (VIRMA, also known as KIAA1429), are involved in the mechanism [20,21,22]. These methyltransferases constitute the methyltransferase complex (MTC). The MTC performs its function by introducing methyl groups to certain adenosine residues in RNA, resulting in m6A modifications [8].
METTL3 is the initial m6A writer identified as possessing the only catalytic subunit that helps methyl transfer happen when it binds to S-Adenosyl Methionine (SAM), a cellular methyl donor. METTL3 is mainly localized in the nucleus, although it is also existing in the cytoplasm [2, 8, 23]. It is pivotal in controlling the cell cycle through the modulation of key gene expression required for cell division. Within the tumor microenvironment, METTL3 contributes to immune evasion and tumor angiogenesis by regulating the secretion of immune cells and cytokines. Additionally, METTL3 has been demonstrated to facilitate the initiation and progression of gynecologic cancers through the modulation of several critical signaling pathways such as the PI3K/Akt and Wnt/β-catenin [17]. METTL14 possesses a SAM-dependent methyltransferase ___domain and an RNA-binding ___domain, which is structurally similar to METTL3 [24]. METTL14 indirectly contributes to the catalytic process by binding to METTL3 and stabilizing its conformation [23]. WTAP is a crucial member of the MTC, which regulates RNA m6A modification by binding to RNA molecules and interacting with proteins. Despite the absence of methyltransferase activity, WTAP facilitates the localization of the METTL3-METTL14 complex to nuclear speckles, thereby enhancing its catalytic activity [20, 25]. The RBM15/15B and KIAA1429 attach to the MTC and guide its correct localization. The former functions as an adaptor protein, attracting MTC to U-rich regions for m6A modification [21], whereas the latter mainly guides m6A modification adjacent the 3ʹ untranslated region (3ʹ UTR) and stop codon [22].
Furthermore, WTAP and ZC3H13 are predominantly localized in the nucleus, where they modulate post-transcriptional modifications of RNA. As essential regulators of m6A methylation, these factors hold potential as novel therapeutic targets for treating gynecological cancers. ZC3H13 plays a critical auxiliary function in the MTC by anchoring and mediating nuclear m6A modification through interaction with WTAP, thereby enhancing the accuracy and efficiency of m6A modification [21, 26]. METTL5 and tRNA methyltransferase activator subunit 112 (TRMT112) mainly involved in rRNA m6A modification. TRMT112 is the only m6A rRNA methyltransferase that contains a conserved ___domain and associates with other methyltransferases to exercise its supplementary impact [27].
The discovery of zinc finger CCHC type containing 4 (ZCCHC4) and METTL16 occurred recently [28, 29]. METTL16 contains a canonical SAM-dependent methyltransferase ___domain and an RNA-binding ___domain. The SAM binding ___domain supplies methyl groups, while the RNA binding ___domain enhances the efficiency and accuracy of m6A modification, hence enhancing the methyltransferase activity of METTL16 [29, 30]. ZCCHC4 possesses multiple CCHC-type zinc finger domains, allowing it to regulate the terminal RNA methylation modification by binding to specific RNA sequences [29, 31, 32]. Moreover, Bawankar et al. led a study on the impact of m6A in Drosophila, which indicates that a factor called HAKAI may affect the biological function of MTC by interacting with MTC subunits through its ubiquitination ___domain [33]. Further investigations are needed to ascertain the exact method and value of HAKAI in m6A modification.
m6A reader
m6A readers are a group of proteins that can recognize and bind to m6A modifications involved in multiple m6A-mediated biological processes, and they play irreplaceable roles in regulating RNA fate. The different binding modes of m6A allow us to divide m6A-binding proteins into two categories. First, RNA recognizes and binds the m6A modification directly, such as the YT521-B homology ___domain (YTH) family, eukaryotic translation initiation factor 3 (eIF3), and the insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs) family.
The YTHDF family protein possesses a highly conserved YTH ___domain, which contains a hydrophobic pocket composed of amino acids and serves as the structural component responsible for recognizing m6A [34, 35]. YTHDF1 is primarily localized in the cytoplasm, where it plays a crucial role in regulating RNA translation process [36]. In gynecologic tumors, YTHDF1 facilitates cancer progression by modulating genes associated with tumor proliferation and invasion, and it may also contribute to chemotherapy resistance. YTHDF2 can recruit mRNA degradation complexes, such as the CCR4-NOT complex, to impact the stability of RNA and accelerate the decline of targeted RNA [37, 38]. YTHDF3 contributes to the regulation of the first two [38]. YTHDC1 promotes regulated splicing by binding Serine/Arginine-Rich Splicing Factor 3 (SRSF3) and Serine/Arginine-Rich Splicing Factor 10 (SRSF10) [39, 40]. YTHDC2 plays a role in m6A modification-mediated nuclear export and RNA degradation [41].
According to pertinent research findings, the four K homology (KH) domains and two RNA recognition motifs (RRMs) of IGF2BPs, which are responsible for RNA recognition and binding, have a substantial impact on cellular stress responses. IGF2BPs are mainly found in the cytoplasm, where they regulate the expression of multiple oncogenes, thereby influencing the progression of reproductive tumors, chemotherapy resistance, and tumor immunity. IGF2BPs directly recognize and bind m6A-modified RNA via their KH ___domain [42]. This direct binding is highly selective and efficient since it specifically identifies and attaches to the m6A modification site.IGF2BPs associated with m6A modification mainly include IGF2BP1, IGF2BP2, and IGF2BP3, which have been demonstrated to contribute to RNA stability [43, 44], translational regulation [44], metabolic regulation [45], embryonic development [46], and cancer-related mechanisms [47], respectively. EIF3 is a multi-component structure consisting of 13 subunits, which plays an essential role in controlling the process of translation. The m6A mutation enables eIF3 binding at the 5ʹUTR region, thereby facilitating translation [48].
Besides, RNA indirectly identifies and binds to m6A modifications, including the heterogeneous nuclear ribonucleoproteins (HNRNPs) family, which consists of HNRNPC, HNRNPG, and HNRNPA2B1. HNRNPs, primarily located in the nucleus but also partially present in the cytoplasm, contribute to the proliferation and migration of tumor cells by modulating the translation of genes associated with the cell cycle [49]. HNRNPs play a critical regulatory role in gynecologic cancers. Targeting HNRNPs could provide a promising therapeutic approach to inhibit the progression of these malignancies. HNRNPs indirectly recognize m6A-modified RNAs through their RRM. m6A modification can change the secondary structure of RNA, increase the binding affinity of HNRNPC to transcripts, and reflect the conversion effect of m6A [50]. HNRNPA2B1 has been demonstrated to bind to m6A-modified RNA and enhance METTL3-dependent alternative splicing processes [51]. Relevant research indicates that HNRNPC/G also participates in alternative splicing of pre-RNAs through various mechanisms, which require specific elucidation in the future [52].
Fragile X Mental Retardation Protein (FMRP) and proline-rich coiled-coil 2 A (Prrc2a) may play a role in the regulation of RNA m6A modification regulatory networks. Researchers have found that they also contribute to neurodevelopment and spermatogenesis [53, 54]. Although these proteins do not belong to the category known as classical m6A reader proteins, they have the potential to help find new ways to treat diseases that are related.
m6A eraser
The m6A erasers refer to a group of demethylases that are accountable for the elimination of the m6A modification. With the combined involvement of Fe2+ and α-ketoglutarate, m6A methylation modification can be eliminated by ALKB homolog 5 (ALKBH5) and fat mass and obesity-associated protein (FTO) [55, 56]. FTO and ALKBH5 are localized predominantly in the nucleus, where they exert demethylation activity through the core catalytic ___domain, most notably the DSBH ___domain [57], recognizing and binding m6A-modified RNA. It has a significant impact on multiple critical stages in RNA metabolism, such as alternative splicing [55], stability [43] etc. Variants in the FTO gene have been reported to be correlated with the progress of cancer [58] and metabolic disorders like obesity [59]. Moreover, ALKBH5 plays a key role in regulating gene expression and germ cells function [56]. Recent researchers have revealed the biological role of ALKBH3 in removing the m6A modification of tRNAs [60]. As technology advances, the role of ALKBH3 in RNA modification will be further illuminated through additional details.
m6A detection technology
Early studies explored m6A modification in RNA by radiolabeling methods such as radioisotope labeling [7]. In addition, enzyme-linked immunosorbent assays (ELISA) utilize anti-m6A antibodies to detect m6A levels in RNA, mainly for quantitative analysis. Dot blot is a rapid method for detecting m6A modification in RNA samples. The specific mechanism is to provide a basis for subsequent finer quantitative analysis by spotting RNA samples on the membrane and detecting them using m6A-specific antibodies or other probes [61].
Traditional immunoassay techniques have advantages in ease of operation and rapid detection for measuring global levels of m6A modification in RNA samples. Mass spectrometry-based methods for the detection of RNA methylation, mainly including two-dimensional thin-layer chromatography (2D-TLC) and liquid chromatography tandem mass spectrometry (LC-MS/MS), are used for quantitative analysis and detection of the presence of m6A modifications [62, 63]. LC-MS/MS involves several steps. First, RNA samples are enzymatically hydrolyzed into nucleotides. Then, the m6A content is analyzed using liquid chromatography separation, followed by UV detection through mass spectrometry [64, 65]. The method is simple in its steps, but it cannot locate sequence information and carries the risk of contamination.
Researchers then created a technique called site-specific cleavage and radiolabeled linkage-assisted extraction and thin-layer chromatography (SCARLET), which combines the benefits of solid phase extraction and thin-layer chromatography to identify methylation at specific sites and make substantial progress in detecting RNA m6A modification [66]. The single-base extension and ligation-based qPCR amplification (SELECT) method is ideal for analyzing specific monomethylation patterns on target RNAs. It allows for the precise and quantitative identification of m6A modifications at the level of individual bases in transcripts that are present in low quantities [67].
High-throughput sequencing technologies have significantly enhanced the methodologies for localizing and quantifying m6A modifications. Methylated RNA Collaborative immunoprecipitation sequencing (MERIP-seq) is the first high-throughput sequencing technique based on m6A antibody immunoprecipitation applied to m6A modification detection, which is mainly suitable for m6A modification fragment detection with sizes of 100 nt to 200 nt [68]. However, the low resolution limits its potential application. m6A-seq has improved detection accuracy over MERIP-seq [69]. The m6A-seq2 method, which was recently updated, enhances the precision and reliability of m6A quantification at site, gene, and sample levels through the integration of multiple m6A immunoprecipitation methods, utilizing mixed samples and barcoding systems [70]. The m6A-LAIC-seq method, which adds a comparison step to adjacent unenriched RNA fragments, achieves quantification of m6A levels within the transcriptome range [71]. A recent study proposed a technique based on m6A modification and transcriptomic sequencing at the single-cell level. The m6A CUT &Tag (m6A-CT) and single-nucleus m6A CUT&Tag (sn-m6A-CT) methods identify m6A-modified RNAs through a CUT&Tag-based approach, offering technical support for an in-depth study of m6A modification and comprehensive transcriptome analysis [72].
Compared to MERIP-seq, photocrosslinking-assisted m6A sequencing (PA-m6A-seq) offers a higher resolution of nearly 23–30 nt. In this method, m6A antibodies and 4-thiouridine (4SU) are mixed to make antibody and m6A-modified RNA cross-linking reactions happen. This is done by shining UV365 mm ultraviolet light on them [73]. However, due to method limitations, it is only suitable for cell culture. m6A single nucleotide resolution crosslinking and immunoprecipitation (mi CLIP) and m6A crosslinking and immunoprecipitation (m6A-CLIP) are also assays for RNA m6A modification by UV254 photocrosslinking, and they achieve high-precision m6A localization at single base resolution [74,75,76]. Combining mi CLIP with high-throughput sequencing (mi CLIP-seq) further enhanced the detection efficiency for m6A.
Following this, researchers have developed several new methods to enhance CLIP technology, including light-activated ribonucleoside-enhanced CLIP (PAR-CLIP) [77], enhanced CLIP (eCLIP) [78], miCLIP2 combined with machine learning [79], and single-end enhanced CLIP (seCLIP-seq) [80]. These techniques compensate for the limitations of previous CLIP approaches and enable more precise and comprehensive studies of RNA-protein complexes. The m6A-cross-linked exoribonuclease sequencing (m6ACE) technology discovered in 2019 is an upgrade to the above technology, which performs accurate quantitative analysis of m6A RNA by combining photocrosslinking, exoribonuclease, and sequencing steps [81]. However, this method is plagued by exorbitant expenses and intricate analysis.
m6A-sensitive RNA-endonuclease-facilitated sequencing (m6A-REF-seq) is a method independent of antibodies, specific endonuclease-based RNA high-throughput sequencing technique that can be used to precisely localize and analyze m6A modifications in RNA [82]. MazF is an endonuclease that can find and cut ACA motifs that have been changed by non-m6A methylation. It can also find m6A sites at a single base and cut them. MazF-coupled RNA sequencing (MAZTER-seq) is also based on ACA sequence-specific cleavage in MazF-based RNA, which allows for measuring m6A levels at the single-base level [83]. Kate D. Meyer proposed a DART-seq method to quantitatively measure genome-wide RNA m6A modification using apolipoprotein B mRNA editing enzyme catalytic subunit 1 (APOBEC1) chimeric m6A-bound YTH domains to deaminate adjacent sites m6A in 2019 [84]. The technique is also antibody-independent and shows a high level of specificity. In 2022, the researchers upgraded the aforementioned methods to detect m6A modification at single-cell resolution [85].
N6-methyladenine labeling sequencing (m6A-label-seq) and m6A-selective allyl chemical labeling (m6A-SEAL) are two new chemical labeling methods that combine high-throughput sequencing methods for finding m6A without antibodies. The former relies on FTO enzymatic activity to transform m6A into N6-hydroxymethyladenosine (hm6A), which then transforms into N6-dithioglycolylmethyladenosine (dm6A) using biotin-labeled dm6A, subsequently undergoing enrichment and additional sequencing analysis [86]. The latter employs a specific chemical reaction to react with the m6A site, introduces an allyl group, and transforms the m6A-modified RNA site into an allyl modification. This process allows for its enrichment and further analysis by antibodies, ultimately determining the distribution and levels of the m6A modification [87].
In 2022, selective allyl chemical marker sequencing (m6A-SAC-seq) specifically labeled the m6A locus using dimethyl transferase MjDim1, and after a series of chemical reactions, the ___location and content of m6A in the transcriptome could be accurately identified [88]. In 2023, Liu et al. proposed glyoxal and nitrite-mediated deamination of unmethylated adenosines (GLORI), a high-throughput sequencing-based single-base detection method. The specific mechanism involves employing a reaction system utilizing glyoxal and nitrite as catalysts that transforms conventional adenosine deamination into inosine (A-to-I). During sequencing, isosine is then read as a G, while m6A modification sites are read as an A. Calculating the proportion of the latter allows us to quantify m6A at the single-base level [89].
Furthermore, nanopore sequencing and single-molecule real-time sequencing (SMRT sequencing) are two techniques to identify and quantify m6A modifications by direct sequencing technology [90, 91]. These methods enable high-resolution detection of m6A at the level of individual molecules, making them valuable tools for conducting detailed investigations into the mechanism and function of m6A modifications in RNA biology. Recently, researchers have enhanced the mentioned methodologies by incorporating machine learning algorithms to improve accuracy [92,93,94].
With technological advances, the detection of m6A modifications has become increasingly precise and efficient. The development of these technologies not only drives advances in epigenomics, but also provides a new perspective for revealing the function of RNA modifications in biological processes. It is worth emphasizing that while high-throughput detection methods offer extensive information on modifications, they are still hindered by challenges related to insufficient sensitivity and specificity. To enhance the reliability of data, researchers can develop more specific antibodies, integrate multiple detection technologies, and implement rigorous quality control measures. Furthermore, advancing data analysis algorithms, including the application of machine learning and artificial intelligence techniques to minimize errors, is a crucial strategy for improving the accuracy of overall analysis. The comprehensive application of these approaches will address current technological limitations and advance m6A research with greater precision.
m6A biological function
m6A and mRNA
m6A in mRNA splicing
Splicing of precursor mRNA (pre-mRNA) is a pivotal stage in gene expression, and m6A modification has a vital impact on alternative splicing. The m6A locus occurs more prevalent in pre-mRNA than in mature mRNA, with approximately four m6A residue levels of methylation present per pre-mRNA [95]. m6A methylation typically occurs in introns and exons undergoing alternative splicing [69]. YTHDC1 recognizes and binds m6A-modified mRNAs, recruiting proteins belonging to the SR (serine/arginine-rich) protein family, such as SRSF3, to drive exon inclusion. Meanwhile, it inhibits other splicing factors, such as SRSF10, to promote exon exclusion, thereby influencing the outcome of alternative splicing [96,97,98]. By binding to YTHDC1, METTL3 can indirectly affect alternative mRNA splicing.
METTL16 affects mRNA splicing by regulating intron SAM synthase retention and maintaining low levels of intracellular SAM [99]. The “m6A switch” mechanism describes the role of m6A modification which alters the local RNA structure to change the RNA molecule from a stable structure to a more easily unraveled structure. This mechanism can affect the accessibility of binding sites for HNRNP family proteins (e.g., HNRNPC and HNRNPG), thereby promoting or inhibiting the skipping process of exons [52, 100]. Serial studies in mouse preadipocytes and human 293 T cell lines have shown that FTO impacts splicing factor binding and splice site selection through its demethylation activity. In addition, demethylation of FTO can indirectly regulate alternative splicing of mRNA by altering the interaction between HNRNP family proteins and mRNA [55, 101].
m6A in mRNA nuclear export
m6A facilitates the process by which mRNA is transported into the cytoplasm for translation through multiple mechanisms. The process of exporting mRNA from the nucleus is done by the transcription-export complex (TREX), with assistance from the MTC. To promote mRNA nuclear export, the MTC recruits TREX to m6A-modified mRNAs and stimulates their interaction with YTHDC1 [41]. YTHDC1 binds to methylated mRNAs and regulates their subcellular localization. It aids the nuclear export of modified mRNAs by interacting with the nuclear export adaptor protein SRSF3 and the export receptor Nuclear RNA Export Factor 1(NXF1). Knockdown of YTHDC1 results in the retention of m6A-modified mRNA within the nucleus [39]. Additionally, FMRP has the ability to attach to the m6A-modified mRNA, thereby physically bind to the export protein Chromosome Maintenance Region 1 (CRM1), which promoting mRNA nuclear export. However, a conditional knockdown of Mettl14 resulted in blockage of the nuclear export process [102, 103]. There are currently few studies on the nuclear export of mRNAs, and the specific mechanism is still unknown.
m6A in mRNA translation
m6A modification is more prevalent in the 5 ʹUTR or 3ʹ UTR of mRNA and affects cyclization of mRNA, thereby promoting translation initiation. The m6A modification in the 5ʹ UTR region has the ability to bind directly to eIF3 and subsequently recognize and bind small ribosomal subunits, initiating cap-independent translation [104]. YTHDF1 enhances translation efficiency and promotes protein production through physical interaction with eIF3. Silencing YTHDF1 results in a down-regulation of protein production of its target mRNA [105]. The interaction of YTHDF3 with YTHDF1 was able to further improve translation efficiency [38]. METTL3-mediated mRNA cyclization is also accomplished by interaction with eIF-associated subunits, and METTL3 enhances the contact of these regions with translation instigation factors to regulate mRNA translation by introducing m6A modifications at the 5 ʹUTR and 3ʹ UTR of mRNAs [104, 106].
The m6A modification located in the 3ʹ UTR region is cap-dependent translation, guided by the interaction between YTHDF1 and YTHDF3 [105]. YTHDF3 stabilizes YTHDF1 binding to mRNA, resulting in enhanced translation [38]. Furthermore, researchers found that YTHDC2 has ATP-dependent RNA helicase activity and enhances target mRNA translational efficiency by encouraging ribosome cycling [107]. Huang et al. proposed that IGF2BPs play a role in regulating stress-related mRNA translation [44]. The m6A modification can also inhibit mRNA translation via related proteins. Interestingly, previous researches have shown that FMRP typically inhibits the translation process of target mRNAs by preventing ribosomal translocation [108, 109]. However, Elias G. Bechara et al. have also proposed that FMRP may positively regulate mRNA translation [110]. As a result, additional investigate is needed to comprehend the precise mechanism of FMRP in regulating of mRNA translation.
m6A in mRNA stability
Various kinds of m6A-reading proteins are the main regulators responsible for regulating mRNA stability. Research has demonstrated that IGF2BPs are crucial for the preservation of mRNA stability. By attaching to target mRNAs, IGF2BPs recruit RNA stabilizers and regulators, such as matrix protein 3 (MATR3) and embryonic lethal abnormal vision 1 (ELAVL1), which act together to protect target mRNAs from degradation [44, 111]. YTHDF2-bound m6A mRNA, on the other hand, induces a decay process through two distinct molecular mechanisms [43]. First, the heat-responsive protein (HRSP12) can physically interact with YTHDF2 as an adaptor protein, causing rapid RNA degradation through the endonuclease cleavage pathway. The existence of HRSP12 binding sites in transcripts is essential for this mechanism [112]. Research has demonstrated that it can also stimulate the degradation of circRNAs with m6A modifications [112]. Second, YTHDF2 directly recruits the deadenylase complex CCR4-NOT, triggering deadenylation and initiating the degradation of m6A-modified mRNAs [37, 113, 114]. YTHDF3 can synergize with YTHDF2 to promote RNA decay [38].
Upstream frameshift 1 protein (UPF1) is a key RNA helicase that has been reported to modulate mRNA stability through the nonsense-mediated mRNA degradation (NMD) pathway [115]. YTHDF2 is also involved in this mechanism, further enhancing mRNA degradation by recruiting UPF1. The interaction reflects the synergistic effect of m6A modification with post-transcriptional regulation. Researchers have also reported that YTHDF1 binds m6A-modified mRNA, forming dynamic concentrating compartments in the cell through a liquid-liquid phase separation mechanism to regulate the translation efficiency of mRNA [116,117,118]. Another reading protein, YTHDC2, has been reported to be able to recruit the exonuclease XRN1 and other RNA degradation complexes, thereby accelerating mRNA degradation [107, 119]. Apart from the aforementioned processes, m6A modification is also crucial for the cytosolic elimination process of non-functional RNAs. RNA degradation complexes degrade m6A by recognizing and labeling nonfunctional RNAs and then binding specific m6A reader proteins [120]. This mechanism guarantees RNA quality control within cells and prevents the accumulation of non-functional RNA that disrupts the normal biological function of cells.
m6A and rRNA
In eukaryotic cells, the m6A modification of ribosomal RNA (rRNA) is crucial for rRNA processing and translation regulation mechanisms [121, 122]. Several m6A regulators are involved in regulating these processes to ensure precise control of rRNA function and efficient intracellular translation processes. m6A modification on rRNA was first identified in humans and other vertebrates 30 years ago. The METTL5 [27, 123,124,125] and ZCCHC4 [31, 126] enzymes are both positioned in the nucleolus and specially change the methylation state of 18S rRNA m6A1832 and 28S rRNA m6A4220. Both of them are crucial in regulating specific cellular processes and the biogenesis of ribosomes.
In addition, recent researchers have also proposed that METTL16 participates in translation-related mechanisms during ribosome biogenesis, thereby improving translational efficiency. This process interacts with Subunits of the eIF3 complex, facilitating its binding to 18S rRNA and controlling the translation initiation process [127]. This process is not influenced by its methyltransferase activity. In bacteria, m6A modification similarly influences the translational function of ribosomes and regulates ribosome assembly [126, 128, 129]. Currently, there is a scarcity of research on the correlation between m6A modification and rRNA, so future investigations are anticipated to further reveal the specific mechanism and provide more information for understanding the widespread role of m6A modification in rRNA.
m6A and lncRNA
At present, the relationship between m6A modification and lncRNAs has garnered significant attention. The m6A modification is vital for regulating the stability and translation processes involved in the biogenesis of lncRNAs, and it affects gene expression through many kinds of complex biological processes. Conversely, lncRNAs can also target m6A-related regulators, which play a crucial role in m6A modification [130,131,132,133,134]. The modification of m6A can alter the structure of local RNAs, thereby inducing RNA-binding protein entry and regulating the function of lncRNAs. For instance, when YTHDC1 recognizes and controls gene silencing caused by long non-coding RNA X-inactivation-specific transcripts (XIST), m6A modification changes the local structure of XIST. This structural change makes it easier for XIST to bind to factors that control silencing and boosts the effect of the X chromosome on silencing [135,136,137]. Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is a lncRNA that is abundantly expressed in the nucleus and up-regulated in neoplastic diseases [138]. The m6A modification regulates MALAT1’s function in splicing by altering its structure, which in turn affects its localization and expression in the nucleus [139]. Correspondingly, MALAT1 affects its own m6A modification level by targeting METTL3 interaction and can also regulate MALAT1 function in regulating gene expression and RNA splicing [137, 139, 140].
