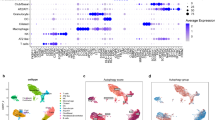
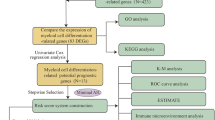
Abstract
Lung cancer is a major cause accounting for cancer-related mortalities, with lung adenocarcinoma (LUAD) being the most prevalent subtype. Given the high clinical and cellular heterogeneities of LUAD, accurate diagnosis and prognosis are crucial to avoid overdiagnosis and overtreatment. Taking full advantage of scRNA-Seq data to resolve the tumor heterogeneities, we explored the overall landscape of LUAD microenvironment. Utilizing the stage-specific tumor cell markers, we have developed highly accurate diagnostic and prognostic models with elevated sensitivity and specificity. The diagnostic model, developed through random forest algorithms with a thirteen-gene signature, achieved an accuracy of 96.4% and an AUC of 0.993. These metrics were further demonstrated by benchmarking with available models and scoring systems in independent cohorts. Concurrently, the prognostic model, formulated via Cox regression with a six-gene signature, effectively predicted overall survival, with elevated risk scores associated with increased fractions of cancer-associated fibroblasts, and higher likelihood of immune escape and T-cell exclusion. Subsequently, two nomograms were developed to predict survival and drug responses, facilitating their integration into clinical practice. Overall, this study underscores the potential of our models for efficient, rapid, and cost-effective diagnosis and prognosis of LUAD, adaptable to multiple expression profiling platforms and quantification methods.

This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 digital issues and online access to articles
118,99 € per year
only 19,83 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The LUAD scRNA-Seq profiles GSE131907 [16] was obtained from GEO. Expression profiles and clinical traits of TCGA-LUAD were obtained from the UCSC Xena browser (https://xenabrowser.net/datapages/). Additionally, microarray datasets GSE7670 [27], GSE102287 [25], GSE30219 [24], GSE19804 [28], GSE10072 [26], GSE31210 [33], GSE13213 [34], and GSE72094 [35] were obtained from GEO. The drug responses and corresponding clinical data were obtained from a previous study [53]. The final diagnostic model is available from the Github repository (https://github.com/univerchen/LUAD).
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209–49.
Spella M, Stathopoulos GT. Immune Resistance in Lung Adenocarcinoma. Cancers (Basel). 2021;13:384.
Senosain MF, Massion PP. Intratumor Heterogeneity in Early Lung Adenocarcinoma. Front Oncol. 2020;10:349.
Seguin L, Durandy M, Feral CC. Lung Adenocarcinoma Tumor Origin: A Guide for Personalized Medicine. Cancers (Basel). 2022;14:1759.
Diaz-Cano SJ. Tumor heterogeneity: mechanisms and bases for a reliable application of molecular marker design. Int J Mol Sci. 2012;13:1951–2011.
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30.
Chatterjee S. Artefacts in histopathology. J Oral Maxillofac Pathol. 2014;18:S111–6.
Hillman H. Limitations of clinical and biological histology. Med Hypotheses. 2000;54:553–64.
Taqi SA, Sami SA, Sami LB, Zaki SA. A review of artifacts in histopathology. J Oral Maxillofac Pathol. 2018;22:279.
D’Ambrosi S, Giannoukakos S, Antunes-Ferreira M, Pedraz-Valdunciel C, Bracht JWP, Potie N, et al. Combinatorial Blood Platelets-Derived circRNA and mRNA Signature for Early-Stage Lung Cancer Detection. Int J Mol Sci. 2023;24:4881.
Ye XD, Zhang N, Jin YX, Xu B, Guo CY, Wang XQ, et al. Dramatically changed immune-related molecules as early diagnostic biomarkers of non-small cell lung cancer. Febs J. 2020;287:783–99.
Freitas C, Sousa C, Machado F, Serino M, Santos V, Cruz-Martins N, et al. The Role of Liquid Biopsy in Early Diagnosis of Lung Cancer. Front Oncol. 2021;11:634316.
Li Y, Sun N, Lu Z, Sun S, Huang J, Chen Z, et al. Prognostic alternative mRNA splicing signature in non-small cell lung cancer. Cancer Lett. 2017;393:40–51.
Li R, Yang YE, Yin YH, Zhang MY, Li H, Qu YQ. Methylation and transcriptome analysis reveal lung adenocarcinoma-specific diagnostic biomarkers. J Transl Med. 2019;17:324.