In a manner that is dependent on m6A, m6A writers regulate the stability of lncRNA. Several researchers suggest that m6A writers play a necessary role in the progress of gynecology-related malignancies. For instance, METTL3 accelerates cervical cancer progression by promoting m6A modification of the lncRNA FOXD2-AS1 and enhancing the stability of FOXD2-AS1 [141]. In a study based on ovarian cancer, it was discovered that the m6A methylation of lncRNA RHPN1-AS1 enlarged the stability and up-regulation of RHPN1-AS by reducing RNA degradation. Additionally, it promoted the development of epithelial ovarian cancer(EOC) by activating the FAK/PI3K/Akt pathway [142]. Related researches have also discovered the specific mechanisms of m6A writer-related regulators and lncRNAs in Acute myeloid leukemia (AML) [143], hepatocellular carcinoma (HCC) [144, 145], gastric cancer [134], breast cancer [146], and nasopharyngeal carcinoma et al. [147, 148].
In general, m6A methylation of lncRNAs reduces RNA degradation and improves their stability. Some lncRNAs, however, have decreased stability after m6A methylation modification. METTL16 regulates HCC progression by inhibiting the stability of RNA transcripts through the interaction of m6A and RAB11B-AS1 lncRNAs [145]. The newly identified regulator ZCCHC4 can also promote colorectal cancer spread through regulating the m6A modification of lncRNAGHRLOS and inhibiting its stability to downregulate gene expression [149].
m6A readers can recognize and attach m6A modifications, which in turn affect the degradation or stabilization of lncRNAs and regulate tumor development. For example, YTHDC1 recognizes and stabilizes the m6A modification of the lncRNATERRA mediated by METTL3, thereby ensuring the stability of the telomere in cancer cells [150]. IGF2BP1 is an important factor in the initiation and progression of gastric cancer, as it promotes the stability of THAP7-AS1 lncRNA [151]. Recent research has also revealed that m6A erasers are involved in the regulation of the biological function of lncRNAs. For example, FTO-mediated m6A modification of lncRNA LINC00022 improves its stability and stimulates cancer cell proliferation by recognizing YTHDF2 [152]. Another demethylase ALKBH5 increases the stability of the lncRNA KCNK15-AS1, inactivates the PI3K/AKT pathway, and upregulates Phosphatase and tensin homolog (PTEN) expression through a m6A-dependent mechanism, thereby preventing tumor cell proliferation and metastasis [153].
Investigating the regulatory mechanism of m6A modification on lncRNAs can provide valuable insights into RNA functions and the role of RNA modifications in various diseases. In the future, more lncRNAs regulated by m6A modification are expected to be discovered. Further research is necessary to ascertain the precise mechanism of lncRNA biosynthesis and the function of m6A modification in this procedure.
m6A and miRNA
microRNAs (miRNAs) are a type of small RNA that are endogenously encoded and single-stranded. They are nearly 22–25 nucleotides in length and are produced from primary transcripts (pri-miRNAs) that are transcribed by RNA polymerase II/III [154, 155]. The 3ʹUTR and regions adjacent to the stop codons have abundant binding sites for miRNAs and m6A [68]. Multiple investigations have revealed that the interplay between m6A modification and miRNAs involves regulatory mechanisms at multiple levels, including miRNA biosynthesis, recognition and degradation of target mRNAs, and mutual regulation between miRNAs and m6A-related factors [156].
DiGeorge syndrome chromosome region 8 (DGCR8) plays a key role in primary miRNA processing [157]. According to Ma et al.‘s study, DGCR8 regulates the processing and maturing of pri-miR-126 by recognizing and binding the m6A modification mediated by METTL14 [158]. DGCR8 can also target and process METTL3 methylation-modified pri-miRNAs, as well as promote miRNA maturation [159, 160]. In contrast to the above results, m6A modification is similarly involved in inhibiting pre-miRNA maturation. An osteoporosis-based study found that METTL3 binds to pre-miR-320 and promotes the m6A modification of this molecule. This mechanism prevents osteoporosis by primarily enhancing the differentiation of osteoblasts through the activation of Runt-related transcription factor 2 (RUNX2) [161]. It was discovered in a tumor-based study that METTL3 and METTL14 act on miR-380-3p through a m6A modification-dependent mechanism. This mechanism targets PTEN deleted on chromosome ten to degrade it, resulting in the motivation of the downstream Akt signaling pathway. Consequently, pancreatic cancer progress is promoted [162].
m6A readers regulate miRNA generation and stability and then regulate miRNA target expression by recognizing and binding m6A-modified RNAs. A study by Lyu et al. utilizing ovarian cancer cell demonstrated that WTAP-dependent m6A methylation stimulates the maturation of miR-200 in a DGCR8-dependent manner, ultimately regulating hexokinase 2 (HK2), a vital enzyme in glycolysis, thereby influencing ovarian tumor metabolism and progression. Researchers have also observed that WTAP-mediated m6A modification participates in the promotion of miRNA maturation processes [163]. Another study demonstrated that METTL3/YTHDF2-dependent m6A methylation regulates the expression of the tumor suppressor gene Vasohibin-1 (VASH1) by modulating the interaction between miR-885-5p and lncRNA MEG3[164]. Through bioinformatics analysis, cell function assays, and in vitro models, the study identified a novel mechanism by which miRNAs contribute to ovarian cancer progression [164]. The nuclear reader HNRNPA2B1 is able to specifically recognize and bind certain m6A-modified pri-miRNAs and promote DGCR8-mediated primary miRNA processing, thereby regulating miRNA maturation [51, 165, 166]. Other research indicated that IGF2BPs protein and HNRNPs family protein can function as oncogenes, binding m6A-modified related miRNAs and participating in their maturation process, thereby promoting the development of prostate cancer, liver cancer, and breast cancer [167,168,169]. Additionally, FTO and ALKBH5 play a significant role in the progression of various kinds of cancer, mainly by controlling the stability of miRNAs and increasing their expression in a m6A-depended way [170,171,172,173].
Notably, both of them possess the capacity to function as either a tumor suppressor or a promoter of cancer advancement. Correspondingly, miRNAs can also target associated m6A-modified mRNAs and participate in regulating their stability, thereby regulating m6A modification [174]. For example, miRNAs have the ability to form RNA-induced silencing complexes (RISC), which is a multiprotein complex with endonuclease activity. MiRNAs modulate gene translation by binding to RISC, which then prompts their binding to the 3ʹ-UTR of m6A-modified target mRNAs, thereby mediating the silencing of target mRNAs [175, 176]. MiR-33a targets METTL3-mediated m6A-dependent mRNA, decreases METTL3 expression and result in declining the growth of lung cancer [177]. In conclusion, m6A modification, facilitated by m6A regulators, significantly influences miRNA synthesis via changing miRNA’s biological functions. Further researches are required to elucidate the complex regulatory mechanisms underlying the relationship between these two processes.
m6A and circRNA
Reverse splicing mechanisms typically form circular RNAs (circRNAs), which are structurally specialised non-coding RNAs that remain unaffected by RNA exonucleases and exhibit greater stability compared to other classes of RNAs [178,179,180]. CircRNAs’ functionality can be regulated by m6A modification through the regulation of their biogenesis, stability, and translational potential [181, 182]. For example, the METTL3/METTL14 complex adds a m6A modification to pre-mRNA to make it easier to cyclize to generate circRNAs [181, 183]. The reading protein YTHDC1 can promote circRNA maturation by selectively enhancing splicing of circRNAs [184]. YTHDC1 is also involved in circRNA transport [185]. Research has found that YTHDC1 silencing leads to circNSUN2 accumulation in the nucleus, confirming this theory [184]. METTL3-mediated m6A modification facilitates the nuclear export process of circHPS5, thereby mediating hepatocellular carcinoma progression [186].
m6A modification can also drive circRNA translation initiation. This process mainly includes two mechanisms. First, YTHDF1 facilitates the translation of circRNA into functional proteins by enhancing the internal ribosome entry site (IRES)-dependent pathway [187, 188]. Second, the m6A modification can directly start the cap-independent translation of circRNAs by bringing in eukaryotic translation initiation complexes through YTHDF3 [181]. However, FTO-mediated m6A modification inhibits circRNA translation. For instance, m6A demethylation of circDDIT4 represses its expression, thereby promoting the development of prostate cancer [181].
Researchers found that reading protein YTHDF2 is able to bind m6A-modified circRNAs and regulate their stability and degradation. For instance, YTHDF2 controls the m6A modification process of circ0003215 by making it break down faster and lowering the amount of circRNA, which helps colorectal cancer spread [189]. METTL3 is responsible for mediating the m6A modification of circMYO1C to enhance its stability by recognizing IGF2BP2 and playing a role in tumor progression [190]. In contrast, some m6A-modified circRNAs could reduce stability. For example, the demethylase ALKBH5 mediates m6A modification of circNRIP1 to suppress tumorigenesis by inhibiting its stability and downregulating the expression of carcinogenesis-related factors [191].
In the meantime, circRNAs can also target related m6A regulators, directly regulate m6A modification of their target mRNAs [192, 193], or indirectly regulate m6A modification through sponge miRNAs. CircRNAs can specifically bind to miRNAs through multiple binding sites, like how water sticks to a sponge. This stops miRNAs from binding to their target mRNAs, which stops them from doing their biological job [194, 195]. CircPUM1, for example, promotes METTL3 expression via sponge miR-590-5p and has an impact on tumor cell proliferation and glycolytic processes [196]. Examining the interaction between circRNAs and m6A modifications facilitates our comprehension of their biological function and potential applications in disease therapy. Several researches have investigated the influence of m6A modification on circRNAs, yet further research is looked-for to fully comprehend the role of circRNAs in methylation modification.
Physiological functions of m6A in female reproductive system
m6A in folliculogenesis and oocyte maturation
The function of m6A modification in folliculogenesis and oocyte maturation is a relatively active area of recent research. During the embryonic stage, primary oocytes within primordial follicles undergo and remain in meiotic prophase I. In preparation for the initial embryonic development, oocytes progressively increase in size and accumulate substantial quantities of mRNA and protein. Oocytes enter metaphase I from meiotic prophase I during follicular maturation, resulting in the formation of secondary oocytes and polar bodies. Secondary oocytes undergo meiotic second division, remain in metaphase II, and complete development prior to ovulation and fertilization [197]. The m6A modification controls the maturation process of oocytes, influences both the stability and translation efficiency of mRNA, regulates the expression of related genes through multiple biological mechanisms involved in precise epigenetic regulation in oocytes.
METTL3 expression is elevated during oogenesis. In a study, siRNA was employed to knockdown Mettl3 in oocytes of the mouse germinalvesicle stage. This caused abnormal spindles, slowed down meiotic progression, reduced the efficacy of mRNA translation, impacted mRNA degradation, and halted oocyte development [198]. Supporting this conclusion, an additional investigation discovered that METTL3 mutations impeded the initial stages of oocyte development and resulted in a substantially lower percentage of fully developed follicles than controls. Intersectin 2 (ITSN2) is a multifunctional protein that is involved in the process of meiotic resumption in oocytes [14]. METTL3 identifies and attaches to ITSN2 and carries out m6A alteration, which can increase its durability and facilitate oocyte development. Specific Mettl3 knockout in mouse oocytes causes meiotic maturation failure [13]. Also, the development of oocytes is halted and adult females become less fertile when mettl3 mutations are detected in zebrafish. This could be due to the global decrease in m6A levels and the blockage of critical genes that are necessary for the production of hormones and gonadotropins [199]. A study conducted on oocytes from pigs revealed that the methylation of m6A by METTL3 and WTAP has the potential to influence the maturation of oocytes and the growth of embryos [200].
The reading proteins YTHDC1 and YTHDC2 are required for oocyte maturation. Seth D. Kasowitz et al. engaged in a mouse study in which the inactivation of YTHDC1 led to mRNA defects in alternative splicing in mouse oocytes. The production of extensive variable polyadenylation altered the 3ʹUTR length, thereby hindering oogenesis [201]. YTHDC2-deficient female mice have small ovaries and lack fertility [202]. YTHDF2 regulates the degradation of maternal mRNA during MII [203]. Knocking down YTHDF2 modifies the amount of transcription of genes that affect oocyte development, thereby affecting the quality of oocytes. In mice, the absence of YTHDF2 results in the failure of oocyte maturation and female-specific infertility. KIAA1429 controls the alternative splicing of genes involved in oocyte maturation. Conditional knockout mice display Kiaa1429 in their oocytes, resulting in female mice becoming infertile [204]. The IGF2BPs family has a role in oocyte maturation by engaging in the m6A methylation process of RNA and enhancing mRNA stability [13]. Additionally, a transcriptome-wide investigation of chicken follicles by Yu et al. indicates that the follicle selection process may be influenced by m6A modification [205].
m6A in embryonic development
Embryonic development commences with fertilization, during which the zygote exhibits a transcriptionally quiescent state. Following this, the maternal-to-zygote transition (MZT) occurs, which involves the activation of the zygote genome (ZGA) and the elimination of maternal storage (RNA and DNA) [206, 207]. The importance of RNA m6A modification in the development of mammalian embryos has been increasingly recognized in a variety of studies [208]. It has been reported that the degradation of maternal mRNA is impeded by the loss of Mettl3 in mouse oocytes, which in turn affects embryonic developmental progression by impeding MZT and zygotic genome activation [198, 209]. m6A methylation can also influence embryonic development by modulating the stability of mammalian embryonic stem cells (mESC). For example, the absence of Mettl3 results in the loss of the self-renewal and differentiation capabilities of rodent embryonic stem cells [210, 211]. Relevant demonstrated that the Mettl3 gene deletion in mammals results in embryonic mortality [212, 213].
Mettl14 deficiency substantially delays early embryonic development and results in embryonic death, primarily by obstructing the process of ectodermal cell differentiation to maturation [214]. Furthermore, a study conducted by Liu et al., which involved the construction of an embryonic mouse model lacking Mettl16, discovered that the absence of Mettl16 resulted in a reduction in the mRNA levels of its methylation target Methionine adenosyltransferase 2 A (MAT2A) and the obstruction of embryonic development. This finding underscores the essential role of Mettl16 during the initial phases of embryonic development [29]. Researchers have also shown that the recently discovered writing protein Mettl5 affects embryogenesis by participating in the m6A modification of rRNA and regulating pluripotency in mouse embryonic stem cells (mESC) [123].
As mentioned above, YTHDC1 not only regulates oogenesis but also participates in the self-renewal and differentiation processes of mouse embryonic stem cells, which affects embryonic development [201]. Studies have shown that YTHDC2-knockout mice survive, but their adult counterparts are infertile [202]. Suppression of the IGF2BP2 protein results in the halting of mouse embryos at the 2-cell stage and hinders the advancement of early embryonic development [215]. A study discovered that the inhibition of YTHDF2 in goat embryonic cells resulted in the prevention of maternal mRNA degradation and impaired zygotic genome activation. It was hypothesized that YTHDF2 influences mRNA clearance by controlling the expression of adenosine deaminase and mRNA decapping enzyme [216]. Studies using mouse and zebrafish have demonstrated that the deletion of the maternal YTHDF2 gene significantly hinders the development of embryos [217, 218].
Furthermore, the demethylase ALKBH5 primarily controls the development of the male testes [219]. FTO in mESC is responsible for the demethylation modification of long interspersed element 1 (LINE1) RNA, which has an impact on embryonic development, growth, and differentiation of embryonic stem cells [220]. To understand the complex mechanisms of gynecology-related diseases, it is essential to conduct a more thorough investigation of the molecular mechanisms of the connection between the m6A modification and embryonic development.
m6A in placental development
The placenta acts as a bridge between the mother and the baby, performing tasks like immunological modulation, nutrition exchange, and stress response. It also plays an essential role in preserving pregnancy and the growth of the fetus [221, 222]. Placental cells, particularly trophoblast cells, are highly proliferative and invasive. The modulation of the maternal immune system and gene expression is facilitated by their involvement in numerous mechanisms, which are designed to ensure the seamless implantation and development of the embryo in utero [223, 224]. Furthermore, the placenta, functioning as a physiological neoplasm, is connected to cancer through multiple processes [225]. The majority of m6A modification sites are situated in introns of human placental tissue and are enriched near stop codons [208, 226, 227]. The methylation modification of m6A significantly influences the biological function of placental cells.
By increasing the stability of placental villous trophoblast cells, the m6A methylation of MYLK mRNA by methyllase-METTL3 can affect their invasiveness. Additionally, it is implicated in the regulation of preeclampsia [228]. YTHDC1 is involved in the m6A methylation modification of circMPP1, which leads to a decrease in its expression and the inhibition of placental function. This process is regulated by both the NF-κB and MAPK3 signaling pathways [229]. Through an analysis of the m6A modification levels at the 5ʹ-UTR of placental mRNA in infants with varying birth weights, one study suggested that m6A modification may regulate placental function and influence fetal growth by participating in translational regulation [208]. Through the development of a BaP-exposed mouse model, Wang et al. discovered that lnc-HZ14 induces early abortion and promotes placental trophoblast pyroptosis by participating in and increasing the METTL3-mediated m6A modification of pyridine ___domain-containing 3 inflammasome (NLRP3) mRNA in the NOD-like receptor (NLR) family [230].
The demethylase ALKBH5 is an important regulator of placental development. Knocking down ALKBH5 can promote trophoblast invasiveness and result in recurrent miscarriage, as it can modify human trophoblast invasiveness by regulating CYR61 mRNA stability [231]. Another study also verified the involvement of ALKBH5 in the progress of preeclampsia by modulating the m6A methylation modification of PPARG mRNA [232]. Placental dysplasia-related proteins and targets are important for the early diagnosis and management of gynecological diseases. Nevertheless, there are limited relevant studies, and their specific mechanisms require further investigation.
m6A in maternal-fetal microenvironment
The maternal-fetal microenvironment is a complex biological interface between the mother and fetus. Its main role is to support the embryo’s growth and development while protecting it from harmful substances and pathogens. Furthermore, the occurrence of related diseases is closely correlated with alterations in the maternal-fetal interface microenvironment [233]. Germ stem cells are implicated in mechanisms related to immune tolerance and fetal development in the maternal-fetal microenvironment. The study of cancer stem cells is provided with a distinctive perspective by the similarity between the maternal-fetal microenvironment and the tumor microenvironment in certain regulatory pathways and mechanisms. The normal development of embryos is ensured by the interaction between multiple important immune cells, which is crucial in the mother and fetus’s mutual equilibrium of immunological tolerance and protection (Fig. 2). Cross-sectional research in these regions offer substantial support for comprehending the operation of the female reproductive system.
A In DCs, m6A methylation regulates the translation of proteases in CD40, CD80, and lysosomes, thereby promoting DC maturation and inhibiting antigen presentation. METTL3 influences the energy metabolism of DC by regulating the expression of HIF-1 and the glycolytic gene LDHA. B by modulating SOCS family mRNA and the IL-7/JAK/STAT5 pathway, METTL3 maintains CD4 + T cell homeostasis. Additionally, it is essential for the development of Tfh cells. METTL14 deficiency results in the blocking of Treg cell differentiation. C In macrophages, METTL3 regulates the tumor microenvironment (TME), disrupts TLR4 and ERK signaling pathways, affects Irakm and Spred2 expression, and inhibits tumor progression. D m6A modification plays an important role in various developmental stages of B cells. E NK cell function is regulated by the degradation of STAT5 mRNA and the expression of SHP-2, which are regulated by YTHDF2 and METTL3, respectively.
m6A and germline stem cells
Female germline stem cells (FGSCs) are a kind of stem cell that are found in the female reproductive system of mammals. They possess the ability to self-renew and develop into fully mature oocytes [234, 235]. The relationship between FGSC and m6A modification has garnered a growing amount of attention in recent years. Zhao et al. performed a comparative investigation of the m6A methylation levels of mRNAs in female mouse germline stem cells and thioguanine and ubarine-resistant (STO) cells using MeRIP-seq. They discovered that the former had significantly higher overall m6A levels than the latter. Subsequent investigations indicated that the proliferation process of FGSC was influenced by the reading protein YTHDF1, and the self-renewal capacity of FGSC was substantially reduced by a specific knockdown of YTHDF1 [236].
In addition, correlational research has suggested that the m6A modification of non-coding RNAs is associated with FGSC. For instance, the nuclear export progression of circGFRα1 is influenced by the m6A modification mediated by methylase METTL14, which in turn regulates GFRα1 expression and influences self-renewal in FGSC [237]. Currently, the specific relationship between m6A modifications and FGSCs is being investigated; however, it is conceivable that m6A modifications may be crucial in the preservation of the self-renewal and differentiation processes of FGSCs. We can anticipate a more comprehensive comprehension of this field as science and technology continue to advance and research continues to deepen.
m6A and cancer stem cells
Cancer stem cells (CSC) are present in various of tumor types and possess stem cell characteristics. They are vital in tumor progression, recurrence, drug resistance, and metastasis of tumors [238,239,240]. The invasiveness of the placenta is somewhat similar to the mechanisms associated with CSCs, which also suggests a potential role for the maternal-fetal microenvironment in the development of tumors. Numerous studies have documented a correlation of m6A and tumor resistance, tumor epithelial-mesenchymal transition (EMT), and self-renewal and differentiation of cancer stem cells [241,242,243]. Currently, there has been a growing interest in the close correlation between CSC and m6A modification. Numerous malignancies including breast cancer [244], lung cancer [245], cutaneous squamous cell carcinoma [246], acute leukemia [247], and colon cancer [111], have been disclosed to play critical functions in m6A modification. Gradually, studies that investigate the relationship between gynecological malignancies and CSC are also being conducted.
A study of cisplatin-resistant ovarian cancer cell lines discovered that YTHDF1 was capable of resisting ovarian cancer cells by improving their CSC-like characteristics and promoting tumor progression by binding to m6A-modified three-part motif protein 29 (TRIM 29) and enhancing its translational efficiency [248]. In cervical cancer, overexpression of TRIM 29 also has a carcinogenic effect, but more researches are required to determine the complete mechanism [249]. Based on an investigation conducted by Chen et al., hypoxia induces the demethylase ALKBH5, which in turn regulates the m6A methylation of the transcription factor SOX2 mRNA and promotes the properties of endometrial cancer stem cells by reducing its methylation level. Hypoxia-inducible factor downregulated the level of ALKBH5 protein, which in turn inhibited the tumorigenicity of CSCs, thereby confirming the aforementioned conclusion [250]. A comprehensive understanding of the characteristics and mechanisms related to CSC could enhance patient survival, reduce tumor recurrence and metastasis, and contribute to a more effective clinical battle against cancer.
m6A modification not only influences the biological behavior of tumor cells but also regulates key processes within the tumor microenvironment (TME), including the hypoxic response, metabolic reprogramming, cell cycle regulation, and immune cell function. In the immune microenvironment, tumor cells depend on glycolysis to sustain their energy supply [251]. A study by Li et al. employed Mettl3-knockdown cervical cancer cell lines (HeLa and SiHa) alongside the human normal cervical epithelial cell line ECT1/E6E7 for in vivo and in vitro assays. The results indicated that YTHDF1 and IGF2BP3-dependent m6A methylation positively regulates glycolysis in cancer cells, thereby promoting tumor growth and progress by enhancing the stability and translation efficiency of PDK4 mRNA [252]. Additionally, another study unveiled that METTL3 regulates CDC25B translation through m6A modification-dependent YTHDF1. This regulation was demonstrated through a comprehensive approach, including in vitro cervical cancer cell cycle synchronization, m6A-seq analysis, in vitro cell proliferation assays, and in vivo nude mouse tumor formation experiments, ultimately promoting cell cycle progression [253]. These findings highlight that m6A modification-related mechanisms hold promise as potential therapeutic targets for cancer in the future.
m6A and DC cells
Dendritic cells (DCs) are antigen-presenting cells (APCs) which perform vital immunoregulatory functions in the maternal-fetal microenvironment and are instrumental in the emergence and development of tumors [254, 255]. Krey et al. developed a transgenic mouse model of DC depletion and discovered that DC deficiency prevented the embryo from implanting and resulted in aberrant vascular development in the uterine decidua. This affected the process of placenta development and the subsequent pregnancy [256]. In the past few years, m6A modification and DCs’ role in the immune microenvironment have gained increasing attention. As early as 2005, researchers proposed that the innate immune response process could be influenced by the m6A modification of RNA, which could reduce DC-mediated Toll-like receptor (TLR) expression [257]. Wang et al. discovered that the methylase Mettl3-mediated m6A modification may boost the expression of associated immune costimulators, increase their translational efficiency, activate DCs, and encourage the initiation of T cells. The TLR4/NF-κB signaling pathway is activated during this process, which results in the production of cytokines. Mettl3 knockdown then impedes this process, which impacts the normal function and maturation processes of DCs [258].
The YTHDF1 reading protein is responsible for the regulation of lysosomal proteases and the facilitation of tumor immune escape. YTHDF1 reduction enhances the capacity of DCs to present tumor antigen by decreasing its antigen consumption [259]. The putative function of YTHDF2 as a tumor immunosuppressive factor is reflected in the mechanism by which it recognizes and binds the m6A modification site of lnc-Dpf3 in DCs, thereby inhibiting DC migration [260]. To summarize, targeting m6A in DCs has great promise for cancer immunotherapy. Furthermore, the association between DC and m6A modification in obstetrics and gynecology is poorly researched, which underscores the need for additional clarification.
m6A and NK cells
Uterine natural killer cells (uNK cells) are a distinct type of immune cell that are present on the endometrium. They are collectively responsible for the preservation of immune homeostasis at the maternal-fetal interface. Former research has shown that the development of the placenta is contingent upon the presence of uNK cells. [261], the growth of the fetus [262], the modulation of inflammation and the immune system [263], and the mechanisms related to tumors [264]. Decidual NK (dNK) cells represent a specific subtype of uNK cell that are predominant in the early maternal-fetal interface of humans [265, 266]. dNK cells also play a critical role in keeping maternal-fetal microenvironment homeostasis. The significance of m6A modification in modulating the activity of NK cells biological function has steadily become an increasingly hot topic, and the interaction between the two in gynecological diseases is also a developing research area.
It has been reported that the process of NK cell maturation is facilitated by the m6A modification mediated by the reading protein YTHDF2. The antitumor properties of NK cells were subsequently inhibited by the specific knockdown of YTHDF2 [264, 267]. The function of METTL3-related m6A modification in regulating NK cell killing activity has also been confirmed [268]. Furthermore, a study conducted a bioinformatics analysis of the m6A target gene cell division cycle 42 effector protein (CDC42EP3) in ovarian cancer and discovered that its down-regulation was associated with NK cell infiltration and the promotion of ovarian cancer progression. This identified a potential association between NK cells and m6A modification in ovarian cancer [269].
In contrast, demethylase FTO has an adverse impact on NK cells, and the specific reduction of FTO results in increased NK cell activation and a decreased risk of melanoma progression [270]. The diagnosis and treatment of gynecological diseases is also likely to be influenced by the specific mechanism of m6A modification in NK cells. Therefore, it is critical to have a comprehensive understanding about this mechanism.
m6A and macrophages
Decidual macrophages are responsible for the primary maintenance of immune tolerance at the maternal-fetal interface during pregnancy by secreting anti-inflammatory cytokines, including IL-10 and CCL2, which counteract the maternal immune system’s attack on the fetus [271]. The critical function of macrophages in immune regulation [272, 273], trophoblast development [274], fetal growth [275], and tumorigenesis [276] has been showed in previous studies. The m6A modification has the capacity to regulate the polarization status (M1 and M2 macrophages) and biological function of macrophages by modulating gene expression mechanisms [277].
Several investigations have confirmed that the methylase METTL3-mediated m6A modification may increase the expression of the transcription factor STAT1, thereby positively influencing M1 macrophage polarization while negatively affecting M2 macrophage polarization [278, 279]. YTHDF2 is also involved in the regulation of m6A modification of STAT1 mRNA and the promotion of its degradation, which in turn promotes the polarization of M1 macrophages. This process is accomplished by interacting with the RNA-binding motif 4 (RBM4) [280]. Nevertheless, a study discovered that lactate could stimulate the production of Mettl3-mediated m6A modification, which then positively regulated the polarization of M2 macrophages and resulted in the invasion of endometriosis [281].
In addition, the interaction between m6A modification and tumor-associated macrophages (TAM) is becoming increasingly significant in the modulation of tumor immunity and drug resistance. For instance, ALKBH5 has the potential to influence the homeostasis of the tumor microenvironment by modulating macrophage polarization and function. Jiang et al. conducted an in vitro simulation of the tumor microenvironment by cultivating ovarian cancer cells and M2 macrophages. They discovered that both demethylase ALKBH5 and TLR4 expression were increased, confirming that ALKBH5-mediated m6A demethylation modification activates the NF-κB pathway, which promotes the invasiveness of ovarian cancer [282].
Furthermore, the significance of the interaction between TAM and the ovarian cancer microenvironment has been elucidated in related studies, which are intended to investigate resistance-associated targets and recurrence markers [283]. In cervical cancer, a study suggested that the glycolytic process is targeted, programmed cell death protein 1 (PD-1) expression is facilitated, and macrophage phagocytic function is inhibited by high expression of the m6A regulator METTL14, which accelerates cancer progression [284].
Therefore, the disclosure of the comprehensive effect of m6A modification on macrophage function could enhance comprehension of the role of macrophages in tumor microenvironment and gynecology-related immune responses, thereby facilitating the development of immunotherapeutic strategies that target m6A modification.
m6A and T cells
The body’s immune balance is maintained by the synergistic action of various subsets of T cells, which are critical factors of the adaptive immune system, in recognizing and responding to pathogens and tumor cells [285]. m6A modification has demonstrated a significant role in the maternal-fetal interface by modulating T cell development, differentiation, and function [286].
Li et al. identified that the methylase METTL3-mediated m6A methylation was capable of targeting the IL-7/STAT5/SOCS pathway and influencing mRNA degradation processes, thereby modulating the proliferation and differentiation of CD4 + T cells [287]. Also, this post-transcriptional regulation, which is activated by METTL3, positively influences the differentiation of T follicular helper (Tfh) cells [288]. Regulation T (Treg) cells are induced at the maternal-fetal interface to safeguard the fetus from immune rejection and are also closely linked to recurrent miscarriage and infertility [289, 290]. A study conducted by Tong et al. discovered that Mettl3 KO mice exhibited infertility as a result of autoimmune defects, and they specifically lost the suppressive function of Tregs in comparison to their WT littermates. This result also serves as confirmation of the essential regulatory function that METTL3 serves in the preservation of the immunosuppressive function of Treg cells [291].
In ovarian cancer, the C3a anaphylatoxin chemotactic receptor (C3AR1) is associated with the activation of T cells and the penetration of immune cells by mediating m6A modification in OC cells, which results in the development of ovarian cancer [292]. An in-depth examination of the molecular mechanisms involved in T cell modification with m6A could aid in the comprehension of the intricate regulatory network of the immune system and potentially offer new strategies and targets for the medical management of maternal-fetal-related diseases.
m6A and B cells
B cells are mainly involved in humoral immunity, which involves the production of specific antibodies to neutralize pathogens and functions by recognizing antigens [293]. Currently, there is a limited number studies examining the correlation between B cells and the maternal-fetal immune microenvironment. According to reports, trophoblast glycoprotein (TPG) is capable of binding B cell receptors and inhibiting their activation, which is crucial for the promotion of maternal-embryonic immune tolerance and the maintenance of normal fetal development [294]. Additionally, a study found that decidual B cells can prevent spontaneous preterm birth by interfering with the PIBF1 pathway, which is regulated by interleukin 33 (IL-33) [295].
Related research has indicated that m6A modification may be a critical factor in the early progress of B cells. For instance, METTL14-mediated m6A methylation is involved in B cell maturation, whereas YTHDF2 reduces interleukin-7 (IL-7)-induced pro-B cell proliferation by binding to m6A modification sites and degrading specific mRNA transcripts, thereby influencing early B cell transformation processes [296]. This process is not regulated by YTHDF1 [296].
Furthermore, the development and prognosis of acute lymphoblastic leukemia [297], human glioma cells [298], and other conditions may be influenced by aberrant methylation modification of m6A in B cells. A study conducted a biological information analysis to screen m6A-related lncRNA prognostic genes in patients with endometrial cancer of the corpus uteri (UCEC). The results indicated that B-cell immune infiltration scores were elevated in the high-risk group than in the low-risk group of the disease, suggesting a possible role for B cells in the progression and prognosis of UCEC [299]. Currently, the investigation of B cells in epigenetic modifications is in its infancy, necessitating the subsequent undertaking of more comprehensive studies on gynecology-related diseases that are founded on the interaction between m6A and B cells.
m6A and female reproductive hormones
Female reproductive hormones are indispensable for the modulation of reproductive system function, the maintenance of reproductive health, and the regulation of the female reproductive cycle [300]. The biological roles of the female reproductive system and the development of related diseases have progressively received attention to the regulation of reproductive hormones by m6A modification. However, there are limited studies in this field. Previous sections have indicated that the suppression of METTL3 has the capacity to modulate the expression of genes that are essential for the synthesis of sex hormones, thereby contributing to oocyte development [199]. Gonadotropin-releasing hormone (GnRH) is a decapeptide hormone secreted by the hypothalamus that affects the progression and function of the female gonad by regulating gonadotropin (LH and FSH) secretion from the anterior pituitary gland.
Pulsatile release of GnRH is indispensable for the preservation of typical reproductive function [301, 302]. A study discovered that the demethylase FTO enhances the stability of Foxp2 mRNA and mediates m6A demethylation, thereby promoting gonadotropin synthesis and secretion by up-regulating the cAMP/PKA signaling pathway [303]. According to a study accompanied by Wan et al., the methylation modification of m6A by METTL3 can be instrumental in the regulation of the mRNA stability of the estrogen response genes Elf3 and Celsr2 and the maintenance of the expression level of the progesterone receptor PR. This modification is vital for the regulation of intrauterine estrogen and progesterone signaling. Knockdown of METTL3 leads to infertility [304]. Another study also confirmed that METTL3-mediated m6A modulates the process of mbryo implantation by affecting progesterone signaling through the regulation of the progesterone receptor gene (Pgr) mRNA translation process [305].
Additionally, Mettl14 is involved in the modulation of embryo implantation through a m6A modification-dependent pathway, a process that involves immune-related mechanisms and estrogen receptor α (ERα) signal transduction pathways [306]. Xue et al. also confirmed that FTO-mediated m6A demethylation is involved in the development of polycystic ovary syndrome by modulating the androgen receptor/protein kinase B (AR/AKT) pathway, which is responsible for affecting androgen levels in granulosa cells and follicular fluid [307]. A thorough investigation of the precise function of m6A modification in the synthesis and secretion of female reproductive hormones can facilitate the exploration of the pathogenesis of reproductive system diseases and the development of innovative strategies for their diagnosis and treatment.
m6A in obstetrics and gynecology-related diseases
The m6A modification is a critical factor in the development of both benign and malignant female reproductive system diseases. By modulating the expression of genes that are linked to malignancies, m6A modification influences the growth and metastasis of cervical cancer (CC), endometrial cancer (EC), choriocarcinoma (CCA), and ovarian cancer (OC) (Fig. 3, Table 1). In the context of benign disorders, m6A modification is implicated in PCOS, primary Ovarian Insufficiency (POI), spontaneous abortion (SA), gestational diabetes (GDM), and preeclampsia (PE) through a variety of underlying mechanisms (Fig. 4). Consequently, comprehensive investigations of m6A modification mechanisms not only enhance the understanding of the pathogenesis of these diseases but also offer robust support for clinical decision-making.
In cervical cancer, m6A modification-associated transcripts consist of mRNA (ACIN1, CENPK, E2F3, GAS5, MYC), circCCDC134, and miR-503-5P. In ovarian cancer, m6A modification-associated transcripts consist of mRNAs (VGLL1, DDR2, Snail, PLAGL2), CircRAB11 and its target-related miRNAs, and lncRNA RMRPs. In endometrial cancer, m6A modification-associated transcripts consist of mRNA (PEG10, PDGFRB, E2F3, FENDRR, IRS1), Circcchd7, miR-3147, and Linc00958. In choriocarcinoma, transcripts associated with m6A modification include HIF1AN, miR-935, and miR-21-5p.
Numerous gynecological disorders, such as endometriosis (EMT), polycystic ovary syndrome (PCOS), premature ovarian failure (POI), spontaneous abortion (SA), gestational diabetes mellitus (GDM), and preeclampsia (PE), have been found to be influenced by m 6 A. In the aforementioned disorders, we describe modifications in the m 6 A methylation-associated enzymes METTL3, METTL14, RBM15, YTHDC1, YTHDF2, IGF2BP2, FTO, and ALKBH5.
m6A in cervical cancer
CC is a public health issue that poses an imperative threat to the reproductive health of women and is the fourth most widespread form of cancer in women worldwide. Cervical malignancy is primarily caused by an insistent infection with high-risk HPV. However, the prevention of cervical cancer has been demonstrated to be effective through regular screening and prophylactic vaccination [308, 309].
m6A-related regulators are crucial to the progression and development of CC. METTL3 regulates the stability and translation of the catalytic subunit of protein phosphatase 6 (PPP6C) through the m6A modification of mRNA, thereby influencing the downstream transcription factor interferon regulatory factor 5 (IRF5) activity and collaboratively regulating the malignant biological behavior of cervical cancer [310]. METTL3 and IGF2BP3 also regulate the expression level of apoptotic chromatin condensation inducer 1 (ACIN1). The specific mechanism by which ACIN1 induces CC invasion may be accomplished through the interaction of these two m6A regulators, thereby up-regulating the expression level of ACIN1 [311].
The role of regulators related to m6A has garnered increasing attention in tumor metabolism, including glycolysis and lipid metabolism. A study by Wang et al. discovered that METTL3 targets and increases the m6A modification of hexokinase 2 mRNA, while recruiting YTHDF1 to recognize the m6A modification sites on the transcript, enhancing mRNA stability and thus promoting the occurrence of CC and glycolysis [312]. METTL14 and IGF2BP3 have the potential to maintain the stability of stearoyl-CoA desaturase (SCD) mRNA through the m6A system, which can influence lipid metabolism processes and provide new therapeutic strategies for CC [313].
Additionally, numerous investigations have successively documented the precise mechanism by which IGF2BP2, ALKBH5, and other compounds regulate the metabolism of cervical malignancies via the m6A mechanism [252, 314, 315]. Zhang et al. discovered that METTL14 and IGF2BP1 mediate the m6A modification of TRIM11 (an E3 ubiquitin ligase) mRNA, thereby increasing its stability and expression levels. The upregulation of TRIM11 promotes the degradation of the PH ___domain and leucine-rich repeat protein phosphatase 1 (PHLPP1), which in turn contributes to the progression of CC [316]. In cervical cancer, the activation of the PI3K/Akt pathway is frequently associated with tumor aggressiveness and poor prognosis [317]. Li et al. demonstrated that METTL14-mediated m6A modification regulates the stability of FTH1 mRNA, influencing the PI3K/Akt signaling pathway and thereby inhibiting the progression of cervical cancer [318].
YTHDF1, a reading protein, identifies and binds the m6A modification on RANBP2 mRNA, thereby improving its translational efficiency and facilitating the development of CC [19]. According to a study investigated by Tao et al., YTHDF2 initially recognizes the m6A modification of Nuclear Receptor Subfamily 4 Group A Member 1 (NR4A1) mRNA, that is mediated by METTL3. Subsequently, it collaborates with the RNA helicase DDX6 to further enhance RNA degradation and decrease NR4A1 expression, thereby propelling CC progression [319]. Another study also demonstrated that YTHDF2 plays a role in regulating the expression of the target gene AXIN1, which indirectly influences the activity of the Wnt/β-catenin signaling pathway. This, in turn, modulates the EMT progression in cervical tumors and affects the sensitivity to chemotherapeutic agents [320]. Huang etal.‘s research in cervical carcinogenesis proved that FTO accelerates tumor progression by eliminating m6A methylation from Bone Morphogenetic Protein 4 (BMP4) mRNA. This mechanism results in a rise in BMP4 protein expression levels, which then inhibits downstream Hippo signaling pathways and activates critical effector molecules [321].
Furthermore, m6A regulators are also implicated in cervical cancer-related mechanisms through ncRNAs. HPV16 E7 gene encodes virus-derived circRNA (circE7) that generates the E7 oncoprotein and is linked to cervical cancer. Related research has indicated that the development and progression of CC may be influenced by the mutual regulation of circE7 and m6A modifications [322]. An investigation accomplished by Liang et al. revealed that the demethylase ALKBH5 was capable of removing the m6A modification of circCCDC134 and improving its stability. Subsequently, circCCDC134 acted carcinogenic in CC by sponge-like adsorbing miR-503-5p and stimulating the expression of hypoxia-inducible factor 1a (HIF-1α) [323]. Huang et al. reported that the m6A modification on precursor miR-193b could facilitate its processing into mature miR-193b through METTL3. Further down-regulation of miR-193b encourages the proliferation and invasion of cervical cancer cells by targeting and promoting CCND1-encoding cyclin D1 (CyclinD1) mRNA expression [324].
YTHDF2 and ALKBH5 interact with the lncRNA Growth Arrest-Specific 5 Antisense 1 (GAS5-AS1) to improve the stability of the tumor suppressor gene Growth Arrest-Specific 5 (GAS5) RNA through a m6A-dependent mechanism, thereby delaying CC invasion and metastasis [325]. By modulating the stability and enhancing the biological function of LRRC75A-AS1, IGF2BP1 contributes to the progression of CC by suppressing the ubiquitinated degradation of the NLR family pyrin ___domain 3 (NLRP3) inflammasome, which is carried out by the ubiquitin ligase SYVN1 [326]. In addition, FTO mediates the m6A methylation modification of lncRNA HOXC13-AS and enhances its stability, thereby upregulating frizzled protein class receptor 6 (FZD6) and further activating Wnt/β-catenin signaling. This process results in the EMT and invasion of CC [327]. Other studies have also detailed the different processes through which m6A modification is implicated in the modulation of CC [325, 328, 329].
In general, m6A modification takes part in cervical cancer through multiple pathways, but the specific mechanism of action needs further exploration.
m6A in ovarian cancer
OC is a gynecological tumor that is high in invasiveness and mortality. OC often presents with non-specific first symptoms, most cases are realized at an advanced stage, and about 70% of cases recurrent at a later stage [330, 331].
Ovarian carcinogenesis is facilitated by m6A regulators through a range of mechanisms. METTL3 expression levels in ovarian cancer displayed an up-regulation trend [332]. A study discovered that the down-regulation of METTL3 resulted in changes in m6A modification levels, which in turn regulated the stability and translation of oncogene protein kinase B (AKT) mRNA, reduced the phosphorylation activity of AKT protein, and inhibited the progression of OC by down-regulating downstream target cyclin D 1 (CCND1) expression levels [17]. METTL3 could also facilitate the maturation of pri-miR-126-5p and mediate its m6A modification. As a result, miR-126-5p targets and upregulates downstream tensin homolog PTEN expression levels, thereby activating the PI3K/Akt/mTOR signaling pathway and contributing to the progression of OC [18]. By maintaining the stability of estigial Like Family Member 1 (VGLL1) and upregulating its expression levels through the m6A system, METTL3 and IGF2BP2 further activate the VGLL1/High Mobility Group AT-hook 1 (HMGA1)/β-catenin axis, resulting in OC metastasis [333].
The impacts of METTL14-associated m6A regulatory mechanisms on OC is complex and may exhibit both pro- and tumor suppressor potential, dependent upon the specific molecular and cellular context [334, 335]. The significant role of m6A-dependent related mechanisms mediated by WTAP, RBM15, KIAA1429, and HNRNP in the development and progression of OC has been confirmed by numerous studies [163, 336,337,338]. A study by Liu et al. found that YTHDF1 is responsible for the m6A modification of EIF3C mRNA, which increases its translational efficacy and contributes to the progression of OC [339]. YTHDF2 contributes to the progression of OC by mediating the m6A modification of BMF mRNA and promoting its degradation, thus affecting the process of apoptosis [340]. Conversely, YTHDC1, an additional YTH family reading protein, functions as an OC suppressor. The precise mechanism of action is primarily regulated by the PIK3R1/STAT3/GANAB axis [341].
A study accomplished by Wang et al. on ovarian cancer suggested that IGF2BPs have the capacity to bind the m6A modification on circNFIX, thereby increasing its stability. The overexpression of circNFIX is implicated in tumor malignancy by up-regulating the expression of the miR-647 target gene interleukin-6 receptor (IL-6R) and activating the downstream JAK1/STAT3 signaling pathway [342]. Meanwhile, this process regulates the programmed death ligand 1 (PD-L1) expression to achieve OC-associated immune escape. The TGF-β signaling pathway is believed to be implicated in drug resistance in tumors. One study suggested that the TGF-β pathway may regulate the expression of MDR1 mRNA by modulating the m6A methylation process of RBM15, thereby influencing the chemoresistance of ovarian cancer cells and impacting the prognosis of OC [343]. Additionally, Huang et al. found that FTO-dependent m6A methylation modification is associated with cisplatin resistance in ovarian cancer, although the specific signaling pathways and underlying mechanisms involved in this process remain to be fully elucidated [344].
Furthermore, The expression levels of snail homologous protein 1 (SNAI1) are reduced by FTO in a m6A-IGF2BP2-dependent manner, thereby inhibiting the invasiveness and metastasis of EOC [345]. According to a study conducted by Lv et al., the demethylase ALKBH5 is responsible for the m6A demethylation of lncRMRP, which in turn promotes the expression level of RMRP and facilitates the development of OC [346]. FTO and ALKBH5 demethylation may affect the expression of genes associated with autophagy in OC and contribute to tumor proliferation and invasion [172, 347].
Follicle-stimulating hormone (FSH) has been demonstrated to be involved in the promotion of EMT in OC during specific instances and is associated with the malignancy of OC. An investigation accomplished by Xu et al. investigated the precise mechanism of this theory. FSH increased the expression of ALKBH5, which removed the m6A modification from Snail mRNA. This resulted in higher Snail expression in a m6A-dependent manner. The process led to reduced expression of E-cadherin and promoted mesenchymal genes like N-cadherin, which contributed to OC metastasis [348].
Multiple mechanisms of signaling involved in the development of OC are regulated by the mentioned multiple m6A regulators in a m6A-dependent manner. Nevertheless, there are significant unresolved questions regarding the role of m6A modification in OC.
m6A in endometrial cancer
EC is an imminent threat to the reproductive health of women, as it is prevalent among Perimenopause and postmenopausal women [349]. Within the past few years, its prevalence has increased.
m6A modification plays a critical role in EC. Liu et al. proposed in a study on EC that METTL3-mediated m6A modification revealed a propensity to be down-regulated, which in turn facilitated the progression of EC by stimulating the AKT pathway and regulating the expression levels of downstream target proteins [350]. The aforementioned mechanisms that drive the malignant effects of EC are also affected by METTL14 mutations. Nevertheless, two additional studies reported conflicting results [351, 352]. Due to this, the biological function of METTL3 in EC remains uncertain.
YTHDF2 and YBX2 have the capacity to collaborate on heat shock protein family A (Hsp70) member 6 (HSPA6) mRNA to regulate HSPA6 expression by modulating its stability, which regulates the survival and progression of cancer cells [353]. Additionally, research by Hong et al. has verified that YTHDF2 decreases EC invasiveness by targeting and downregulating the expression of insulin receptor substrate 1 (IRS1) via the m6A system [354]. The recognition and binding of m6A-modified paternally expressed gene 10 (PEG10) mRNA by IGF2BP1 promotes the expression of PEG10 protein by increasing its stability, which promotes the progression of EC [355]. Numerous investigations confirmed the precise mechanism by which IGF2BP family proteins induce EC invasion [356,357,358]. Wang et al. proposed that WTAP and IGF2BP3 may collaboratively regulate the expression of the downstream early growth response factor-1 (EGR1)/PTEN axis through m6A methylation. This regulatory mechanism plays a crucial role in modulating the characteristics and progression of EC stem cells [359].
Furthermore, the m6A modification of ncRNAs has been identified as a critical factor in the mechanisms of EC development. For instance, circCHD7 attracts the reader protein IGF2BP2 to bind and enhance the stability of platelet-derived growth factor receptor β (PDGFRB) mRNA through m6A modification. This process further activates the downstream JAK/STAT signaling pathway by upregulating PDGFRB expression and affecting EC progression [360]. In a study of endometrial cancer investigated by Wang et al., it was discovered that LINC00958 attaches to IGF2BP3, which in turn promotes the stability of E2F3 mRNA and boosts the expression of E2F3 gene levels through a m6A methylation mechanism. Consequently, it promotes the invasiveness of EC [361].
FTO activates the Wnt signaling pathway, which in turn leads to EC invasion, by inhibiting HOXB13 mRNA degradation via a m6A modification-dependent pathway [362]. ALKBH5, another demethylase, further facilitates the activation of the insulin-like growth factor 1 receptor (IGF1R) signaling pathway by improving the stability and translational efficiency of IGF1R mRNA by targeting m6A modification [363]. This mechanism results in the proliferation and invasion of EC. In addition, ALKBH5 is a factor which regulates the expression of tumor stemness genes(e.g., SOX2) and impacts the pathogenicity of EC [250].
The invasiveness of EC is influenced by a variety of m6A regulators, which regulate and target the expression of key genes and the activity of signaling pathways. In the future, more studies will identify the rigorous mechanism of m6A modification in the advancement of EC.
m6A in choriocarcinoma
CCA is a rare and highly aggressive malignant tumor that arises from prenatal trophoblastic tissue. It has a very low occurrence rate but carries a nearly 100% mortality rate unless specific chemotherapeutic drugs are used [364].
The field of m6A modification in CCA is undergoing accelerated development. The most of current investigations on the association between CCA and m6A modification involve mechanisms related to ncRNAs. Lu et al. conducted a study which demonstrated that miR-373-3p, an EMT inhibitor, inhibited CCA invasion by downregulating the TGFβ signaling pathway [365]. MiR935 has been reported to be meaningfully upregulated in CCA tissues [366]. Additionally, a study discovered that METTL3 induces the maturation of miR-935 and mediates its m6A modification, thereby increasing its expression level. Following this, miR-935 targets and inhibits the production of the gap junction protein GJA1, which causes CCA cells to proliferate and invade [367]. Also, Ye et al. confirmed that METTL3 enhances the level of miR-21-5p through the m6A mechanism, which activates the hypoxia-inducible factor asparagine hydroxylase (HIF1A) and further up-regulates the expression of pro-angiogenic factor (VEGF). This increases the blood supply to tumor cells, thereby driving CCA growth and metastasis [368].
Studies on how CCA interacts with other m6A regulators are not available at this time. In the future, we anticipate that additional research will reveal the comprehensive mechanism of m6A modification in CCA, thereby generating novel concepts for the diagnosis and treatment of this kind of disease.
m6A in adenomyosis and endometriosis
Adenomyosis is a common gynecological condition which is characterized by the invasion of the myometrium by endometrial glands and stroma, resulting in myometrial hypertrophy. Dysmenorrhea and irregular vaginal hemorrhage are the primary characteristics of the disease, greatly affect the quality of life of those impacted [369]. According to published research, m6A regulators may regulate immune responses and cell adhesion during the development of adenomyosis [370]. While there is a scarcity of research on m6A modification in adenomyosis, there is some sign that it may be influential in the endometrium and associated diseases. Zhai et al. confirmed that the METTL3-mediated m6A modification was capable of targeting and regulating the expression of critical genes, including insulin-like growth factor 1 (IGF1) and the detoxifying enzyme DDT, that influenced the invasiveness of adenomyosis. This was achieved by analyzing endometrium-associated gene expression profiles in patients with adenomyosis in the GEO database [370].
EMT is a multipart condition that is initiated by abnormal growth-related mechanisms in the endometrium, some of which are presently unknown [371]. According to a study accompanied by Li et al., bioinformatics analysis indicated that six m6A regulators (METTL3, YTHDF2, YTHDF3, FTO, HNRNPC, and HNRNPA2B1) were differentially expressed among ectopic endometrium and controls. Among them, the growth and invasion of ectopic endometrial cells may be regulated by changes in the expression of HNRNPA2B1 and HNRNPC, which may influence immune mechanisms in endometriosis [372]. Furthermore, another study showed that the down-regulation of methylase METTL3 further impacted DGCR8-mediated splicing processes by reducing the m6A modification level of pri-miR126. This decrease in m6A modification level led to a decrease in the generation of mature miRNA126, which consequently facilitated the migration and invasion of endometriosis [373].
The aforementioned studies offer preliminary insights into the investigation of m6A modification and the correlation between endometriosis and adenomyosis. To cultivate a thorough comprehension of the precise mechanisms of the interaction between the two, additional in vitro verification in the future is required.
m6A in POI
POI may be influenced by the activity of m6A regulators in regulating germ cell metabolism and function. Cao et al. discovered that the secretion of interleukin 1β (IL-1β) by theca cells was upregulated by decreased levels of m6A modification, which in turn promoted the development of POI by knocking down METTL3 [374]. Pathogenic variants in the reading protein YTHDC2 may influence the normal progression of the meiotic process and contribute to the development of POI [375]. FTO-mediated m6A demethylation has the potential to improve the stability of circBRCA1, which subsequently binds miR-642a-5p and exerts a sponge-like effect. This prevents the interaction of circBRCA1 with the transcription factor (FOXO1) and contributes to the prevention of POI [376]. This conclusion was substantiated by Ding et al. [377].
Furthermore, it has been demonstrated that chemical toxicants, including cyclophosphamide (CTX) and 4-vinylcyclohexene diepoxide (VCD), can impair ovarian function and induce POI. In a rat model of VCD-induced ovarian dysfunction, ALKBH5 acts on YAP mRNA in a m6A-dependent mechanism to further upregulate YAP expression and affect ovarian function [378]. In a human granulosa cell model and a mouse model of POI, investigators discovered that the m6A modification levels in cells or tissues increased substantially in a concentration-dependent manner after CTX treatment, thereby driving the progression of POI [379]. The direction and focus of future research are the specific mechanisms or relevant targets implicated in m6A affecting POI.
m6A in PCOS
Hyperandrogenism, insulin resistance, and ovulatory dysfunction are frequently encountered in PCOS [380]. The pathogenesis of PCOS is significantly influenced by the m6A methylation modification of mRNA. It has been demonstrated that the activity and protein of insulin-related signal transduction pathways, such as the PI3K/AKT pathway, appear to influence the pathogenesis of PCOS in murine PCOS models through the m6A mechanism [15]. Nevertheless, specific studies are still in the preliminary phases of development.
In addition, various studies have previously shown a correlation between FTO gene abnormalities and the mechanisms that lead to the development of PCOS [381, 382]. FTO expression is positively correlated with androgen levels, and androgen excess is one of the significant manifestations of PCOS. Xue et al. accomplished a study that verified the precise mechanism of FTO-mediated m6A demethylation modification in PCOS-induced androgen excess. Through the m6A mechanism, FTO enhances the stability of key enzymes involved in androgen synthesis, including CYP17A1 and HSD3B2, and up-regulates their expression levels, thereby contributing to androgen synthesis. The inhibition of FTO contributes to a decrease in the expression of these enzymes and the androgen receptor (AR), which in turn reduces the AR/AKT signaling activity and effectively reduces androgen production and secretion, thereby improving the hyperandrogenic state [383]. Another study suggested that FTO could also contribute to insulin resistance in PCOS by removing the m6A modification from the membrane-associated protein FLOT2 (Flotillin-2) mRNA. This would reduce its degradation, increase its stability, and up-regulate FLOT2 expression levels [384].
Moreover, a study of m6A modification in luteinized granulosa cells (GC) from PCOS patients found that YTHDF2 mediated m6A modification of Forkhead Box O3 (FOXO3) mRNA and participated in its degradation in GC from controls group. However, this mechanism was absent in PCOS group, indicating that there is dysregulation of FOXO3 transcription by m6A regulators in PCOS, which may affect pathological development of PCOS [385]. In the future, these discoveries will offer novel concepts for elucidating the fundamental mechanism of PCOS pathogenesis.
m6A in spontaneous abortion
SA is a frequent complication of pregnancy, and its mechanism is intricate. Numerous researches have verified the association between m6A methylation levels and miscarriage [386, 387]. Huang et al. hypothesized that the m6A modification of Zinc Finger and BTB Domain Containing 4 (ZBTB4) mRNA by METTL3 may be implicated in recurrent miscarriage-associated mechanisms by regulating ZBTB4 expression [388]. Another investigation determined that the reading protein HnRNPC regulates the expression of 5-methyltetrahydrofolate homocysteine methyltransferase (MTHFR) through the m6A mechanism and may induce recurrent miscarriage [389].
The correlation between abortion and benzene exposure was progressively confirmed. Benzo (a) pyrene-7,8-dihydrodiol-9,10-epoxy (BPDE) is a group of chemicals that have reproductive toxicity. They can impact trophoblast function through a variety of pathways, which may result in abortion. According to a study conducted by Xu et al., the methylase METTL14 was upregulated by BPDE exposure. Thus, increased its stability and promoted its expression level by adding m6A modification to lnc-HZ01 and MXD1 mRNA. Consequently, a positive self-feedback loop mechanism was established between the two, which hindered trophoblast proliferation and resulted in abortion [390]. In addition, two additional studies have shown that the up-regulation of lnc-HZ14 [230] and lncRNA-HZ09 [391] is a result of the up-regulation of BPDE exposure, respectively. These mechanisms are related to abortion pathogenesis.
Multiple mechanisms are employed by the demethylase ALKBH5 to induce abortion-associated diseases. Li et al. hypothesis that ALKBH5 could impede trophoblast invasion by increasing the stability of CYR61 mRNA and mediating m6A modification [231]. Another study, which utilized bioinformatics analysis, indicates that the dysregulation of the VEGF signaling pathway may result from ALKBH5 overexpression. This dysregulation impacts the function of macrophages in the endometrium and promotes recurrent spontaneous abortion [392]. In contrast, Zheng et al. discovered that ALKBH5 is capable of up-regulating the expression levels of transcription factors SMAD1 and SMAD5 through the m6A demethylation mechanism, thereby promoting trophoblast viability and preventing spontaneous abortion [393].
Abnormalities of the maternal-fetal interface are frequently linked to spontaneous abortion. FTO-mediated m6A modification processes have been reported to be crucial for the preservation of normal maternal-fetal interfaces. The downregulation of FTO has the potential to result in maternal-fetal-related morbidity, including spontaneous abortion [394]. In research concerning the pathogenesis of unexplained abortion, investigators have also discovered that the FTO-related m6A modification mechanism regulates the MEG3-TGF-β signaling pathway, thereby influencing chorionic invasiveness and inducing abortion [395].
Investigating the relationship between the mechanism of abortion and m6A will aid in comprehending the molecular underpinnings of abortion and may yield novel clinical preventive and therapeutic strategies.
m6A in preeclampsia
As previously mentioned, the mechanisms by which m6A regulators regulate the invasion and migration of placental trophoblasts are involved. However, the progression of PE may be influenced by aberrant trophoblastic invasion. Related research has shown a strong correlation between the development of PE and the upregulation of m6A modification levels [208, 396]. In contrast, there are also indications that the levels of m6A modification are downregulated in PE. Complex mechanisms associated with the METTL3-myosin light chain kinase (MYLK) axis and the WTAP/IGF2BP1/high mobility group nucleosome binding ___domain 3 (HMGN3) axis are implicated in this discovery [228, 397]. Further investigation of its impact on m6A modification levels will assist in the identification of the pathogenesis of PE.
In a study conducted by Chen, it was suggested that METTL3 and YTHDF2 reduce the level of transmembrane BAX inhibitor 6 (TMBIM6) by regulating the m6A modification of TMBIM6 mRNA and promoting its degradation. This process results in the dysfunction of trophoblasts and the development of PE [398]. Newly published research has also discovered that METTL3 is implicated in the progress of PE by influencing the iron apoptotic mechanism of trophoblasts. It regulates the m6A modification of Acyl-CoA Synthase Long Chain Family Member 4 (ACSL4) mRNA through a specific mechanism [399].
Furthermore, in HTR-8/SVneo cells, the up-regulation of METTL14 alters the stability and translation of FOXO3a mRNA by increasing m6A modification. This results in increased FOXO3a expression, which in turn leads to PE development [400]. One study discovered that the overexpression of the methylase RBM15 reduced CD82 expression and facilitated trophoblastic invasion and PE development by increasing the levels of m6A modification, promoting YTHDF2 recognition and binding to CD82 mRNA, and enhancing its degradation [401]. In contrast, ALKBH5 has the potential to alleviate the pathological process of PE by increasing the expression level of PLAC8 by eliminating the m6A modification on placenta-specific 8 (PLAC8) mRNA [402].
The ncRNAs are also regulated by the m6A regulator to influence trophoblast function in PE through a variety of mechanisms. Wang et al. confirmed that METTL3 mediates the m6A modification of lncRNA HOXD-AS1 and maintains its stability. Subsequently, the up-regulated HOXD-AS1 recruits miR-135a and exerts a sponge-like effect, relieving its inhibition of the target gene anti-apoptotic protein β-repein transducat-containing protein (β-TRCP) mRNA. This contributes to the upregulation of β-TRCP expression, stimulation of the NF-κB signaling pathway, and an impact on PE development [403]. Similarly, the circSETD2/miR-181a-5p/Myeloid Cell Leukemia 1 (MCL1) axis, which is mediated by METTL3, is also implicated in the development of PE [404].
METTL14 and IGF2BP3 may also be responsible for the regulation of circPAPPA2 m6A methylation stability and the enhancement of its expression, which can impact the progression of PE and placental function [405]. Finally, the regulation of angiogenesis-related gene expression in trophoblasts may be facilitated by the IGF2BP2-mediated m6A modification of linc01116 RNA, which offers a novel perspective on the development of PE [406].
m6A in GDM
The manipulation of insulin sensitivity, inflammatory response, placental function, and cellular metabolism is the fundamental mechanism of m6A modification in GDM [407,408,409]. One study suggests that METTL3-dependent m6A modification enhances the stability of hsa_circ_0072380, potentially influencing the pathogenesis of GDM by regulating pathways or gene expression associated with these factors [410]. Furthermore, Ning et al. elucidated the pivotal role of m6A modification in placental development in GDM, demonstrating that FTO regulates the fusion process of trophoblast cells by modulating the stability and expression of SIK1 mRNA [411]. A study that utilized GEO data indicated that m6A-modified LINC00667 may be involved in the development of GDM by modulating the expression of downstream GDM-related target genes in response to the recognition and binding of YTHDF3 [412]. This conclusion must, however, be verified in in vivo or in vitro models.
In high glucose-cultured HTR8/SVneo cells, METTL14 inhibits FOXO1 expression, enhances trophoblast function, increases downstream miR-497-5p expression levels, and mediates XIST silencing through the m6A mechanism, thereby impeding the progression of GDM [413]. Chen et al. discovered that chemokine ligand 5 (CCL5) levels were generally higher in GDM patients. Conversely, METTL14 possibly influenced the progression of GDM by mediating m6A modification of CCL5 and affecting its stability, which may have resulted in the promotion of proliferation, migration, and apoptosis of trophoblasts [414]. In addition, Fang et al. discovered that RBM15 regulates CLDN4 expression through m6A modification, which subsequently affects hepatic glucose and lipid metabolism in the progeny of GDM mice, resulting in insulin resistance [415].
Currently, there are insufficient studies on the function of m6A modification in GDM regulation, and the precise mechanism is unclear. Therefore, further studies are necessary. Future research should aim to validate these preliminary findings through more sufficient in vivo and in vitro experiments, alongside large-scale, multicenter clinical studies, to further elucidate the biological functions and underlying mechanisms of m6A modification in GDM. Such studies will provide a more robust and reliable foundation for a comprehensive understanding of the role of m6A modification in the pathogenesis and progression of GDM.
Treatment and prognosis
The m6A modification mechanism in various of gynecological tumors has been progressively elucidated, which is anticipated to promote the creation of innovative therapeutic strategies and enhance the prognosis of patients. Furthermore, the potential of m6A modifiers as biomarkers for the early diagnosis and prognosis of cancer is increasingly recognized and investigated. While traditional tumor markers (e.g., CA125, HE4,SCC) and diagnostic methods (e.g., HPV detection, TCT) have played an important role in the early detection and monitoring of gynecological cancers, their clinical applicability is constrained by limitations such as low sensitivity, poor specificity, and insufficient capacity for dynamic monitoring. Future advancements in quantifying and localizing specific m6A modification levels and loci within blood exosomal RNA and tissue mRNA hold promise for overcoming these challenges. Such approaches could provide powerful tools for the early and accurate diagnosis, biomarker discovery, and the personalization of treatment strategies for gynecological malignancies. However, the clinical application of m6A modification in early detection remains largely in the basic research phase, with no existing clinical studies to date. Thus, further standardization and rigorous clinical validation will be necessary to translate these findings into clinical practice.
In CC, m6A methylation levels are significantly correlated with cancer progression and survival. The levels of ZC3H13, YTHDF1, and YTHDC1 expression in cervical squamous cell carcinoma (CESC) were found to significantly correlate with the prognosis of CESC by bioinformatics analysis [416]. These proteins may also be useful targets for cancer therapy. According to Wang et al., the aforementioned conclusions were partially corroborated by the findings of a biological analysis study of CC patients in the GEO and TCGA databases. The study revealed that RBM15 is a prognostic biomarker in CC, and ZC3H13 may impact overall survival (OS) in CC patients [417]. Therefore, further mechanistic investigations are necessary to validate these conclusions in the future.
A study confirmed that YTHDF1 induces the expression of RANBP2 (RAN-binding protein 2) through a m6A-dependent mechanism, thereby facilitating CC metastasis [19]. The m6A modification of low-density lipoprotein receptor-related protein 6 (LRP6) mRNA is mediated by YTHDF3, which subsequently promotes the expression of LRP6 protein and activates downstream signaling pathways. This process is responsible for the invasion of CC and the metastasis of lymph nodes [418]. YTHDFs proteins may have a significant impact on CC treatment, as suggested by relevant studies.
A study on the mechanism of CESC resistance revealed that FTO, through eliminating m6A alteration on β-catenin mRNA, increasing its stability, and up-regulating expression levels, might boost patient chemoresistance and lessen the lethal effect of chemotherapeutic medicines on cancer cells [419]. Centromere protein K (CENPK) is also a contributing factor to recurrence and drug resistance in CC. A study discovered that the Wnt/β-catenin signaling pathway was activated by methylase ZC3H13, which increased the level of CENPK mRNA and mediated the m6A methylation modification process. This process resulted in the progression of CC and development of chemoresistance [420]. In cancer management, early detection of potential recurrence, assessment of patient response to treatment, and prediction of prognosis are crucial for optimizing therapeutic strategies and improving patient survival and quality of life. Therefore, dynamic monitoring of m6A modifier expression levels, coupled with the development of predictive models based on these markers, could provide a more comprehensive evaluation of treatment responses, recurrence risks, and long-term prognoses. Additionally, clinicians could stratify patients into high-risk and low-risk groups based on m6A modification factor expression, which would offer a foundation for personalized treatment plans. Currently, most in vivo and in vitro studies primarily focus on the expression levels of m6A modification-related molecules and their correlation with patient prognosis. However, further large-scale cohort studies are required to validate these findings, ensuring a robust clinical translation of m6A methylation modifications in reproductive system diseases.
The results of numerous bioinformatics-related researches have indicated that m6A modifier regulators have significant prognostic potential in OC [421,422,423,424,425,426,427,428]. A study based on OC patients in the TCGA database revealed that high expression of WTAP, YTHDF1/2/3, FTO, and ALKBH1/5, while low expression level of METTL14 may be associated with poor prognosis in OC [425]. Li et al.‘s study indicates that VIRMA, IGF2BP1, and HNRNPA2B1 may be correlated to the prognosis of OC. The risk prediction model has been found to be effective based on the aforementioned conclusions. In addition, the investigators employed the miRbase database to screen upstream miRNA targets of the aforementioned genes and discovered that miR-196b-5p may target IGF2BP1, modulate the expression level of downstream PTEN protein, and lead to OC progression [422]. Zhu et al. additionally demonstrated the predictive role of KIAA1429 and YTHDC2 in OC [426]. Nevertheless, these findings are only suitable for use as a preliminary reference, as they are not supported by confirmatory in vitro experiments.
Yu et al. conducted an experimental study on high-grade serous ovarian cancer (HGSOC) that confirmed elevated WTAP expression as a poor predictor of HGSOC [429]. This research partially corroborates the conclusions of Han et al. [425]. Sun et al. also discovered that ALKBH5 overexpression may be involved in pertinent mechanisms that mediate OC lymph node metastasis [430]. A study conducted by Ye et al. was innovative in that it combined machine learning algorithms to screen m6A modification-related lncRNAs in serous ovarian cancer (SOC) and establish a predictive model (m6A-LRM). The study found that METTL3 down-regulation may be involved in regulating RP11-508M8.1 (one of the selected lncRNAs) and promoting OC cell invasion [428]. The model demonstrated a high degree of predictive accuracy for the treatment effect of SOC and OS in the subsequent validation trial.
Platinum medications serve as the foundation of initial OC treatment. A study on cisplatin resistance in OC patients discovered that METTL3 increased the stability of lncRNA RHPN1 antisense RNA 1 (lncRNA RHPN1-AS1) in a m6A-dependent mechanism. This, in turn, resulted in increased cisplatin resistance in OC patients by upregulating the downstream PI3K/AKT pathway [431]. Another study also demonstrated that the METTL3/YTHDF2 axis also downregulates interferon-inducible RNA helicase 1 (IFFO1) expression by mediating the m6A modification of IFFO1 mRNA and promoting its degradation, leading to cisplatin resistance and metastasis in ovarian cancer [432].
Furthermore, Yuan et al. verified that the RBM15 expression level can predict paclitaxel resistance in OC patients. The specific mechanism is affected by the transforming growth factor-β (TGF-β)/RBM15/multidrug resistance protein 1 (MDR1) axis, in which RBM15 regulates MDR1 expression through the m6A mechanism, thereby affecting the effect of chemotherapy [343]. Specific mechanisms of ALKBH5 [433], FTO [434], and YTHDF1 [248, 435] in OC cisplatin resistance, as well as METTL3 [436] in OC carboplatin resistance, have also been reported in numerous studies.
Given the m6A regulators previously mentioned, there is an increasing interest in the investigation of targeted agents that are linked to OC resistance. For instance, AE-848 has the capacity to function as a targeted agent for OC therapy by destabilizing IGF2BP3-targeted mRNA, thereby impeding the growth and progression of OC cells [437]. Sulfonamides (Sul) are potential targets for OC therapy and may influence the process of apoptosis through the METTL3-mediated m6A mechanism [438]. As prospective biomarkers and therapeutic targets in OC, FTO and ALKBH5 may be implicated in PARP inhibitor (PARPi)-related biological mechanisms [439]. However, research on targeted therapies related to m6A modification has yet to reach the clinical trial stage. Furthermore, most existing studies rely on animal and cellular models, which do not adequately reflect the complexity of human tumors, particularly in terms of the tumor microenvironment, immune response, and treatment response. The intricate nature of m6A modification mechanisms, combined with tumor heterogeneity and individual variability, makes it challenging for current models to fully capture the dynamics of tumor growth and metastasis. Moreover, existing studies lack an in-depth exploration of long-term tumor progression and the mechanisms underlying drug resistance, which significantly restricts the clinical translation and application of m6A modification research findings. Future research should concentrate on this topic in order to elucidate the molecular mechanisms which may aid in the development of clinical treatments.
The regulatory mechanisms of pertinent prognostic biomarkers in EC are also influenced by the m6A modification and its regulators. Based on CpG sites in the m6A regulator, specific prognostic prediction models have been developed in related studies that are effective in foreseeing the prognosis of patients with EC [440]. Zhai et al. hypothesized that FTO, YTHDF1 and RBM15 are important factors in the prognosis of EC patients in a study that utilized the TCGA dataset of EC patients [205]. An additional experimental study conducted recently also verified that the elevated METTL3, METTL14, FTO, HNRNPC, and HNRNPA2B1 expression levels are associated with a shorter OS in EC patient [351]. In addition, Zhan et al. discovered that METTL3 and YTHDF2 are involved in the m6A modification process of NLRC5 mRNA, which upregulates its stability and inhibits degradation. This results in an increase in NLRC5 expression levels, a reduction in NLRC5-mediated immune surveillance, and a participation in EC progression [441]. The IGF2BP1 is involved in the process of EC cell metastasis, as it binds to and promotes the expression and accumulation of TRKB protein in a m6A-dependent manner [358].
Moreover, two studies accomplished by Shi [442] et al. and Shan [443] et al. demonstrated a correlation between the expression levels of m6A-related lncRNA and the survival of ECs, as well as immune-related mechanisms. Consequently, it is conceivable that the m6A regulators listed above may be promising targets in the development of EC, and the inhibitors or activators that are generated may have potential regulatory effects on the treatment and prognosis of EC patients.
Limited research has been conducted on the topic of EC treatment and resistance. It has been reported that METTL14 and YTHDF2 are indirectly implicated in the progression of iron death by influencing the methylation process of protein arginine methyltransferase (PRMT) through the m6A mechanism. However, the aforementioned processes are impeded by PRMT inhibition, which enhances the sensitivity of endometrial cancer cells to iron death and offers a novel perspective on EC treatment and drug resistance research [444]. In summary, clinicians will be able to detect and treat gynecological malignancies at an early period, thereby enhancing the survival rate of patients, by conducting a thorough analysis of the high-risk factors associated with their occurrence and development.
Additionally, treatment of other female reproductive system diseases and m6A-related mechanisms. In a rat model of PCOS, Zhang et al. found that Cangfu Daotan Decoction (CDD) methylation of m6A decreased the activity of the Wnt/β-catenin pathway, thereby improving the recovery of ovarian function [445]. Another study suggested that butyric acid supplementation could alleviate the symptoms of PCOS by inhibiting the expression of METTL3 and reducing the stability and expression of FOSL2 mRNA. This would down-regulate the expression of NLRP3 and further decrease the expression of downstream inflammatory factors [446].
There are inflammatory factors that contribute to the development of ovarian senescence, whereas melatonin has a potent antioxidant effect. Zhu et al. demonstrated that melatonin mitigates the decline in ovarian function by regulating the expression of the m6A regulator YTHDF2 and reducing its mediated mRNA degradation, further blocking the MAPK-NF-κB signaling pathway and achieves anti-inflammatory objectives [447]. Interesting, it has been shown that melatonin is also essential for the protection of pregnancy by modulating inflammatory and apoptosis-related pathways through the m6A mechanism [448]. Finally, Chen et al. confirmed that m6A expression was significantly elevated in trophoblasts of GDM patients with obesity compared to non-obese individuals. Hesperidin was able to indicate its potential role in the treatment of GDM by participating in the regulation of autophagy and m6A methylation levels [449]. In the future, more comprehensive research is needed to investigate the specific mechanism of m6A modification in the treatment of female reproductive system diseases.
The development of drugs targeting m6A modification regulators represents a cutting-edge frontier in the field of RNA epigenetics. To date, clinical trials of related therapeutics have progressed relatively slowly, with the majority of researches still in the basic research or early-phase clinical trial stages. Currently, m6A-modifying regulators under investigation include METTL3 inhibitors such as STM2457 [450] and STC-15, as well as FTO inhibitors like MO-I-500 and R-2-hydroxyglutaric acid (R-2HG) [451], which have shown potential for treating various solid tumors and related diseases. These compounds are anticipated to emerge as novel therapeutic agents in the RNA epigenetics ___domain. Notably, a clinical trial (NCT05584111) evaluating the METTL3 inhibitor STC-15 in advanced solid tumors is underway, with preliminary results expected by 2024. With an increasing understanding of the molecular mechanisms underlying m6A modification and the continuous evolution of drug development technologies, it is projected that more m6A-targeted therapies will advance into clinical trials, potentially offering new treatment avenues for obstetric and gynecological diseases.
Conclusion and future perspectives
The m6A modification is a highly prevalent methylation modification in RNA that is critical for cellular physiology and pathology. In this study, we present a comprehensive overview of m6A-related regulators and their biological function-related mechanisms, the development history of m6A detection methods, the various roles that m6A modification plays in the female reproductive system, and their possible connections to associated diseases.
Additionally, the m6A modification has a multifaceted effect on the development of related maladies and the health of female reproductive systems. It regulates t the maturation and development of reproductive cells, the synthesis and signal transduction of reproductive hormones, the maintenance of maternal-fetal microenvironment homeostasis, and the mechanisms related to embryonic development. This perspective provides obstetricians and gynecologists with a fresh understanding of the diseases in question. Currently, the primary bottlenecks and challenges in the field of m6A modification include the unclear and complex regulatory mechanisms, the experimental model is not fully representative, and the limited accuracy of detection technologies. Although m6A modification has shown potential in the occurrence, progression, and targeted therapy of various diseases, its clinical application as a biomarker remains in the early stages. There is also a lack of m6A-targeted drugs or therapeutic strategies, and numerous technical and biological hurdles must still be overcome for successful clinical translation. The gap between basic research and clinical application is hindered by both technical and practical obstacles. To address these challenges, it is essential for basic research to focus on standardizing m6A detection methods and improving their accuracy. Additionally, collaboration with clinical practice should be strengthened through large-scale, multi-center studies to validate findings and facilitate the integration of m6A modification research into clinical settings. Moreover, future research should explore the interactions between m6A modification and other epigenetic modifications, such as DNA methylation and histone modifications, particularly in gynecological cancers. Investigating how these epigenetic changes synergistically or mutually regulate gene expression and cellular function will provide deeper insights into the mechanisms driving disease initiation and progression, thereby offering a more comprehensive theoretical foundation for early diagnosis, prognosis evaluation, and targeted therapies.
This study has several limitations. Many of the studies related to m6A modification and obstetric and gynecological diseases included in this review have small sample sizes and primarily focus on cell lines and small animal models, which are not supported by large-scale clinical data. While these experimental models provide essential insights into the biological mechanisms of m6A modification, they may not fully capture the complexity and diversity of human diseases. Additionally, small sample studies are prone to statistical biases that could affect the reliability of the findings. Therefore, future research should prioritize the collection of clinical samples, particularly through multi-center, large-scale clinical cohort studies, to more comprehensively assess the clinical relevance of m6A modification in the onset, progression, and prognosis of diseases of the female reproductive system.
In addition, while relevant studies have highlighted the potential role of m6A modification in diseases of the female reproductive system, particularly cancer, the underlying molecular mechanisms and the complex regulatory networks involved remain not fully elucidated. One example is that the scarcity of researches on the regulatory mechanisms of m6A modification on immune cells and reproductive cells in various pathological conditions. Additionally, the regulatory mechanisms of the m6A regulator are complex, as they may serve distinct functions in various diseases. Moreover, the regulatory mechanism of the m6A mechanism in specific gynecological diseases (e.g., PCOS, endometriosis, GDM, etc.) has not been thoroughly examined.
Therefore, future research should focus on elucidating the detailed molecular mechanisms of m6A methylation modification in regulating gene expression, and investigate how these processes influence the beginning and progression of female reproductive system diseases. Additionally, it is crucial to explore strategies for enhancing the specificity and precision of targeted therapies, addressing potential drug resistance issues that may arise during treatment. This will provide theoretical support for the development of individualized treatment plans and facilitate the translation of m6A modification mechanisms into clinical practice, ultimately advancing therapy in clinical settings.
Data availability
All data in this review are publicly available.
References
Frye M, Jaffrey SR, Pan T, Rechavi G, Suzuki T. RNA modifications: what have we learned and where are we headed? Nat Rev Genet. 2016;17:365–72.
Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK, et al. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018;46:D303–d7.
Meyer KD, Jaffrey SR. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat Rev Mol Cell Biol. 2014;15:313–26.
Li X, Xiong X, Wang K, Wang L, Shu X, Ma S, et al. Transcriptome-wide mapping reveals reversible and dynamic N(1)-methyladenosine methylome. Nat Chem Biol. 2016;12:311–6.
Chen YS, Yang WL, Zhao YL, Yang YG. Dynamic transcriptomic m(5) C and its regulatory role in RNA processing. Wiley Interdiscip Rev RNA. 2021;12:e1639.
Guzzi N, Muthukumar S, Cieśla M, Todisco G, Ngoc PCT, Madej M, et al. Pseudouridine-modified tRNA fragments repress aberrant protein synthesis and predict leukaemic progression in myelodysplastic syndrome. Nat Cell Biol. 2022;24:299–306.
Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci USA. 1974;71:3971–5.
Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169:1187–200.
Saleeby E, Brindis CD. Women, reproductive health, and health reform. JAMA. 2011;306:1256–7.
Zhao J, Kong Y, Xiang Y, Yang J. The research landscape of the quality of life or psychological impact on gynecological cancer patients: A bibliometric analysis. Front Oncol. 2023;13:1115852.
De Graaff AA, Van Lankveld J, Smits LJ, Van Beek JJ, Dunselman GA. Dyspareunia and depressive symptoms are associated with impaired sexual functioning in women with endometriosis, whereas sexual functioning in their male partners is not affected. Hum Reprod. 2016;31:2577–86.
Sun X, Lu J, Li H, Huang B. The role of m(6)a on female reproduction and fertility: from gonad development to ovarian aging. Front Cell Dev Biol. 2022;10:884295.
Mu H, Zhang T, Yang Y, Zhang D, Gao J, Li J, et al. METTL3-mediated mRNA N(6)-methyladenosine is required for oocyte and follicle development in mice. Cell Death Dis. 2021;12:989.
Zhang J, Ma R, Li L, Wang L, Hou X, Han L, et al. Intersectin 2 controls actin cap formation and meiotic division in mouse oocytes through the Cdc42 pathway. Faseb j. 2017;31:4277–85.
Zou L, Li W, Xu D, Zhu S, Jiang B. Alteration of the N(6)-methyladenosine methylation landscape in a mouse model of polycystic ovary syndrome. J Ovarian Res. 2023;16:157.
Sun X, Zhang Y, Hu Y, An J, Li L, Wang Y, et al. Decreased expression of m(6)A demethylase FTO in ovarian aging. Arch Gynecol Obstet. 2021;303:1363–9.
Liang S, Guan H, Lin X, Li N, Geng F, Li J. METTL3 serves an oncogenic role in human ovarian cancer cells partially via the AKT signaling pathway. Oncol Lett. 2020;19:3197–204.
Bi X, Lv X, Liu D, Guo H, Yao G, Wang L, et al. METTL3-mediated maturation of miR-126-5p promotes ovarian cancer progression via PTEN-mediated PI3K/Akt/mTOR pathway. Cancer Gene Ther. 2021;28:335–49.
Wang H, Luo Q, Kang J, Wei Q, Yang Y, Yang D, et al. YTHDF1 aggravates the progression of cervical cancer through m(6)A-mediated up-regulation of RANBP2. Front Oncol. 2021;11:650383.
Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177–89.
Knuckles P, Lence T, Haussmann IU, Jacob D, Kreim N, Carl SH, et al. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m(6)A machinery component Wtap/Fl(2)d. Genes Dev. 2018;32:415–29.
Yue Y, Liu J, Cui X, Cao J, Luo G, Zhang Z, et al. VIRMA mediates preferential m(6)A mRNA methylation in 3’UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018;4:10.
Wang P, Doxtader KA, Nam Y. Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol Cell. 2016;63:306–17.
Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10:93–5.
Schwartz S, Mumbach MR, Jovanovic M, Wang T, Maciag K, Bushkin GG, et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5’ sites. Cell Rep. 2014;8:284–96.
Wen J, Lv R, Ma H, Shen H, He C, Wang J, et al. Zc3h13 regulates nuclear RNA m(6)A methylation and mouse embryonic stem cell self-renewal. Mol Cell. 2018;69:1028–38.e6.
van Tran N, Ernst FGM, Hawley BR, Zorbas C, Ulryck N, Hackert P, et al. The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res. 2019;47:7719–33.
Warda AS, Kretschmer J, Hackert P, Lenz C, Urlaub H, Höbartner C, et al. Human METTL16 is a N(6)-methyladenosine (m(6)A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 2017;18:2004–14.
Mendel M, Chen KM, Homolka D, Gos P, Pandey RR, McCarthy AA, et al. Methylation of structured RNA by the m(6)A Writer METTL16 is essential for mouse embryonic development. Mol Cell. 2018;71:986–1000.e11.
Aoyama T, Yamashita S, Tomita K. Mechanistic insights into m6A modification of U6 snRNA by human METTL16. Nucleic Acids Res. 2020;48:5157–68.
Ren W, Lu J, Huang M, Gao L, Li D, Wang GG, et al. Structure and regulation of ZCCHC4 in m(6)A-methylation of 28S rRNA. Nat Commun. 2019;10:5042.
Pinto R, Vågbø CB, Jakobsson ME, Kim Y, Baltissen MP, O’Donohue MF, et al. The human methyltransferase ZCCHC4 catalyses N6-methyladenosine modification of 28S ribosomal RNA. Nucleic Acids Res. 2020;48:830–46.
Bawankar P, Lence T, Paolantoni C, Haussmann IU, Kazlauskiene M, Jacob D, et al. Hakai is required for stabilization of core components of the m(6)A mRNA methylation machinery. Nat Commun. 2021;12:3778.
Zhang Z, Theler D, Kaminska KH, Hiller M, de la Grange P, Pudimat R, et al. The YTH ___domain is a novel RNA binding ___domain. J Biol Chem. 2010;285:14701–10.
Hazra D, Chapat C, Graille M. m6A mRNA destiny: chained to the rhYTHm by the YTH-containing proteins. Genes (Basel). 2019;10:49.
Rauch S, He C, Dickinson BC. Targeted m(6)A reader proteins to study epitranscriptomic regulation of single RNAs. J Am Chem Soc. 2018;140:11974–81.
Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, et al. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun. 2016;7:12626.
Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, et al. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 2017;27:315–28.
Roundtree IA, Luo GZ, Zhang Z, Wang X, Zhou T, Cui Y, et al. YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. Elife. 2017;6:e31311.
Roundtree IA, He C. Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Trends Genet. 2016;32:320–1.
Lesbirel S, Viphakone N, Parker M, Parker J, Heath C, Sudbery I, et al. The m(6)A-methylase complex recruits TREX and regulates mRNA export. Sci Rep. 2018;8:13827.
Nielsen J, Christiansen J, Lykke-Andersen J, Johnsen AH, Wewer UM, Nielsen FC. A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Mol Cell Biol. 1999;19:1262–70.
Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–20.
Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20:285–95.
Bechara R, Amatya N, Bailey RD, Li Y, Aggor FEY, Li DD, et al. The m(6)A reader IMP2 directs autoimmune inflammation through an IL-17- and TNFα-dependent C/EBP transcription factor axis. Sci Immunol. 2021;6:eabd1287.
Mamsen LS, Zafeiri A, Bøtkjær JA, Hardlei JR, Ernst E, Oxvig C, et al. Expression of the insulin-like growth factor system in first- and second-trimester human embryonic and fetal gonads. J Clin Endocrinol Metab. 2020;105:e3157–68.
Sun CY, Cao D, Du BB, Chen CW, Liu D. The role of Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs) as m(6)A readers in cancer. Int J Biol Sci. 2022;18:2744–58.
Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, et al. 5’ UTR m(6)A promotes cap-independent translation. Cell. 2015;163:999–1010.
Wang YC, Chang KC, Lin BW, Lee JC, Lai CH, Lin LJ, et al. The EGF/hnRNP Q1 axis is involved in tumorigenesis via the regulation of cell cycle-related genes. Exp Mol Med. 2018;50:1–14.
Zhou, Shi KI, Lyu H, Wylder R, Matuszek AC, Pan JN, et al. Regulation of co-transcriptional Pre-mRNA Splicing by m(6)A through the low-complexity protein hnRNPG. Mol Cell. 2019;76:70–81.e9.
Alarcón CR, Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie SF. HNRNPA2B1 is a mediator of m(6)A-dependent nuclear RNA processing events. Cell. 2015;162:1299–308.
Liu N, Zhou KI, Parisien M, Dai Q, Diatchenko L, Pan T. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 2017;45:6051–63.
Tan X, Zheng C, Zhuang Y, Jin P, Wang F. The m6A reader PRRC2A is essential for meiosis I completion during spermatogenesis. Nat Commun. 2023;14:1636.
Chen YS, Dong J, Tan W, Liu H, Zhang SM, Zou J, et al. The potential role of ribonucleic acid methylation in the pathological mechanisms of fragile X syndrome. Behav Brain Res. 2023;452:114586.
Bartosovic M, Molares HC, Gregorova P, Hrossova D, Kudla G, Vanacova S. N6-methyladenosine demethylase FTO targets pre-mRNAs and regulates alternative splicing and 3’-end processing. Nucleic Acids Res. 2017;45:11356–70.
Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18–29.
Tsujikawa K, Koike K, Kitae K, Shinkawa A, Arima H, Suzuki T, et al. Expression and sub-cellular localization of human ABH family molecules. J Cell Mol Med. 2007;11:1105–16.
Guo J, Zhao L, Duan M, Yang Z, Zhao H, Liu B, et al. Demethylases in tumors and the tumor microenvironment: Key modifiers of N(6)-methyladenosine methylation. Biomed Pharmacother. 2024;174:116479.
Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–94.
Ueda Y, Ooshio I, Fusamae Y, Kitae K, Kawaguchi M, Jingushi K, et al. AlkB homolog 3-mediated tRNA demethylation promotes protein synthesis in cancer cells. Sci Rep. 2017;7:42271.
Nagarajan A, Janostiak R, Wajapeyee N. Dot blot analysis for measuring global N(6)-methyladenosine modification of RNA. Methods Mol Biol. 2019;1870:263–71.
Munns TW, Podratz KC, Katzman PA. Separation of methylated bases of ribonucleic acid by two-dimensional thin-layer chromatography. J Chromatogr. 1973;76:401–6.
Bodi Z, Fray RG. Detection and quantification of N (6)-methyladenosine in messenger RNA by TLC. Methods Mol Biol. 2017;1562:79–87.
Thüring K, Schmid K, Keller P, Helm M. LC-MS analysis of methylated RNA. Methods Mol Biol. 2017;1562:3–18.
Wang Y, Jia G. Detection methods of epitranscriptomic mark N6-methyladenosine. Essays Biochem. 2020;64:967–79.
Liu N, Parisien M, Dai Q, Zheng G, He C, Pan T. Probing N6-methyladenosine RNA modification status at single nucleotide resolution in mRNA and long noncoding RNA. Rna. 2013;19:1848–56.
Xiao Y, Wang Y, Tang Q, Wei L, Zhang X, Jia G. An elongation- and ligation-based qPCR amplification method for the radiolabeling-free detection of locus-specific N(6) -methyladenosine modification. Angew Chem Int Ed Engl. 2018;57:15995–6000.
Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell. 2012;149:1635–46.
Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–6.
Dierks D, Garcia-Campos MA, Uzonyi A, Safra M, Edelheit S, Rossi A, et al. Multiplexed profiling facilitates robust m6A quantification at site, gene and sample resolution. Nat Methods. 2021;18:1060–7.
Molinie B, Wang J, Lim KS, Hillebrand R, Lu ZX, Van Wittenberghe N, et al. m(6)A-LAIC-seq reveals the census and complexity of the m(6)A epitranscriptome. Nat Methods. 2016;13:692–8.
Hamashima K, Wong KW, Sam TW, Teo JHJ, Taneja R, Le MTN, et al. Single-nucleus multiomic mapping of m(6)A methylomes and transcriptomes in native populations of cells with sn-m6A-CT. Mol Cell. 2023;S1097–2765:00649–4.
Chen K, Lu Z, Wang X, Fu Y, Luo GZ, Liu N, et al. High-resolution N(6) -methyladenosine (m(6) A) map using photo-crosslinking-assisted m(6) A sequencing. Angew Chem Int Ed Engl. 2015;54:1587–90.
Grozhik AV, Linder B, Olarerin-George AO, Jaffrey SR. Mapping m(6)A at individual-nucleotide resolution using crosslinking and immunoprecipitation (miCLIP). Methods Mol Biol. 2017;1562:55–78.
Ke S, Alemu EA, Mertens C, Gantman EC, Fak JJ, Mele A, et al. A majority of m6A residues are in the last exons, allowing the potential for 3’ UTR regulation. Genes Dev. 2015;29:2037–53.
Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE, Jaffrey SR. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods. 2015;12:767–72.
Maatz H, Kolinski M, Hubner N, Landthaler M. Transcriptome-wide Identification of RNA-binding Protein Binding Sites Using Photoactivatable-Ribonucleoside-Enhanced Crosslinking Immunoprecipitation (PAR-CLIP). Curr Protoc Mol Biol. 2017;118:27.6.1–6.19.
Van Nostrand EL, Pratt GA, Shishkin AA, Gelboin-Burkhart C, Fang MY, Sundararaman B, et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat Methods. 2016;13:508–14.
Körtel N, Rücklé C, Zhou Y, Busch A, Hoch-Kraft P, Sutandy FXR, et al. Deep and accurate detection of m6A RNA modifications using miCLIP2 and m6Aboost machine learning. Nucleic Acids Res. 2021;49:e92.
Blue SM, Yee BA, Pratt GA, Mueller JR, Park SS, Shishkin AA, et al. Transcriptome-wide identification of RNA-binding protein binding sites using seCLIP-seq. Nat Protoc. 2022;17:1223–65.
Koh CWQ, Goh YT, Goh WSS. Atlas of quantitative single-base-resolution N(6)-methyl-adenine methylomes. Nat Commun. 2019;10:5636.
Zhang Z, Chen LQ, Zhao YL, Yang CG, Roundtree IA, Zhang Z, et al. Single-base mapping of m(6)A by an antibody-independent method. Sci Adv. 2019;5:eaax0250.
Garcia-Campos MA, Edelheit S, Toth U, Safra M, Shachar R, Viukov S, et al. Deciphering the “m(6)A Code” via Antibody-Independent Quantitative Profiling. Cell. 2019;178:731–47.e16.
Meyer KD. DART-seq: an antibody-free method for global m(6)A detection. Nat Methods. 2019;16:1275–80.
Tegowski M, Flamand MN, Meyer KD. scDART-seq reveals distinct m(6)A signatures and mRNA methylation heterogeneity in single cells. Mol Cell. 2022;82:868–78.e10.
Shu X, Cao J, Cheng M, Xiang S, Gao M, Li T, et al. A metabolic labeling method detects m(6)A transcriptome-wide at single base resolution. Nat Chem Biol. 2020;16:887–95.
Wang Y, Xiao Y, Dong S, Yu Q, Jia G. Antibody-free enzyme-assisted chemical approach for detection of N(6)-methyladenosine. Nat Chem Biol. 2020;16:896–903.
Hu L, Liu S, Peng Y, Ge R, Su R, Senevirathne C, et al. m(6)A RNA modifications are measured at single-base resolution across the mammalian transcriptome. Nat Biotechnol. 2022;40:1210–9.
Liu C, Sun H, Yi Y, Shen W, Li K, Xiao Y, et al. Absolute quantification of single-base m(6)A methylation in the mammalian transcriptome using GLORI. Nat Biotechnol. 2023;41:355–66.
Garalde DR, Snell EA, Jachimowicz D, Sipos B, Lloyd JH, Bruce M, et al. Highly parallel direct RNA sequencing on an array of nanopores. Nat Methods. 2018;15:201–6.
Chen Y, Wu Y, Liu L, Feng J, Zhang T, Qin S, et al. Study of the whole genome, methylome and transcriptome of Cordyceps militaris. Sci Rep. 2019;9:898.
Teng H, Stoiber M, Bar-Joseph Z, Kingsford C. Detecting m6A RNA modification from nanopore sequencing using a semi-supervised learning framework. bioRxiv. 2024.
Hassan D, Ariyur A, Daulatabad SV, Mir Q, Janga SC. Nm-Nano: a machine learning framework for transcriptome-wide single-molecule mapping of 2´-O-methylation (Nm) sites in nanopore direct RNA sequencing datasets. RNA Biol. 2024;21:1–15.
Hendra C, Pratanwanich PN, Wan YK, Goh WSS, Thiery A, Göke J. Detection of m6A from direct RNA sequencing using a multiple instance learning framework. Nat Methods. 2022;19:1590–8.
Salditt-Georgieff M, Jelinek W, Darnell JE, Furuichi Y, Morgan M, Shatkin A. Methyl labeling of HeLa cell hnRNA: a comparison with mRNA. Cell. 1976;7:227–37.
Lence T, Akhtar J, Bayer M, Schmid K, Spindler L, Ho CH, et al. m(6)A modulates neuronal functions and sex determination in Drosophila. Nature. 2016;540:242–7.
Haussmann IU, Bodi Z, Sanchez-Moran E, Mongan NP, Archer N, Fray RG, et al. m(6)A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature. 2016;540:301–4.
Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, et al. Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol Cell. 2016;61:507–19.
Pendleton KE, Chen B, Liu K, Hunter OV, Xie Y, Tu BP, et al. The U6 snRNA m(6)A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell. 2017;169:824–35.e14.
Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560–4.
Zhao X, Yang Y, Sun BF, Shi Y, Yang X, Xiao W, et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014;24:1403–19.
Edens BM, Vissers C, Su J, Arumugam S, Xu Z, Shi H, et al. FMRP modulates neural differentiation through m(6)A-dependent mRNA nuclear export. Cell Rep. 2019;28:845–54.e5.
Hsu PJ, Shi H, Zhu AC, Lu Z, Miller N, Edens BM, et al. The RNA-binding protein FMRP facilitates the nuclear export of N(6)-methyladenosine-containing mRNAs. J Biol Chem. 2019;294:19889–95.
Lin S, Choe J, Du P, Triboulet R, Gregory RI. The m(6)A methyltransferase METTL3 promotes translation in human cancer cells. Mol Cell. 2016;62:335–45.
Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, et al. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161:1388–99.
Choe J, Lin S, Zhang W, Liu Q, Wang L, Ramirez-Moya J, et al. mRNA circularization by METTL3-eIF3h enhances translation and promotes oncogenesis. Nature. 2018;561:556–60.
Kretschmer J, Rao H, Hackert P, Sloan KE, Höbartner C, Bohnsack MT. The m(6)A reader protein YTHDC2 interacts with the small ribosomal subunit and the 5’-3’ exoribonuclease XRN1. Rna. 2018;24:1339–50.
Edupuganti RR, Geiger S, Lindeboom RGH, Shi H, Hsu PJ, Lu Z, et al. N(6)-methyladenosine (m(6)A) recruits and repels proteins to regulate mRNA homeostasis. Nat Struct Mol Biol. 2017;24:870–8.
Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–61.
Bechara EG, Didiot MC, Melko M, Davidovic L, Bensaid M, Martin P, et al. A novel function for fragile X mental retardation protein in translational activation. PLoS Biol. 2009;7:e16.
Li T, Hu PS, Zuo Z, Lin JF, Li X, Wu QN, et al. METTL3 facilitates tumor progression via an m(6)A-IGF2BP2-dependent mechanism in colorectal carcinoma. Mol Cancer. 2019;18:112.
Park OH, Ha H, Lee Y, Boo SH, Kwon DH, Song HK, et al. Endoribonucleolytic cleavage of m(6)A-containing RNAs by RNase P/MRP complex. Mol Cell. 2019;74:494–507.e8.
Zheng D, Ezzeddine N, Chen CY, Zhu W, He X, Shyu AB. Deadenylation is prerequisite for P-body formation and mRNA decay in mammalian cells. J Cell Biol. 2008;182:89–101.
Lee Y, Choe J, Park OH, Kim YK. Molecular mechanisms driving mRNA degradation by m(6)A modification. Trends Genet. 2020;36:177–88.
Boo SH, Ha H, Lee Y, Shin MK, Lee S, Kim YK. UPF1 promotes rapid degradation of m(6)A-containing RNAs. Cell Rep. 2022;39:110861.
Ries RJ, Zaccara S, Klein P, Olarerin-George A, Namkoong S, Pickering BF, et al. m(6)A enhances the phase separation potential of mRNA. Nature. 2019;571:424–8.
Fu Y, Zhuang X. m(6)A-binding YTHDF proteins promote stress granule formation. Nat Chem Biol. 2020;16:955–63.
Zhang JZ, Mehta S, Zhang J. Liquid-liquid phase separation: a principal organizer of the cell’s biochemical activity architecture. Trends Pharm Sci. 2021;42:845–56.
Wojtas MN, Pandey RR, Mendel M, Homolka D, Sachidanandam R, Pillai RS. Regulation of m(6)A Transcripts by the 3’→5’ RNA Helicase YTHDC2 Is Essential for a Successful Meiotic Program in the Mammalian Germline. Mol Cell. 2017;68:374–87.e12.
Tan L, Cheng W, Liu F, Wang DO, Wu L, Cao N, et al. Positive natural selection of N6-methyladenosine on the RNAs of processed pseudogenes. Genome Biol. 2021;22:180.
Starr JL, Fefferman R. The occurrence of methylated bases in ribosomal ribonucleic acid of escherichia COLI K12 W-6. J Biol Chem. 1964;239:3457–61.
Hayashi Y, Osawa S, Miura K. The methyl groups in ribosomal RNA from Escherichia coli. Biochim Biophys Acta. 1966;129:519–31.
Xing M, Liu Q, Mao C, Zeng H, Zhang X, Zhao S, et al. The 18S rRNA m(6) A methyltransferase METTL5 promotes mouse embryonic stem cell differentiation. EMBO Rep. 2020;21:e49863.
Ignatova VV, Stolz P, Kaiser S, Gustafsson TH, Lastres PR, Sanz-Moreno A, et al. The rRNA m(6)A methyltransferase METTL5 is involved in pluripotency and developmental programs. Genes Dev. 2020;34:715–29.
Sepich-Poore C, Zheng Z, Schmitt E, Wen K, Zhang ZS, Cui XL, et al. The METTL5-TRMT112 N(6)-methyladenosine methyltransferase complex regulates mRNA translation via 18S rRNA methylation. J Biol Chem. 2022;298:101590.
Ma H, Wang X, Cai J, Dai Q, Natchiar SK, Lv R, et al. N(6-)Methyladenosine methyltransferase ZCCHC4 mediates ribosomal RNA methylation. Nat Chem Biol. 2019;15:88–94.
Xue M, Dong L, Zhang H, Li Y, Qiu K, Zhao Z, et al. METTL16 promotes liver cancer stem cell self-renewal via controlling ribosome biogenesis and mRNA translation. J Hematol Oncol. 2024;17:7.
Golovina AY, Dzama MM, Osterman IA, Sergiev PV, Serebryakova MV, Bogdanov AA, et al. The last rRNA methyltransferase of E. coli revealed: the yhiR gene encodes adenine-N6 methyltransferase specific for modification of A2030 of 23S ribosomal RNA. RNA. 2012;18:1725–34.
Sergiev PV, Serebryakova MV, Bogdanov AA, Dontsova OA. The ybiN gene of Escherichia coli encodes adenine-N6 methyltransferase specific for modification of A1618 of 23 S ribosomal RNA, a methylated residue located close to the ribosomal exit tunnel. J Mol Biol. 2008;375:291–300.
Chen DH, Zhang JG, Wu CX, Li Q. Non-coding RNA m6A modification in cancer: mechanisms and therapeutic targets. Front Cell Dev Biol. 2021;9:778582.
Yan J, Huang X, Zhang X, Chen Z, Ye C, Xiang W, et al. LncRNA LINC00470 promotes the degradation of PTEN mRNA to facilitate malignant behavior in gastric cancer cells. Biochem Biophys Res Commun. 2020;521:887–93.
Wang Y, Lu JH, Wu QN, Jin Y, Wang DS, Chen YX, et al. LncRNA LINRIS stabilizes IGF2BP2 and promotes the aerobic glycolysis in colorectal cancer. Mol Cancer. 2019;18:174.
Cui X, Wang Z, Li J, Zhu J, Ren Z, Zhang D, et al. Cross talk between RNA N6-methyladenosine methyltransferase-like 3 and miR-186 regulates hepatoblastoma progression through Wnt/β-catenin signalling pathway. Cell Prolif. 2020;53:e12768.
Lai X, Wei J, Gu XZ, Yao XM, Zhang DS, Li F, et al. Dysregulation of LINC00470 and METTL3 promotes chemoresistance and suppresses autophagy of chronic myelocytic leukaemia cells. J Cell Mol Med. 2021;25:4248–59.
Yang L, Froberg JE, Lee JT. Long noncoding RNAs: fresh perspectives into the RNA world. Trends Biochem Sci. 2014;39:35–43.
Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M, et al. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537:369–73.
Nesterova TB, Wei G, Coker H, Pintacuda G, Bowness JS, Zhang T, et al. Systematic allelic analysis defines the interplay of key pathways in X chromosome inactivation. Nat Commun. 2019;10:3129.
Jin D, Guo J, Wu Y, Du J, Yang L, Wang X, et al. m(6)A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the MALAT1-miR-1914-3p-YAP axis to induce NSCLC drug resistance and metastasis. J Hematol Oncol. 2021;14:32.
Zhou KI, Parisien M, Dai Q, Liu N, Diatchenko L, Sachleben JR, et al. N(6)-methyladenosine modification in a long noncoding RNA hairpin predisposes its conformation to protein binding. J Mol Biol. 2016;428:822–33.
Wang X, Liu C, Zhang S, Yan H, Zhang L, Jiang A, et al. N(6)-methyladenosine modification of MALAT1 promotes metastasis via reshaping nuclear speckles. Dev Cell. 2021;56:702–15.e8.
Ji F, Lu Y, Chen S, Lin X, Yu Y, Zhu Y, et al. m(6)A methyltransferase METTL3-mediated lncRNA FOXD2-AS1 promotes the tumorigenesis of cervical cancer. Mol Ther Oncolytics. 2021;22:574–81.
Wang J, Ding W, Xu Y, Tao E, Mo M, Xu W, et al. Long non-coding RNA RHPN1-AS1 promotes tumorigenesis and metastasis of ovarian cancer by acting as a ceRNA against miR-596 and upregulating LETM1. Aging (Albany NY). 2020;12:4558–72.
Jin J, Fu L, Hong P, Feng W. MALAT-1 regulates the AML progression by promoting the m6A modification of ZEB1. Acta Biochim Pol. 2023;70:37–43.
Kong H, Sun J, Zhang W, Zhang H, Li H. Long intergenic non-protein coding RNA 1273 confers sorafenib resistance in hepatocellular carcinoma via regulation of methyltransferase 3. Bioengineered. 2022;13:3108–21.
Dai YZ, Liu YD, Li J, Chen MT, Huang M, Wang F, et al. METTL16 promotes hepatocellular carcinoma progression through downregulating RAB11B-AS1 in an m(6)A-dependent manner. Cell Mol Biol Lett. 2022;27:41.
Sun T, Wu Z, Wang X, Wang Y, Hu X, Qin W, et al. LNC942 promoting METTL14-mediated m(6)A methylation in breast cancer cell proliferation and progression. Oncogene. 2020;39:5358–72.
Li ZX, Zheng ZQ, Yang PY, Lin L, Zhou GQ, Lv JW, et al. WTAP-mediated m(6)A modification of lncRNA DIAPH1-AS1 enhances its stability to facilitate nasopharyngeal carcinoma growth and metastasis. Cell Death Differ. 2022;29:1137–51.
Zheng ZQ, Li ZX, Zhou GQ, Lin L, Zhang LL, Lv JW, et al. Long Noncoding RNA FAM225A Promotes Nasopharyngeal Carcinoma Tumorigenesis and Metastasis by Acting as ceRNA to Sponge miR-590-3p/miR-1275 and Upregulate ITGB3. Cancer Res. 2019;79:4612–26.
Chen K, Zhang J, Meng L, Kong L, Lu M, Wang Z, et al. The epigenetic downregulation of LncGHRLOS mediated by RNA m6A methylase ZCCHC4 promotes colorectal cancer tumorigenesis. J Exp Clin Cancer Res. 2024;43:44.
Chen L, Zhang C, Ma W, Huang J, Zhao Y, Liu H. METTL3-mediated m6A modification stabilizes TERRA and maintains telomere stability. Nucleic Acids Res. 2022;50:11619–34.
Liu HT, Zou YX, Zhu WJ, Sen L, Zhang GH, Ma RR, et al. lncRNA THAP7-AS1, transcriptionally activated by SP1 and post-transcriptionally stabilized by METTL3-mediated m6A modification, exerts oncogenic properties by improving CUL4B entry into the nucleus. Cell Death Differ. 2022;29:627–41.
Cui Y, Zhang C, Ma S, Li Z, Wang W, Li Y, et al. RNA m6A demethylase FTO-mediated epigenetic up-regulation of LINC00022 promotes tumorigenesis in esophageal squamous cell carcinoma. J Exp Clin Cancer Res. 2021;40:294.
He Y, Yue H, Cheng Y, Ding Z, Xu Z, Lv C, et al. ALKBH5-mediated m(6)A demethylation of KCNK15-AS1 inhibits pancreatic cancer progression via regulating KCNK15 and PTEN/AKT signaling. Cell Death Dis. 2021;12:1121.
Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610.
Lin YH. MicroRNA networks modulate oxidative stress in cancer. Int J Mol Sci. 2019;20:4497.
Han X, Guo J, Fan Z. Interactions between m6A modification and miRNAs in malignant tumors. Cell Death Dis. 2021;12:598.
Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, et al. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901.
Ma JZ, Yang F, Zhou CC, Liu F, Yuan JH, Wang F, et al. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N(6) -methyladenosine-dependent primary MicroRNA processing. Hepatology. 2017;65:529–43.
Alarcón CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N6-methyladenosine marks primary microRNAs for processing. Nature. 2015;519:482–5.
Peng W, Li J, Chen R, Gu Q, Yang P, Qian W, et al. Upregulated METTL3 promotes metastasis of colorectal Cancer via miR-1246/SPRED2/MAPK signaling pathway. J Exp Clin Cancer Res. 2019;38:393.
Yan G, Yuan Y, He M, Gong R, Lei H, Zhou H, et al. m(6)A methylation of precursor-miR-320/RUNX2 controls osteogenic potential of bone marrow-derived mesenchymal stem cells. Mol Ther Nucleic Acids. 2020;19:421–36.
Jiang Z, Song X, Wei Y, Li Y, Kong D, Sun J. N(6)-methyladenosine-mediated miR-380-3p maturation and upregulation promotes cancer aggressiveness in pancreatic cancer. Bioengineered. 2022;13:14460–71.
Lyu Y, Zhang Y, Wang Y, Luo Y, Ding H, Li P, et al. HIF-1α regulated WTAP overexpression promoting the Warburg effect of ovarian cancer by m6A-dependent manner. J Immunol Res. 2022;2022:6130806.
Li Y, Lou S, Zhang J, Zhao S, Lou G. m(6)A methylation-mediated regulation of LncRNA MEG3 suppresses ovarian cancer progression through miR-885-5p and the VASH1 pathway. J Transl Med. 2024;22:113.
Sun M, Shen Y, Jia G, Deng Z, Shi F, Jing Y, et al. Activation of the HNRNPA2B1/miR-93-5p/FRMD6 axis facilitates prostate cancer progression in an m6A-dependent manner. J Cancer. 2023;14:1242–56.
Sun K, Chen L, Li Y, Huang B, Yan Q, Wu C, et al. METTL14-dependent maturation of pri-miR-17 regulates mitochondrial homeostasis and induces chemoresistance in colorectal cancer. Cell Death Dis. 2023;14:148.
Park S, Yang HD, Seo JW, Nam JW, Nam SW. hnRNPC induces isoform shifts in miR-21-5p leading to cancer development. Exp Mol Med. 2022;54:812–24.
Wang Z, Tong D, Han C, Zhao Z, Wang X, Jiang T, et al. Blockade of miR-3614 maturation by IGF2BP3 increases TRIM25 expression and promotes breast cancer cell proliferation. EBioMedicine. 2019;41:357–69.
Zhou Y, Huang T, Siu HL, Wong CC, Dong Y, Wu F, et al. IGF2BP3 functions as a potential oncogene and is a crucial target of miR-34a in gastric carcinogenesis. Mol Cancer. 2017;16:77.
Zhou G, Yan K, Liu J, Gao L, Jiang X, Fan Y. FTO promotes tumour proliferation in bladder cancer via the FTO/miR-576/CDK6 axis in an m6A-dependent manner. Cell Death Discov. 2021;7:329.
Azhati B, Reheman A, Dilixiati D, Rexiati M. FTO-stabilized miR-139-5p targets ZNF217 to suppress prostate cancer cell malignancies by inactivating the PI3K/Akt/mTOR signal pathway. Arch Biochem Biophys. 2023;741:109604.
Zhu H, Gan X, Jiang X, Diao S, Wu H, Hu J. ALKBH5 inhibited autophagy of epithelial ovarian cancer through miR-7 and BCL-2. J Exp Clin Cancer Res. 2019;38:163.
Jin D, Guo J, Wu Y, Yang L, Wang X, Du J, et al. m(6)A demethylase ALKBH5 inhibits tumor growth and metastasis by reducing YTHDFs-mediated YAP expression and inhibiting miR-107/LATS2-mediated YAP activity in NSCLC. Mol Cancer. 2020;19:40.
Chen T, Hao YJ, Zhang Y, Li MM, Wang M, Han W, et al. m(6)A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell Stem Cell. 2015;16:289–301.
Park MS, Araya-Secchi R, Brackbill JA, Phan HD, Kehling AC, Abd El-Wahab EW, et al. Multidomain convergence of argonaute during RISC assembly correlates with the formation of internal water clusters. Mol Cell. 2019;75:725–40.e6.
Liang XH, Nichols JG, Hsu CW, Vickers TA, Crooke ST. mRNA levels can be reduced by antisense oligonucleotides via no-go decay pathway. Nucleic Acids Res. 2019;47:6900–16.
Du M, Zhang Y, Mao Y, Mou J, Zhao J, Xue Q, et al. MiR-33a suppresses proliferation of NSCLC cells via targeting METTL3 mRNA. Biochem Biophys Res Commun. 2017;482:582–9.
Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20:675–91.
Patop IL, Wüst S, Kadener S. Past, present, and future of circRNAs. EMBO J. 2019;38:e100836.
Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381–8.
Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 2017;27:626–41.
Di Timoteo G, Dattilo D, Centrón-Broco A, Colantoni A, Guarnacci M, Rossi F, et al. Modulation of circRNA metabolism by m(6)A modification. Cell Rep. 2020;31:107641.
Zhou C, Molinie B, Daneshvar K, Pondick JV, Wang J, Van Wittenberghe N, et al. Genome-wide maps of m6A circRNAs identify widespread and cell-type-specific methylation patterns that are distinct from mRNAs. Cell Rep. 2017;20:2262–76.
He RZ, Jiang J, Luo DX. m6A modification of circNSUN2 promotes colorectal liver metastasis. Genes Dis. 2021;8:6–7.
Patil DP, Pickering BF, Jaffrey SR. Reading m(6)A in the transcriptome: m(6)A-binding proteins. Trends Cell Biol. 2018;28:113–27.
Rong D, Wu F, Lu C, Sun G, Shi X, Chen X, et al. m6A modification of circHPS5 and hepatocellular carcinoma progression through HMGA2 expression. Mol Ther Nucleic Acids. 2021;26:637–48.
An Y, Duan H. The role of m6A RNA methylation in cancer metabolism. Mol Cancer. 2022;21:14.
Coots RA, Liu XM, Mao Y, Dong L, Zhou J, Wan J, et al. m(6)A Facilitates eIF4F-Independent mRNA Translation. Mol Cell. 2017;68:504–14.e7.
Chen B, Hong Y, Gui R, Zheng H, Tian S, Zhai X, et al. N6-methyladenosine modification of circ_0003215 suppresses the pentose phosphate pathway and malignancy of colorectal cancer through the miR-663b/DLG4/G6PD axis. Cell Death Dis. 2022;13:804.
Guan H, Tian K, Luo W, Li M. m(6)A-modified circRNA MYO1C participates in the tumor immune surveillance of pancreatic ductal adenocarcinoma through m(6)A/PD-L1 manner. Cell Death Dis. 2023;14:120.
Ji X, Lv C, Huang J, Dong W, Sun W, Zhang H. ALKBH5-induced circular RNA NRIP1 promotes glycolysis in thyroid cancer cells by targeting PKM2. Cancer Sci. 2023;114:2318–34.
Wei W, Sun J, Zhang H, Xiao X, Huang C, Wang L, et al. Circ0008399 interaction with WTAP promotes assembly and activity of the m(6)A methyltransferase complex and promotes cisplatin resistance in bladder cancer. Cancer Res. 2021;81:6142–56.
Huang Q, Guo H, Wang S, Ma Y, Chen H, Li H, et al. A novel circular RNA, circXPO1, promotes lung adenocarcinoma progression by interacting with IGF2BP1. Cell Death Dis. 2020;11:1031.
Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17:272–83.
Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–8.
Li M, Wang Q, Zhang X, Yan N, Li X. CircPUM1 promotes cell growth and glycolysis in NSCLC via up-regulating METTL3 expression through miR-590-5p. Cell Cycle. 2021;20:1279–94.
Li R, Albertini DF. The road to maturation: somatic cell interaction and self-organization of the mammalian oocyte. Nat Rev Mol Cell Biol. 2013;14:141–52.
Sui X, Hu Y, Ren C, Cao Q, Zhou S, Cao Y, et al. METTL3-mediated m(6)A is required for murine oocyte maturation and maternal-to-zygotic transition. Cell Cycle. 2020;19:391–404.
Xia H, Zhong C, Wu X, Chen J, Tao B, Xia X, et al. Mettl3 mutation disrupts gamete maturation and reduces fertility in zebrafish. Genetics. 2018;208:729–43.
Wang YK, Yu XX, Liu YH, Li X, Liu XM, Wang PC, et al. Reduced nucleic acid methylation impairs meiotic maturation and developmental potency of pig oocytes. Theriogenology. 2018;121:160–7.
Kasowitz SD, Ma J, Anderson SJ, Leu NA, Xu Y, Gregory BD, et al. Nuclear m6A reader YTHDC1 regulates alternative polyadenylation and splicing during mouse oocyte development. PLoS Genet. 2018;14:e1007412.
Hsu PJ, Zhu Y, Ma H, Guo Y, Shi X, Liu Y, et al. Ythdc2 is an N(6)-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017;27:1115–27.
Ivanova I, Much C, Di Giacomo M, Azzi C, Morgan M, Moreira PN, et al. The RNA m(6)A reader YTHDF2 is essential for the post-transcriptional regulation of the maternal transcriptome and oocyte competence. Mol Cell. 2017;67:1059–67.e4.
Hu Y, Ouyang Z, Sui X, Qi M, Li M, He Y, et al. Oocyte competence is maintained by m(6)A methyltransferase KIAA1429-mediated RNA metabolism during mouse follicular development. Cell Death Differ. 2020;27:2468–83.
Fan Y, Zhang C, Zhu G. Profiling of RNA N6-methyladenosine methylation during follicle selection in chicken ovary. Poult Sci. 2019;98:6117–24.
Schulz KN, Harrison MM. Mechanisms regulating zygotic genome activation. Nat Rev Genet. 2019;20:221–34.
Asami M, Lam BYH, Ma MK, Rainbow K, Braun S, VerMilyea MD, et al. Human embryonic genome activation initiates at the one-cell stage. Cell Stem Cell. 2022;29:209–16.e4.
Taniguchi K, Kawai T, Kitawaki J, Tomikawa J, Nakabayashi K, Okamura K, et al. Epitranscriptomic profiling in human placenta: N6-methyladenosine modification at the 5’-untranslated region is related to fetal growth and preeclampsia. Faseb j. 2020;34:494–512.
Wu Y, Xu X, Qi M, Chen C, Li M, Yan R, et al. N(6)-methyladenosine regulates maternal RNA maintenance in oocytes and timely RNA decay during mouse maternal-to-zygotic transition. Nat Cell Biol. 2022;24:917–27.
Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol. 2014;16:191–8.
Batista PJ, Molinie B, Wang J, Qu K, Zhang J, Li L, et al. mA RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014;15:707–19.
Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, et al. Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science. 2015;347:1002–6.
Cao Z, Zhang L, Hong R, Li Y, Wang Y, Qi X, et al. METTL3-mediated m6A methylation negatively modulates autophagy to support porcine blastocyst development. ‡ Biol Reprod 2021;104:1008–21.
Meng TG, Lu X, Guo L, Hou GM, Ma XS, Li QN, et al. Mettl14 is required for mouse postimplantation development by facilitating epiblast maturation. Faseb j. 2019;33:1179–87.
Liu HB, Muhammad T, Guo Y, Li MJ, Sha QQ, Zhang CX, et al. RNA-binding protein IGF2BP2/IMP2 is a critical maternal activator in early zygotic genome activation. Adv Sci (Weinh). 2019;6:1900295.
Deng M, Chen B, Liu Z, Cai Y, Wan Y, Zhang G, et al. YTHDF2 regulates maternal transcriptome degradation and embryo development in goat. Front Cell Dev Biol. 2020;8:580367.
Zhao BS, Wang X, Beadell AV, Lu Z, Shi H, Kuuspalu A, et al. m(6)A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature. 2017;542:475–8.
Li M, Zhao X, Wang W, Shi H, Pan Q, Lu Z, et al. Ythdf2-mediated m(6)A mRNA clearance modulates neural development in mice. Genome Biol. 2018;19:69.
Landfors M, Nakken S, Fusser M, Dahl JA, Klungland A, Fedorcsak P. Sequencing of FTO and ALKBH5 in men undergoing infertility work-up identifies an infertility-associated variant and two missense mutations. Fertil Steril. 2016;105:1170–9.e5.
Wei J, Yu X, Yang L, Liu X, Gao B, Huang B, et al. FTO mediates LINE1 m(6)A demethylation and chromatin regulation in mESCs and mouse development. Science. 2022;376:968–73.
Li H, Huang Q, Liu Y, Garmire LX. Single cell transcriptome research in human placenta. Reproduction. 2020;160:R155–r67.
Costa MA. The endocrine function of human placenta: an overview. Reprod Biomed Online. 2016;32:14–43.
Megli CJ, Coyne CB. Infections at the maternal-fetal interface: an overview of pathogenesis and defence. Nat Rev Microbiol. 2022;20:67–82.
Evans J, Salamonsen LA, Winship A, Menkhorst E, Nie G, Gargett CE, et al. Fertile ground: human endometrial programming and lessons in health and disease. Nat Rev Endocrinol. 2016;12:654–67.
Burki TK. Placenta can provide new insights on the genetics of cancer. Lancet Oncol. 2021;22:591.
Xiao S, Cao S, Huang Q, Xia L, Deng M, Yang M, et al. The RNA N(6)-methyladenosine modification landscape of human fetal tissues. Nat Cell Biol. 2019;21:651–61.
She J, Tan K, Liu J, Cao S, Li Z, Peng Y, et al. The Alteration of m(6)A Modification at the Transcriptome-Wide Level in Human Villi During Spontaneous Abortion in the First Trimester. Front Genet. 2022;13:861853.
Zhao J, Ding H, Ding J, Shi X, He Y, Zhu H, et al. The m(6)A methyltransferase METTL3 promotes trophoblast cell invasion by regulating MYLK expression. Placenta. 2022;129:1–6.
Wang D, Guan H, Xia Y. YTHDC1 maintains trophoblasts function by promoting degradation of m6A-modified circMPP1. Biochem Pharm. 2023;210:115456.
Wang R, Xu X, Yang J, Chen W, Zhao J, Wang M, et al. BPDE exposure promotes trophoblast cell pyroptosis and induces miscarriage by up-regulating lnc-HZ14/ZBP1/NLRP3 axis. J Hazard Mater. 2023;455:131543.
Li XC, Jin F, Wang BY, Yin XJ, Hong W, Tian FJ. The m6A demethylase ALKBH5 controls trophoblast invasion at the maternal-fetal interface by regulating the stability of CYR61 mRNA. Theranostics. 2019;9:3853–65.
Guo Y, Song W, Yang Y. Inhibition of ALKBH5-mediated m(6) A modification of PPARG mRNA alleviates H/R-induced oxidative stress and apoptosis in placenta trophoblast. Environ Toxicol. 2022;37:910–24.
Yao Y, Ye Y, Chen J, Zhang M, Cai X, Zheng C. Maternal-fetal immunity and recurrent spontaneous abortion. Am J Reprod Immunol. 2024;91:e13859.
Zou K, Yuan Z, Yang Z, Luo H, Sun K, Zhou L, et al. Production of offspring from a germline stem cell line derived from neonatal ovaries. Nat Cell Biol. 2009;11:631–6.
Lehmann R. Germline stem cells: origin and destiny. Cell Stem Cell. 2012;10:729–39.
Zhao X, Tian GG, Fang Q, Pei X, Wang Z, Wu J. Comparison of RNA m(6)A and DNA methylation profiles between mouse female germline stem cells and STO cells. Mol Ther Nucleic Acids. 2021;23:431–9.
Li X, Tian G, Wu J. Novel circGFRα1 promotes self-renewal of female germline stem cells mediated by m(6)A writer METTL14. Front Cell Dev Biol. 2021;9:640402.
Prager BC, Xie Q, Bao S, Rich JN. Cancer stem cells: the architects of the tumor ecosystem. Cell Stem Cell. 2019;24:41–53.
Batlle E, Clevers H. Cancer stem cells revisited. Nat Med. 2017;23:1124–34.
Barbato L, Bocchetti M, Di Biase A, Regad T. Cancer stem cells and targeting strategies. Cells. 2019;8:926.
Hong K. Emerging function of N6-methyladenosine in cancer. Oncol Lett. 2018;16:5519–24.
Chen B, Li Y, Song R, Xue C, Xu F. Functions of RNA N6-methyladenosine modification in cancer progression. Mol Biol Rep. 2019;46:1383–91.
Zhang SY, Zhang SW, Fan XN, Zhang T, Meng J, Huang Y. FunDMDeep-m6A: identification and prioritization of functional differential m6A methylation genes. Bioinformatics. 2019;35:i90–i8.
Zhang C, Samanta D, Lu H, Bullen JW, Zhang H, Chen I, et al. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m6A-demethylation of NANOG mRNA. Proc Natl Acad Sci USA. 2016;113:E2047–56.
Shi Y, Fan S, Wu M, Zuo Z, Li X, Jiang L, et al. YTHDF1 links hypoxia adaptation and non-small cell lung cancer progression. Nat Commun. 2019;10:4892.
Zhou R, Gao Y, Lv D, Wang C, Wang D, Li Q. METTL3 mediated m(6)A modification plays an oncogenic role in cutaneous squamous cell carcinoma by regulating ΔNp63. Biochem Biophys Res Commun. 2019;515:310–7.
Paris J, Morgan M, Campos J, Spencer GJ, Shmakova A, Ivanova I, et al. Targeting the RNA m(6)A Reader YTHDF2 Selectively Compromises Cancer Stem Cells in Acute Myeloid Leukemia. Cell Stem Cell. 2019;25:137–48.e6.
Hao L, Wang JM, Liu BQ, Yan J, Li C, Jiang JY, et al. m6A-YTHDF1-mediated TRIM29 upregulation facilitates the stem cell-like phenotype of cisplatin-resistant ovarian cancer cells. Biochim Biophys Acta Mol Cell Res. 2021;1868:118878.
Xu R, Hu J, Zhang T, Jiang C, Wang HY. TRIM29 overexpression is associated with poor prognosis and promotes tumor progression by activating Wnt/β-catenin pathway in cervical cancer. Oncotarget. 2016;7:28579–91.
Chen G, Liu B, Yin S, Li S, Guo Y, Wang M, et al. Hypoxia induces an endometrial cancer stem-like cell phenotype via HIF-dependent demethylation of SOX2 mRNA. Oncogenesis. 2020;9:81.
Lundø K, Trauelsen M, Pedersen SF, Schwartz TW. Why Warburg Works: Lactate controls immune evasion through GPR81. Cell Metab. 2020;31:666–8.
Li Z, Peng Y, Li J, Chen Z, Chen F, Tu J, et al. N(6)-methyladenosine regulates glycolysis of cancer cells through PDK4. Nat Commun. 2020;11:2578.
Li H, Zhong Y, Cao G, Shi H, Liu Y, Li L, et al. METTL3 promotes cell cycle progression via m(6)A/YTHDF1-dependent regulation of CDC25B translation. Int J Biol Sci. 2022;18:3223–36.
Morante-Palacios O, Fondelli F, Ballestar E, Martínez-Cáceres EM. Tolerogenic dendritic cells in autoimmunity and inflammatory diseases. Trends Immunol. 2021;42:59–75.
Diamond MS, Lin JH, Vonderheide RH. Site-dependent immune escape due to impaired dendritic cell cross-priming. Cancer Immunol Res. 2021;9:877–90.
Krey G, Frank P, Shaikly V, Barrientos G, Cordo-Russo R, Ringel F, et al. In vivo dendritic cell depletion reduces breeding efficiency, affecting implantation and early placental development in mice. J Mol Med (Berl). 2008;86:999–1011.
Karikó K, Buckstein M, Ni H, Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–75.
Wang H, Hu X, Huang M, Liu J, Gu Y, Ma L, et al. Mettl3-mediated mRNA m(6)A methylation promotes dendritic cell activation. Nat Commun. 2019;10:1898.
Han D, Liu J, Chen C, Dong L, Liu Y, Chang R, et al. Anti-tumour immunity controlled through mRNA m(6)A methylation and YTHDF1 in dendritic cells. Nature. 2019;566:270–4.
Liu J, Zhang X, Chen K, Cheng Y, Liu S, Xia M, et al. CCR7 chemokine receptor-inducible lnc-Dpf3 restrains dendritic cell migration by inhibiting HIF-1α-mediated glycolysis. Immunity. 2019;50:600–15.e15.
Croy BA, Di Santo JP, Greenwood JD, Chantakru S, Ashkar AA. Transplantation into genetically alymphoid mice as an approach to dissect the roles of uterine natural killer cells during pregnancy-a review. Placenta. 2000;21:S77–80. Suppl A
Fu B, Zhou Y, Ni X, Tong X, Xu X, Dong Z, et al. Natural killer cells promote fetal development through the secretion of growth-promoting factors. Immunity. 2017;47:1100–13.e6.
Fu B, Li X, Sun R, Tong X, Ling B, Tian Z, et al. Natural killer cells promote immune tolerance by regulating inflammatory TH17 cells at the human maternal-fetal interface. Proc Natl Acad Sci USA. 2013;110:E231–40.
Li X, Ma S, Deng Y, Yi P, Yu J. Targeting the RNA m(6)A modification for cancer immunotherapy. Mol Cancer. 2022;21:76.
Koopman LA, Kopcow HD, Rybalov B, Boyson JE, Orange JS, Schatz F, et al. Human decidual natural killer cells are a unique NK cell subset with immunomodulatory potential. J Exp Med. 2003;198:1201–12.
Vento-Tormo R, Efremova M, Botting RA, Turco MY, Vento-Tormo M, Meyer KB, et al. Single-cell reconstruction of the early maternal-fetal interface in humans. Nature. 2018;563:347–53.
Ma S, Yan J, Barr T, Zhang J, Chen Z, Wang LS, et al. The RNA m6A reader YTHDF2 controls NK cell antitumor and antiviral immunity. J Exp Med. 2021;218:e20210279.
Song H, Song J, Cheng M, Zheng M, Wang T, Tian S, et al. METTL3-mediated m(6)A RNA methylation promotes the anti-tumour immunity of natural killer cells. Nat Commun. 2021;12:5522.
Yan Y, Liang Q, Xu Z, Yi Q. Integrative bioinformatics and experimental analysis revealed down-regulated CDC42EP3 as a novel prognostic target for ovarian cancer and its roles in immune infiltration. PeerJ. 2021;9:e12171.
Kim SM, Oh SC, Lee SY, Kong LZ, Lee JH, Kim TD. FTO negatively regulates the cytotoxic activity of natural killer cells. EMBO Rep. 2023;24:e55681.
Liu S, Diao L, Huang C, Li Y, Zeng Y, Kwak-Kim JYH. The role of decidual immune cells on human pregnancy. J Reprod Immunol. 2017;124:44–53.
Engert S, Rieger L, Kapp M, Becker JC, Dietl J, Kämmerer U. Profiling chemokines, cytokines and growth factors in human early pregnancy decidua by protein array. Am J Reprod Immunol. 2007;58:129–37.
Nagamatsu T, Schust DJ. The immunomodulatory roles of macrophages at the maternal-fetal interface. Reprod Sci. 2010;17:209–18.
Abumaree MH, Stone PR, Chamley LW. The effects of apoptotic, deported human placental trophoblast on macrophages: possible consequences for pregnancy. J Reprod Immunol. 2006;72:33–45.
True H, Blanton M, Sureshchandra S, Messaoudi I. Monocytes and macrophages in pregnancy: the good, the bad, and the ugly. Immunol Rev. 2022;308:77–92.
Li M, Yang Y, Xiong L, Jiang P, Wang J, Li C. Metabolism, metabolites, and macrophages in cancer. J Hematol Oncol. 2023;16:80.
Du J, Liao W, Liu W, Deb DK, He L, Hsu PJ, et al. N(6)-adenosine methylation of Socs1 mRNA is required to sustain the negative feedback control of macrophage activation. Dev Cell. 2020;55:737–53.e7.
Liu Y, Liu Z, Tang H, Shen Y, Gong Z, Xie N, et al. The N(6)-methyladenosine (m(6)A)-forming enzyme METTL3 facilitates M1 macrophage polarization through the methylation of STAT1 mRNA. Am J Physiol Cell Physiol. 2019;317:C762–c75.
Lei H, He M, He X, Li G, Wang Y, Gao Y, et al. METTL3 induces bone marrow mesenchymal stem cells osteogenic differentiation and migration through facilitating M1 macrophage differentiation. Am J Transl Res. 2021;13:4376–88.
Huangfu N, Zheng W, Xu Z, Wang S, Wang Y, Cheng J, et al. RBM4 regulates M1 macrophages polarization through targeting STAT1-mediated glycolysis. Int Immunopharmacol. 2020;83:106432.
Gou Y, Wang H, Wang T, Wang H, Wang B, Jiao N, et al. Ectopic endometriotic stromal cells-derived lactate induces M2 macrophage polarization via Mettl3/Trib1/ERK/STAT3 signalling pathway in endometriosis. Immunology. 2023;168:389–402.
Jiang Y, Wan Y, Gong M, Zhou S, Qiu J, Cheng W. RNA demethylase ALKBH5 promotes ovarian carcinogenesis in a simulated tumour microenvironment through stimulating NF-κB pathway. J Cell Mol Med. 2020;24:6137–48.
Jazwinska DE, Kulawiec DG, Zervantonakis IK. Cancer-mesothelial and cancer-macrophage interactions in the ovarian cancer microenvironment. Am J Physiol Cell Physiol. 2023;325:C721–c30.
Wang B, Mao Z, Ye J, Jiao X, Zhang T, Wang Q, et al. Glycolysis induced by METTL14 is essential for macrophage phagocytosis and phenotype in cervical cancer. J Immunol. 2024;212:723–36.
Legut M, Gajic Z, Guarino M, Daniloski Z, Rahman JA, Xue X, et al. A genome-scale screen for synthetic drivers of T cell proliferation. Nature. 2022;603:728–35.
Qi H, Kastenmüller W, Germain RN. Spatiotemporal basis of innate and adaptive immunity in secondary lymphoid tissue. Annu Rev Cell Dev Biol. 2014;30:141–67.
Li HB, Tong J, Zhu S, Batista PJ, Duffy EE, Zhao J, et al. m(6)A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature. 2017;548:338–42.
Yao Y, Yang Y, Guo W, Xu L, You M, Zhang YC, et al. METTL3-dependent m(6)A modification programs T follicular helper cell differentiation. Nat Commun. 2021;12:1333.
Balasundaram P, Farhana A. Immunology at the Maternal-Fetal Interface. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2023.
Aluvihare VR, Kallikourdis M, Betz AG. Regulatory T cells mediate maternal tolerance to the fetus. Nat Immunol. 2004;5:266–71.
Tong J, Cao G, Zhang T, Sefik E, Amezcua Vesely MC, Broughton JP, et al. m(6)A mRNA methylation sustains Treg suppressive functions. Cell Res. 2018;28:253–6.
Huang J, Zhou L, Deng K. Prognostic marker C3AR1 is associated with ovarian cancer cell proliferation and immunosuppression in the tumor microenvironment. J Ovarian Res. 2023;16:64.
Laumont CM, Nelson BH. B cells in the tumor microenvironment: Multi-faceted organizers, regulators, and effectors of anti-tumor immunity. Cancer Cell. 2023;41:466–89.
Rizzuto G, Brooks JF, Tuomivaara ST, McIntyre TI, Ma S, Rideaux D, et al. Establishment of fetomaternal tolerance through glycan-mediated B cell suppression. Nature. 2022;603:497–502.
Huang B, Faucette AN, Pawlitz MD, Pei B, Goyert JW, Zhou JZ, et al. Interleukin-33-induced expression of PIBF1 by decidual B cells protects against preterm labor. Nat Med. 2017;23:128–35.
Zheng Z, Zhang L, Cui XL, Yu X, Hsu PJ, Lyu R, et al. Control of early B cell development by the RNA N(6)-methyladenosine methylation. Cell Rep. 2020;31:107819.
Mäkinen A, Nikkilä A, Haapaniemi T, Oksa L, Mehtonen J, Vänskä M, et al. IGF2BP3 associates with proliferative phenotype and prognostic features in B-cell acute lymphoblastic leukemia. Cancers (Basel). 2021;13:1505.
Yin D, Kong C, Chen M. Effect of hnRNPA2/B1 on the proliferation and apoptosis of glioma U251 cells via the regulation of AKT and STAT3 pathways. Biosci Rep. 2020;40:BSR20190318.
Xie BF, Xia Y, Lin DH, Lian B, Zhang ML, Liu L, et al. Pan-cancer gene analysis of m6A modification and immune infiltration in uterine corpus endometrial carcinoma. Comput Intell Neurosci. 2022;2022:6530884.
MacLean JA, 2nd, Hayashi K. Progesterone actions and resistance in gynecological disorders. Cells. 2022;11:647.
Ortmann O, Weiss JM, Diedrich K. Gonadotrophin-releasing hormone (GnRH) and GnRH agonists: mechanisms of action. Reprod Biomed Online. 2002;5:1–7.
Greenblatt RB, Mahesh VB. PItuitary-ovarian relationships. Metabolism. 1965;14:320–6.
Wang HQ, Ma YR, Zhang YX, Wei FH, Zheng Y, Ji ZH, et al. GnRH-driven FTO-mediated RNA m(6)A modification promotes gonadotropin synthesis and secretion. BMC Biol. 2024;22:104.
Wan S, Sun Y, Zong J, Meng W, Yan J, Chen K, et al. METTL3-dependent m(6)A methylation facilitates uterine receptivity and female fertility via balancing estrogen and progesterone signaling. Cell Death Dis. 2023;14:349.
Zheng ZH, Zhang GL, Jiang RF, Hong YQ, Zhang QY, He JP, et al. METTL3 is essential for normal progesterone signaling during embryo implantation via m(6)A-mediated translation control of progesterone receptor. Proc Natl Acad Sci USA. 2023;120:e2214684120.
Kobayashi R, Kawabata-Iwakawa R, Terakawa J, Sugiyama M, Morita S, Horii T, et al. Aberrant activation of estrogen receptor-α signaling in Mettl14-deficient uteri impairs embryo implantation. Faseb j. 2023;37:e23093.
Jing YX, Li HX, Yue F, Li YM, Yu X, He JJ, et al. N6-methyladenosine demethylase FTO related to hyperandrogenism in PCOS via AKT pathway. Gynecol Endocrinol. 2023;39:2276167.
de Sanjose S, Quint WG, Alemany L, Geraets DT, Klaustermeier JE, Lloveras B, et al. Human papillomavirus genotype attribution in invasive cervical cancer: a retrospective cross-sectional worldwide study. Lancet Oncol. 2010;11:1048–56.
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49.
Hou PX, Fan Q, Zhang Q, Liu JJ, Wu Q. M6A-induced transcription factor IRF5 contributes to the progression of cervical cancer by upregulating PPP6C. Clin Exp Pharm Physiol. 2024;51:e13868.
Su C, Zhang Y, Chen P, Yang W, Du J, Zhang D. Methyltransferase-like 3 induces the development of cervical cancer by enhancing insulin-like growth factor 2 mRNA-binding proteins 3-mediated apoptotic chromatin condensation inducer 1 mRNA stability. Bioengineered. 2022;13:7034–48.
Wang Q, Guo X, Li L, Gao Z, Su X, Ji M, et al. N(6)-methyladenosine METTL3 promotes cervical cancer tumorigenesis and Warburg effect through YTHDF1/HK2 modification. Cell Death Dis. 2020;11:911.
Han C, Hu C, Liu T, Sun Y, Hu F, He Y, et al. IGF2BP3 enhances lipid metabolism in cervical cancer by upregulating the expression of SCD. Cell Death Dis. 2024;15:138.
Hu C, Liu T, Han C, Xuan Y, Jiang D, Sun Y, et al. HPV E6/E7 promotes aerobic glycolysis in cervical cancer by regulating IGF2BP2 to stabilize m(6)A-MYC expression. Int J Biol Sci. 2022;18:507–21.
Zhen L, Pan W. ALKBH5 inhibits the SIRT3/ACC1 axis to regulate fatty acid metabolism via an m6A-IGF2BP1-dependent manner in cervical squamous cell carcinoma. Clin Exp Pharm Physiol. 2023;50:380–92.
Zhang P, Tang Y, Zhao J, Yang J, Chen Y, Gong Y, et al. TRIM11 regulated by m6A modification promotes the progression of cervical cancer by PHLPP1 ubiquitination. Neoplasma. 2023;70:659–69.
Lin B, Wang Q, Wang X, Wei H, Nie X, Li L, et al. Expression of variant isoforms of the tyrosine kinase SYK differentially regulates cervical cancer progression through PI3K/AKT pathway. Sci Rep. 2024;14:29080.
Li L, Zeng J, He S, Yang Y, Wang C. METTL14 decreases FTH1 mRNA stability via m6A methylation to promote sorafenib-induced ferroptosis of cervical cancer. Cancer Biol Ther. 2024;25:2349429.
Yu T, Wu F, Jia Y, Zhang X, Qi X, Jin Z, et al. RNA N(6)-methyladenosine modification mediates downregulation of NR4A1 to facilitate malignancy of cervical cancer. Cell Biosci. 2022;12:207.
Wu M, Chen G, Liao X, Xiao L, Zheng J. YTHDF2 interference suppresses the EMT of cervical cancer cells and enhances cisplatin chemosensitivity by regulating AXIN1. Drug Dev Res. 2022;83:1190–200.
Huang J, Yang J, Zhang Y, Lu D, Dai Y. FTO promotes cervical cancer cell proliferation, colony formation, migration and invasion via the regulation of the BMP4/Hippo/YAP1/TAZ pathway. Exp Cell Res. 2023;427:113585.
Zhao J, Lee EE, Kim J, Yang R, Chamseddin B, Ni C, et al. Transforming activity of an oncoprotein-encoding circular RNA from human papillomavirus. Nat Commun. 2019;10:2300.
Liang L, Zhu Y, Li J, Zeng J, Wu L. ALKBH5-mediated m6A modification of circCCDC134 facilitates cervical cancer metastasis by enhancing HIF1A transcription. J Exp Clin Cancer Res. 2022;41:261.
Huang C, Liang J, Lin S, Wang D, Xie Q, Lin Z, et al. N(6)-methyladenosine associated silencing of miR-193b promotes cervical cancer aggressiveness by targeting CCND1. Front Oncol. 2021;11:666597.
Wang X, Zhang J, Wang Y. Long noncoding RNA GAS5-AS1 suppresses growth and metastasis of cervical cancer by increasing GAS5 stability. Am J Transl Res. 2019;11:4909–21.
Sui H, Shi C, Yan Z, Chen J, Man L, Wang F. LRRC75A-AS1 drives the epithelial-mesenchymal transition in cervical cancer by binding IGF2BP1 and inhibiting SYVN1-mediated NLRP3 ubiquitination. Mol Cancer Res. 2024;22:1075–1087.
Wang T, Li W, Ye B, Zhang S, Lei X, Zhang D. FTO-stabilized lncRNA HOXC13-AS epigenetically upregulated FZD6 and activated Wnt/β-catenin signaling to drive cervical cancer proliferation, invasion, and EMT. J buon. 2021;26:1279–91.
Zhang Y, Wang D, Wu D, Zhang D, Sun M. Long Noncoding RNA KCNMB2-AS1 stabilized by N(6)-methyladenosine modification promotes cervical cancer growth through acting as a competing endogenous RNA. Cell Transpl. 2020;29:963689720964382.
Ji F, Lu Y, Chen S, Yu Y, Lin X, Zhu Y, et al. IGF2BP2-modified circular RNA circARHGAP12 promotes cervical cancer progression by interacting m(6)A/FOXM1 manner. Cell Death Discov. 2021;7:215.
Baert T, Ferrero A, Sehouli J, O’Donnell DM, González-Martín A, Joly F, et al. The systemic treatment of recurrent ovarian cancer revisited. Ann Oncol. 2021;32:710–25.
Govindarajan M, Wohlmuth C, Waas M, Bernardini MQ, Kislinger T. High-throughput approaches for precision medicine in high-grade serous ovarian cancer. J Hematol Oncol. 2020;13:134.
Hua W, Zhao Y, Jin X, Yu D, He J, Xie D, et al. METTL3 promotes ovarian carcinoma growth and invasion through the regulation of AXL translation and epithelial to mesenchymal transition. Gynecol Oncol. 2018;151:356–65.
Li H, Cai L, Pan Q, Jiang X, Zhao J, Xiang T, et al. N(6)-methyladenosine-modified VGLL1 promotes ovarian cancer metastasis through high-mobility group AT-hook 1/Wnt/β-catenin signaling. iScience. 2024;27:109245.
Li Y, Peng H, Jiang P, Zhang J, Zhao Y, Feng X, et al. Downregulation of methyltransferase-like 14 promotes ovarian cancer cell proliferation through stabilizing TROAP mRNA. Front Oncol. 2022;12:824258.
Wei YS, Yao DS, Li L, Lu Y, Yang XM, Zhang WG. [Expression of METTL14 in epithelial ovarian cancer and the effect on cell proliferation, invasion and migration of A2780 and SKOV3 cells]. Zhonghua Fu Chan Ke Za Zhi. 2022;57:46–56.
Fu Y, Jia XC. WTAP-mediated N6-methyladenosine modification on EGR3 in different types of epithelial ovarian cancer. J Biol Regul Homeost Agents. 2020;34:1505–12.
Gan L, Zhao S, Gao Y, Qi Y, Su M, Wang A, et al. N6-methyladenosine methyltransferase KIAA1429 promoted ovarian cancer aerobic glycolysis and progression through enhancing ENO1 expression. Biol Direct. 2023;18:64.
Wu Q, Li G, Gong L, Cai J, Chen L, Xu X, et al. Identification of miR-30c-5p as a tumor suppressor by targeting the m(6) A reader HNRNPA2B1 in ovarian cancer. Cancer Med. 2023;12:5055–70.
Liu T, Wei Q, Jin J, Luo Q, Liu Y, Yang Y, et al. The m6A reader YTHDF1 promotes ovarian cancer progression via augmenting EIF3C translation. Nucleic Acids Res. 2020;48:3816–31.
Xu F, Li J, Ni M, Cheng J, Zhao H, Wang S, et al. FBW7 suppresses ovarian cancer development by targeting the N(6)-methyladenosine binding protein YTHDF2. Mol Cancer. 2021;20:45.
Wang X, Chen Q, Bing Z, Zhou S, Xu Z, Hou Y, et al. Low expression of m6A reader YTHDC1 promotes progression of ovarian cancer via PIK3R1/STAT3/GANAB axis. Int J Biol Sci. 2023;19:4672–88.
Jung SY, Riew TR, Yun HH, Lim JH, Hwang JW, Jung SW, et al. Skeletal muscle-specific bis depletion leads to muscle dysfunction and early death accompanied by impairment in protein quality control. Int J Mol Sci. 2023;24:9635.
Yuan J, Guan W, Li X, Wang F, Liu H, Xu G. RBM15-mediating MDR1 mRNA m(6)A methylation regulated by the TGF-β signaling pathway in paclitaxel-resistant ovarian cancer. Int J Oncol. 2023;63:112.
Huang H, Zhao G, Cardenas H, Valdivia AF, Wang Y, Matei D. N6-Methyladenosine RNA Modifications Regulate the Response to Platinum Through Nicotinamide N-methyltransferase. Mol Cancer Ther. 2023;22:393–405.
Sun M, Zhang X, Bi F, Wang D, Zhou X, Li X, et al. FTO inhibits epithelial ovarian cancer progression by destabilising SNAI1 mRNA through IGF2BP2. Cancers (Basel). 2022;14:5218.
Lyu Y, Wang Y, Ding H, Li P. Hypoxia-induced m6A demethylase ALKBH5 promotes ovarian cancer tumorigenicity by decreasing methylation of the lncRNA RMRP. Am J Cancer Res. 2023;13:4179–91.
Zhang Z, Zhu H, Hu J. CircRAB11FIP1 promoted autophagy flux of ovarian cancer through DSC1 and miR-129. Cell Death Dis. 2021;12:219.
Xu X, Zhuang X, Yu H, Li P, Li X, Lin H, et al. FSH induces EMT in ovarian cancer via ALKBH5-regulated Snail m6A demethylation. Theranostics. 2024;14:2151–66.
Colombo N, Creutzberg C, Amant F, Bosse T, González-Martín A, Ledermann J, et al. ESMO-ESGO-ESTRO consensus conference on endometrial cancer: diagnosis, treatment and follow-up. Ann Oncol. 2016;27:16–41.
Liu J, Eckert MA, Harada BT, Liu SM, Lu Z, Yu K, et al. m(6)A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat Cell Biol. 2018;20:1074–83.
Ralser DJ, Condic M, Klümper N, Ellinger J, Staerk C, Egger EK, et al. Comprehensive immunohistochemical analysis of N6-methyladenosine (m6A) writers, erasers, and readers in endometrial cancer. J Cancer Res Clin Oncol. 2023;149:2417–24.
Ma Z, Li Q, Liu P, Dong W, Zuo Y. METTL3 regulates m6A in endometrioid epithelial ovarian cancer independently of METTl14 and WTAP. Cell Biol Int. 2020;44:2524–31.
Cai Y, Li N, Li H. YBX2 modulates mRNA stability via interaction with YTHDF2 in endometrial cancer cells. Exp Cell Res. 2023;427:113586.
Hong L, Pu X, Gan H, Weng L, Zheng Q. YTHDF2 inhibit the tumorigenicity of endometrial cancer via downregulating the expression of IRS1 methylated with m(6)A. J Cancer. 2021;12:3809–18.
Zhang L, Wan Y, Zhang Z, Jiang Y, Gu Z, Ma X, et al. IGF2BP1 overexpression stabilizes PEG10 mRNA in an m6A-dependent manner and promotes endometrial cancer progression. Theranostics. 2021;11:1100–14.
Ruan P, Wang S, Yang C, Huang X, Sun P, Tan A. m(6)A mRNA methylation regulates the ERK/NF-κB/AKT signaling pathway through the PAPPA/IGFBP4 axis to promote proliferation and tumor formation in endometrial cancer. Cell Biol Toxicol. 2023;39:1611–26.
Xue T, Liu X, Zhang M, E Q, Liu S, Zou M, et al. PADI2-catalyzed MEK1 citrullination activates ERK1/2 and promotes IGF2BP1-mediated SOX2 mRNA stability in endometrial cancer. Adv Sci (Weinh). 2021;8:2002831.
Tian J, Cheng H, Wang N, Wang C. SLERT, as a novel biomarker, orchestrates endometrial cancer metastasis via regulation of BDNF/TRKB signaling. World J Surg Oncol. 2023;21:27.
Wang B, Wang Y, Wang W, Wang Z, Zhang Y, Pan X, et al. WTAP/IGF2BP3 mediated m6A modification of the EGR1/PTEN axis regulates the malignant phenotypes of endometrial cancer stem cells. J Exp Clin Cancer Res. 2024;43:204.
Shi R, Zhao R, Shen Y, Wei S, Zhang T, Zhang J, et al. IGF2BP2-modified circular RNA circCHD7 promotes endometrial cancer progression via stabilizing PDGFRB and activating JAK/STAT signaling pathway. Cancer Gene Ther. 2024;31:1221–1236.
Wang C, Kong F, Ma J, Miao J, Su P, Yang H, et al. IGF2BP3 enhances the mRNA stability of E2F3 by interacting with LINC00958 to promote endometrial carcinoma progression. Cell Death Discov. 2022;8:279.
Azzam SK, Alsafar H, Sajini AA. FTO m6A demethylase in obesity and cancer: implications and underlying molecular mechanisms. Int J Mol Sci. 2022;23:3800.
Pu X, Gu Z, Gu Z. ALKBH5 regulates IGF1R expression to promote the proliferation and tumorigenicity of endometrial cancer. J Cancer. 2020;11:5612–22.
Froeling FE, Seckl MJ. Gestational trophoblastic tumours: an update for 2014. Curr Oncol Rep. 2014;16:408.
Lu Y, Li X, Zuo Y, Xu Q, Liu L, Wu H, et al. miR-373-3p inhibits epithelial-mesenchymal transition via regulation of TGFβR2 in choriocarcinoma. J Obstet Gynaecol Res. 2021;47:2417–32.
Peng X, Zhang Z, Mo Y, Liu J, Wang S, Liu H. Bioinformatics analysis of choriocarcinoma-related MicroRNA-transcription factor-target gene regulatory networks and validation of key miRNAs. Onco Targets Ther. 2021;14:3903–19.
Wang W, Shi J, Zheng L. METTL3 promotes choriocarcinoma progression by activating the miR-935/GJA1 pathway in an m6A-dependent manner. Am J Reprod Immunol. 2023;90:e13791.
Ye K, Li L, Wu B, Wang D. METTL3 m6A-dependently promotes miR-21-5p maturation to accelerate choriocarcinoma progression via the HIF1AN-induced inactivation of the HIF1A/VEGF pathway. Genes Genomics. 2022;44:1311–22.
Bird CC, McElin TW, Manalo-Estrella P. The elusive adenomyosis of the uterus-revisited. Am J Obstet Gynecol. 1972;112:583–93.
Zhai J, Li S, Sen S, Opoku-Anane J, Du Y, Chen ZJ, et al. m(6)A RNA methylation regulators contribute to eutopic endometrium and myometrium dysfunction in adenomyosis. Front Genet. 2020;11:716.
Bulun SE, Yilmaz BD, Sison C, Miyazaki K, Bernardi L, Liu S, et al. Endometriosis. Endocr Rev. 2019;40:1048–79.
Jiang L, Zhang M, Wu J, Wang S, Yang X, Yi M, et al. Exploring diagnostic m6A regulators in endometriosis. Aging (Albany NY). 2020;12:25916–38.
Li X, Xiong W, Long X, Dai X, Peng Y, Xu Y, et al. Inhibition of METTL3/m6A/miR126 promotes the migration and invasion of endometrial stromal cells in endometriosis. † Biol Reprod 2021;105:1221–33.
Cao M, Yuan C, Chen X, He G, Chen T, Zong J, et al. METTL3 deficiency leads to ovarian insufficiency due to IL-1β overexpression in theca cells. Free Radic Biol Med. 2024;222:72–84.
McGlacken-Byrne SM, Del Valle I, Quesne Stabej PL, Bellutti L, Garcia-Alonso L, Ocaka LA, et al. Pathogenic variants in the human m6A reader YTHDC2 are associated with primary ovarian insufficiency. JCI Insight. 2022;7:e154671.
Sohi GK, Farooqui N, Mohan A, Rajagopalan KS, Xing L, Zhu XY, et al. The impact of hypoxia preconditioning on mesenchymal stem cells performance in hypertensive kidney disease. Stem cell Res Ther. 2024;15:162.
Ding C, Zou Q, Ding J, Ling M, Wang W, Li H, et al. Increased N6-methyladenosine causes infertility is associated with FTO expression. J Cell Physiol. 2018;233:7055–66.
Li Y, Li M, Liu J, Nie G, Yang H. Altered m6A modification is involved YAP-mediated apoptosis response in 4-vinylcyclohexene diepoxide induced ovotoxicity. Ecotoxicol Environ Saf. 2023;262:115192.
Huang B, Ding C, Zou Q, Wang W, Li H. Cyclophosphamide regulates N6-methyladenosine and m6A RNA enzyme levels in human granulosa cells and in ovaries of a premature ovarian aging mouse model. Front Endocrinol (Lausanne). 2019;10:415.
Kostroun KE, Goldrick K, Mondshine JN, Robinson RD, Mankus E, Reddy S, et al. Impact of updated international diagnostic criteria for the diagnosis of polycystic ovary syndrome. F S Rep. 2023;4:173–8.
Liu AL, Xie HJ, Xie HY, Liu J, Yin J, Hu JS, et al. Association between fat mass and obesity associated (FTO) gene rs9939609 A/T polymorphism and polycystic ovary syndrome: a systematic review and meta-analysis. BMC Med Genet. 2017;18:89.
Tan S, Scherag A, Janssen OE, Hahn S, Lahner H, Dietz T, et al. Large effects on body mass index and insulin resistance of fat mass and obesity associated gene (FTO) variants in patients with polycystic ovary syndrome (PCOS). BMC Med Genet. 2010;11:12.
Zhou S, Hua R, Quan S. N6-methyladenosine regulator-mediated methylation modification patterns and immune infiltration characterization in Polycystic Ovary Syndrome (PCOS). J Ovarian Res. 2023;16:73.
Zhou L, Han X, Li W, Wang N, Yao L, Zhao Y, et al. N6-methyladenosine demethylase FTO induces the dysfunctions of ovarian granulosa cells by upregulating flotillin 2. Reprod Sci. 2022;29:1305–15.
Zhang S, Deng W, Liu Q, Wang P, Yang W, Ni W. Altered m(6) A modification is involved in up-regulated expression of FOXO3 in luteinized granulosa cells of non-obese polycystic ovary syndrome patients. J Cell Mol Med. 2020;24:11874–82.
Luo Y, Chen J, Cui Y, Fang F, Zhang Z, Hu L, et al. Transcriptome-wide high-throughput m(6) A sequencing of differential m(6) A methylation patterns in the decidual tissues from RSA patients. Faseb j. 2023;37:e22802.
Meng J, Li X, Liu W, Xiao Y, Tang H, Wu Y, et al. The role of vitamin D in the prevention and treatment of SARS-CoV-2 infection: A meta-analysis of randomized controlled trials. Clin Nutr. 2023;42:2198–206.
Huang N, Gao Y, Zhang M, Guo L, Qin L, Liao S, et al. METTL3-Mediated m(6)A RNA Methylation of ZBTB4 Interferes With Trophoblast Invasion and Maybe Involved in RSA. Front Cell Dev Biol. 2022;10:894810.
Zhang M, Huang N, Gao Y, Feng Z, Kang B, Guo H, et al. HNRNPC mediated m(6)A methylation of 5-methyltetrahydrofolate-homocysteine methyltransferase and involved in the occurrence of RSA. J Reprod Immunol. 2023;160:104160.
Xu Z, Tian P, Guo J, Mi C, Liang T, Xie J, et al. Lnc-HZ01 with m6A RNA methylation inhibits human trophoblast cell proliferation and induces miscarriage by up-regulating BPDE-activated lnc-HZ01/MXD1 positive feedback loop. Sci Total Environ. 2021;776:145950.
Dai M, Huang W, Huang X, Ma C, Wang R, Tian P, et al. BPDE, the migration and invasion of human trophoblast cells, and occurrence of miscarriage in humans: roles of a novel lncRNA-HZ09. Environ Health Perspect. 2023;131:17009.
Zhao Y, Sun J, Jin L. The N6-methyladenosine regulator ALKBH5 mediated stromal cell-macrophage interaction via VEGF signaling to promote recurrent spontaneous abortion: a bioinformatic and in vitro study. Int J Mol Sci. 2022;23:15819.
Zheng Q, Yang F, Gan H, Jin L. Hypoxia induced ALKBH5 prevents spontaneous abortion by mediating m(6)A-demethylation of SMAD1/5 mRNAs. Biochim Biophys Acta Mol Cell Res. 2022;1869:119316.
Qiu W, Zhou Y, Wu H, Lv X, Yang L, Ren Z, et al. RNA demethylase FTO mediated RNA m(6)A modification is involved in maintaining maternal-fetal interface in spontaneous abortion. Front Cell Dev Biol. 2021;9:617172.
Zhang J, Liu X, Gao Y. FTO protein regulates the TGF-β signalling pathway through RNA N6-methyladenosine modification to induce unexplained recurrent spontaneous abortion. Febs j. 2024;291:1545–59.
Gu Y, Chu X, Morgan JA, Lewis DF, Wang Y. Upregulation of METTL3 expression and m6A RNA methylation in placental trophoblasts in preeclampsia. Placenta. 2021;103:43–9.
Bian Y, Li J, Shen H, Li Y, Hou Y, Huang L, et al. WTAP dysregulation-mediated HMGN3-m6A modification inhibited trophoblast invasion in early-onset preeclampsia. Faseb j. 2022;36:e22617.
Chen Y, Liu X, Li L, He X, Zheng F, Zhang Y, et al. Methyltransferase-like 3 aggravates endoplasmic reticulum stress in preeclampsia by targeting TMBIM6 in YTHDF2-dependent manner. Mol Med. 2023;29:19.
Wang Y, Zhang G, Gao Y, Zhang X, Qi H. METTL3 promotes trophoblast ferroptosis in preeclampsia by stabilizing the ACSL4 m(6)A modification. Exp Cell Res. 2024;437:113990.
Fan W, Zhou W, Yan Q, Peng Y, Wang H, Kong C, et al. Upregulation of METTL14 contributes to trophoblast dysfunction by elevating FOXO3a expression in an m(6)A-dependent manner. Placenta. 2022;124:18–27.
You G, Li Z, Li L, Xu C. Overexpression of RBM15 modulated the effect of trophoblast cells by promoting the binding ability between YTHDF2 and the CD82 3’UTR to decrease the expression of CD82. Heliyon. 2024;10:e30702.
Zhang Y, Guo X, Chen Z, Guo R. Low m6A modification-mediated upregulation of PLAC8 promotes trophoblast cell invasion and migration in preeclampsia. Eur J Med Res. 2023;28:466.
Wang L, Shi L, Zhou B, Hong L, Gong H, Wu D. METTL3-mediated lncRNA HOXD-AS1 stability regulates inflammation, and the migration and invasion of trophoblast cells via the miR-135a/ β-TRCP axis. Noncoding RNA Res. 2024;9:12–23.
Wang D, Guan H, Wang Y, Song G, Xia Y. N6-methyladenosine modification in trophoblasts promotes circSETD2 expression, inhibits miR-181a-5p, and elevates MCL1 transcription to reduce apoptosis of trophoblasts. Environ Toxicol. 2023;38:422–35.
Zhang Y, Yang H, Long Y, Zhang Y, Chen R, Shi J, et al. circRNA N6-methyladenosine methylation in preeclampsia and the potential role of N6-methyladenosine-modified circPAPPA2 in trophoblast invasion. Sci Rep. 2021;11:24357.
Zhang T, Tang X, Zhu Y, Wang C, Jiang Z, Yang N, et al. IGF2BP2 enhances LincRNA01116 stability via m(6) A: A potential biomarker and therapeutic target for patients with pre-eclampsia. J Cell Biochem. 2023;124:239–53.
Wang J, Wang K, Liu W, Cai Y, Jin H. m6A mRNA methylation regulates the development of gestational diabetes mellitus in Han Chinese women. Genomics. 2021;113:1048–56.
Meng J, Liu W, Wu Y, Xiao Y, Tang H, Gao S. Is it necessary to wear compression stockings and how long should they be worn for preventing post thrombotic syndrome? A meta-analysis of randomized controlled trials. Thromb Res. 2023;225:79–86.
Meng J, Tang H, Xiao Y, Liu W, Wu Y, Xiong Y, et al. Appropriate thromboprophylaxis strategy for COVID-19 patients on dosage, antiplatelet therapy, outpatient, and postdischarge prophylaxis: a meta-analysis of randomized controlled trials. Int J Surg. 2024;110:3910–22.
Zhang S, Cai S, Ye L, Shen L, Zhu C, Huang J, et al. METTL3 mediates m6A modification of hsa_circ_0072380 to regulate the progression of gestational diabetes mellitus. Gene. 2024;931:148894.
Ning J, Yan J, Wang S, Cui Z, Xue Y, Juan J, et al. Demethylase FTO-mediated m6A modification of SIK1 modulates placental cytotrophoblast syncytialization in type 2 diabetes mellitus. iScience. 2024;27:109900.
Du R, Bai Y, Li L. Biological networks in gestational diabetes mellitus: insights into the mechanism of crosstalk between long non-coding RNA and N(6)-methyladenine modification. BMC Pregnancy Childbirth. 2022;22:384.
Li Y, Liu Y, Yao X, Wang H, Shi Z, He M. METTL14-mediated lncRNA XIST silencing alleviates GDM progression by facilitating trophoblast cell proliferation and migration via the miR-497-5p/FOXO1 axis. J Biochem Mol Toxicol. 2024;38:e23621.
Chen Y, Lv G, Du X, Yang F, Zhao Z. Fentanyl promoted the growth of placenta trophoblast cells through regulating the METTL14 mediated CCL5 levels. Biol Pharm Bull. 2023;46:1797–804.
Fang J, Wu X, He J, Zhang H, Chen X, Zhang H, et al. RBM15 suppresses hepatic insulin sensitivity of offspring of gestational diabetes mellitus mice via m6A-mediated regulation of CLDN4. Mol Med. 2023;29:23.
Pan J, Xu L, Pan H. Development and validation of an m6A RNA methylation regulator-based signature for prognostic prediction in cervical squamous cell carcinoma. Front Oncol. 2020;10:1444.
Wang S, Ding B, Wang S, Yan W, Xia Q, Meng D, et al. Gene signature of m(6)A RNA regulators in diagnosis, prognosis, treatment, and immune microenvironment for cervical cancer. Sci Rep. 2022;12:17667.
Zhong S, Guo Q, Chen X, Luo X, Long Y, Chong T, et al. The inhibition of YTHDF3/m(6)A/LRP6 reprograms fatty acid metabolism and suppresses lymph node metastasis in cervical cancer. Int J Biol Sci. 2024;20:916–36.
Zhou S, Bai ZL, Xia D, Zhao ZJ, Zhao R, Wang YY, et al. FTO regulates the chemo-radiotherapy resistance of cervical squamous cell carcinoma (CSCC) by targeting β-catenin through mRNA demethylation. Mol Carcinog. 2018;57:590–7.
Lin X, Wang F, Chen J, Liu J, Lin YB, Li L, et al. N(6)-methyladenosine modification of CENPK mRNA by ZC3H13 promotes cervical cancer stemness and chemoresistance. Mil Med Res. 2022;9:19.
Fan L, Lin Y, Lei H, Shu G, He L, Yan Z, et al. A newly defined risk signature, consisting of three m(6)A RNA methylation regulators, predicts the prognosis of ovarian cancer. Aging (Albany NY). 2020;12:18453–75.
Li Q, Ren CC, Chen YN, Yang L, Zhang F, Wang BJ, et al. A risk score model incorporating three m6A RNA methylation regulators and a related network of miRNAs-m6A regulators-m6A target genes to predict the prognosis of patients with ovarian cancer. Front Cell Dev Biol. 2021;9:703969.
Wei Q, Yang D, Liu X, Zhao H, Yang Y, Xu J, et al. Exploration of the role of m(6) A RNA methylation regulators in malignant progression and clinical prognosis of ovarian cancer. Front Genet. 2021;12:650554.
Zhang C, Liu J, Guo H, Hong D, Ji J, Zhang Q, et al. m6A RNA methylation regulators were associated with the malignancy and prognosis of ovarian cancer. Bioengineered. 2021;12:3159–76.
Han X, Liu J, Cheng G, Cui S. Gene signatures and prognostic values of m6A RNA methylation regulators in ovarian cancer. Cancer Control. 2020;27:1073274820960460.
Zhu W, Zhao L, Kong B, Liu Y, Zou X, Han T, et al. The methylation modification of m6A regulators contributes to the prognosis of ovarian cancer. Ann Transl Med. 2022;10:59.
Zheng P, Li N, Zhan X. Ovarian cancer subtypes based on the regulatory genes of RNA modifications: Novel prediction model of prognosis. Front Endocrinol (Lausanne). 2022;13:972341.
Ye L, Tong X, Pan K, Shi X, Xu B, Yao X, et al. Identification of potential novel N6-methyladenosine effector-related lncRNA biomarkers for serous ovarian carcinoma: a machine learning-based exploration in the framework of 3P medicine. Front Pharm. 2024;15:1351929.
Yu HL, Ma XD, Tong JF, Li JQ, Guan XJ, Yang JH. WTAP is a prognostic marker of high-grade serous ovarian cancer and regulates the progression of ovarian cancer cells. Onco Targets Ther. 2019;12:6191–201.
Sun R, Yuan L, Jiang Y, Wan Y, Ma X, Yang J, et al. ALKBH5 activates FAK signaling through m6A demethylation in ITGB1 mRNA and enhances tumor-associated lymphangiogenesis and lymph node metastasis in ovarian cancer. Theranostics. 2023;13:833–48.
Cui S. METTL3-mediated m6A modification of lnc RNA RHPN1-AS1 enhances cisplatin resistance in ovarian cancer by activating PI3K/AKT pathway. J Clin Lab Anal. 2022;36:e24761.
Zhang Y, Qiu JG, Jia XY, Ke Y, Zhang MK, Stieg D, et al. METTL3-mediated N6-methyladenosine modification and HDAC5/YY1 promote IFFO1 downregulation in tumor development and chemo-resistance. Cancer Lett. 2023;553:215971.
Nie S, Zhang L, Liu J, Wan Y, Jiang Y, Yang J, et al. ALKBH5-HOXA10 loop-mediated JAK2 m6A demethylation and cisplatin resistance in epithelial ovarian cancer. J Exp Clin Cancer Res. 2021;40:284.
Chen L, Gao W, Lin L, Sha C, Li T, Chen Q, et al. A methylation- and immune-related lncRNA signature to predict ovarian cancer outcome and uncover mechanisms of chemoresistance. J Ovarian Res. 2023;16:186.
Yin X, Zhao S, Zhang M, Xing J, Zhou J, Gao W, et al. m6A-modified RIPK4 facilitates proliferation and cisplatin resistance in epithelial ovarian cancer. Gynecol Oncol. 2024;180:99–110.
Lin YB, Xu BH. N6-methyladenosine methyltransferase METTL3 enhances PTGER2 expression to increase ovarian cancer stemness and chemoresistance. Front Biosci (Landmark Ed). 2023;28:199.
Shu C, Gu MH, Zeng C, Shao WG, Li HY, Ma XH, et al. Small-molecule exhibits anti-tumor activity by targeting the RNA m(6)A reader IGF2BP3 in ovarian cancer. Am J Cancer Res. 2023;13:4888–902.
Yu HY, Yang L, Liu YC, Yu AJ. Sulforaphene suppressed cell proliferation and promoted apoptosis of COV362 cells in endometrioid ovarian cancer. PeerJ. 2023;11:e16308.
Fukumoto T, Zhu H, Nacarelli T, Karakashev S, Fatkhutdinov N, Wu S, et al. N(6)-methylation of adenosine of FZD10 mRNA contributes to PARP inhibitor resistance. Cancer Res. 2019;79:2812–20.
Zhang X, Pang X, Huang Y, Qian S. A seven-m6A regulator-related CpG site-based prognostic signature for endometrial carcinoma. Med (Baltim). 2021;100:e26648.
Liang D, Feng Y, Zandkarimi F, Wang H, Zhang Z, Kim J, et al. Ferroptosis surveillance independent of GPX4 and differentially regulated by sex hormones. Cell. 2023;186:2748–64.e22.
Shi R, Wang Z, Zhang J, Yu Z, An L, Wei S, et al. N6-methyladenosine-related long noncoding RNAs as potential prognosis biomarkers for endometrial cancer. Int J Gen Med. 2021;14:8249–62.
Shan L, Lu Y, Xiang CC, Zhu X, Zuo ED, Cheng X. Identification of five m6A-related lncRNA genes as prognostic markers for endometrial cancer based on TCGA database. J Immunol Res. 2022;2022:2547029.
Wang Y, Wang C, Guan X, Ma Y, Zhang S, Li F, et al. PRMT3-mediated arginine methylation of METTL14 promotes malignant progression and treatment resistance in endometrial carcinoma. Adv Sci (Weinh). 2023;10:e2303812.
Zhang Y, Zhou H, Ding C. The ameliorative effect of CangFu Daotan Decoction on polycystic ovary syndrome of rodent model is associated with m6A methylation and Wnt/β-catenin pathway. Gynecol Endocrinol. 2023;39:2181637.
Liu K, He X, Huang J, Yu S, Cui M, Gao M, et al. Short-chain fatty acid-butyric acid ameliorates granulosa cells inflammation through regulating METTL3-mediated N6-methyladenosine modification of FOSL2 in polycystic ovarian syndrome. Clin Epigenetics. 2023;15:86.
Zhu R, Ji X, Wu X, Chen J, Li X, Jiang H, et al. Melatonin antagonizes ovarian aging via YTHDF2-MAPK-NF-κB pathway. Genes Dis. 2022;9:494–509.
Luo XY, Zhao YZ, Peng JZ, Hu Y, Yang WJ, Xu A. Punch grafting can be used as a supplementary treatment for perilesional halo after melanocyte grafting for vitiligo. J Eur Acad Dermatol Venereol. 2024.
Chen Q, Hu K, Shi J, Li H, Li W. Hesperidin inhibits methylation and autophagy in LPS and high glucose-induced human villous trophoblasts. Biochem Biophys Res Commun. 2023;671:278–85.
Zhao R, Chen J, Wang Y, Xiao H, Mei P, Lin W, et al. Prognostic roles of dysregulated METTL3 protein expression in cancers and potential anticancer value by inhibiting METTL3 function. Fundam Clin Pharm. 2024;38:924–39.
Zhao H, Meng L, Du P, Liao X, Mo X, Gong M, et al. IDH1 mutation produces R-2-hydroxyglutarate (R-2HG) and induces mir-182-5p expression to regulate cell cycle and tumor formation in glioma. Biol Res. 2024;57:30.
Su C, Zhang Y, Chen P, Yang W, Du J, Zhang D. Methyltransferase-like 3 induces the development of cervical cancer by enhancing insulin-like growth factor 2 mRNA-binding proteins 3-mediated apoptotic chromatin condensation inducer 1 mRNA stability. Bioengineered. 2022;13:7034–7048.
Author information
Authors and Affiliations
Contributions
YF and PL were involved in the conception of the study. PL, YL, and HM were involved in writing the article. PL, JZ, QZ and RY were involved in the production of figures. All authors critically revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, P., Lin, Y., Ma, H. et al. Epigenetic regulation in female reproduction: the impact of m6A on maternal-fetal health. Cell Death Discov. 11, 43 (2025). https://doi.org/10.1038/s41420-025-02324-z
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41420-025-02324-z