Sun L, Zhang Z, Yao Y, Li WY, Gu J. Analysis of expression differences of immune genes in non-small cell lung cancer based on TCGA and ImmPort data sets and the application of a prognostic model. Ann Transl Med. 2020;8:550.
Kim N, Kim HK, Lee K, Hong Y, Cho JH, Choi JW, et al. Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma. Nat Commun. 2020;11:2285.
Ma KY, Schonnesen AA, Brock A, Van Den Berg C, Eckhardt SG, Liu Z, et al. Single-cell RNA sequencing of lung adenocarcinoma reveals heterogeneity of immune response-related genes. JCI Insight. 2019;4:121387.
Zavidij O, Haradhvala NJ, Mouhieddine TH, Sklavenitis-Pistofidis R, Cai S, Reidy M, et al. Single-cell RNA sequencing reveals compromised immune microenvironment in precursor stages of multiple myeloma. Nat Cancer. 2020;1:493–506.
Lu T, Yang X, Shi Y, Zhao M, Bi G, Liang J, et al. Single-cell transcriptome atlas of lung adenocarcinoma featured with ground glass nodules. Cell Discov. 2020;6:69.
Olsen TK, Baryawno N. Introduction to Single-Cell RNA Sequencing. Curr Protoc Mol Biol. 2018;122:e57.
Hao Y, Hao S, Andersen-Nissen E, Mauck WM 3rd, Zheng S, et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–3587.e29.
Team RC. R: A language and environment for statistical computing. 2013.
Liaw A, Wiener M. Classification and regression by randomForest. R N. 2002;2:18–22.
Rousseaux S, Debernardi A, Jacquiau B, Vitte AL, Vesin A, Nagy-Mignotte H, et al. Ectopic activation of germline and placental genes identifies aggressive metastasis-prone lung cancers. Sci Transl Med. 2013;5:186ra66.
Mitchell KA, Zingone A, Toulabi L, Boeckelman J, Ryan BM. Comparative Transcriptome Profiling Reveals Coding and Noncoding RNA Differences in NSCLC from African Americans and European Americans. Clin Cancer Res. 2017;23:7412–25.
Landi MT, Dracheva T, Rotunno M, Figueroa JD, Liu H, Dasgupta A, et al. Gene expression signature of cigarette smoking and its role in lung adenocarcinoma development and survival. PLoS One. 2008;3:e1651.
Su LJ, Chang CW, Wu YC, Chen KC, Lin CJ, Liang SC, et al. Selection of DDX5 as a novel internal control for Q-RT-PCR from microarray data using a block bootstrap re-sampling scheme. BMC Genomics. 2007;8:140.
Lu TP, Tsai MH, Lee JM, Hsu CP, Chen PC, Lin CW, et al. Identification of a novel biomarker, SEMA5A, for non-small cell lung carcinoma in nonsmoking women. Cancer Epidemiol Biomark Prev. 2010;19:2590–7.
Sheng M, Xie X, Wang J, Gu W. A Pathway-Based Strategy to Identify Biomarkers for Lung Cancer Diagnosis and Prognosis. Evol Bioinform Online. 2019;15:1176934319838494.
Zhang BZ, Wang YD, Zhou XZ, Zhang Z, Ju HY, Diao XQ, et al. Construction of a Prognostic and Early Diagnosis Model for LUAD Based on Necroptosis Gene Signature and Exploration of Immunotherapy Potential. Cancers. 2022;14:5153.
Chen Q, Wang XY, Hu J. Systematically integrative analysis identifies diagnostic and prognostic candidates and small-molecule drugs for lung adenocarcinoma. Transl Cancer Res. 2021;10:3619–46.
Cai SH, Guo XT, Huang CJ, Deng YJ, Du LD, Liu WY, et al. Integrative analysis and experiments to explore angiogenesis regulators correlated with poor prognosis, immune infiltration and cancer progression in lung adenocarcinoma. J Transl Med. 2021;19:361.
Okayama H, Kohno T, Ishii Y, Shimada Y, Shiraishi K, Iwakawa R, et al. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res. 2012;72:100–11.
Tomida S, Takeuchi T, Shimada Y, Arima C, Matsuo K, Mitsudomi T, et al. Relapse-related molecular signature in lung adenocarcinomas identifies patients with dismal prognosis. J Clin Oncol. 2009;27:2793–9.
Schabath MB, Welsh EA, Fulp WJ, Chen L, Teer JK, Thompson ZJ, et al. Differential association of STK11 and TP53 with KRAS mutation-associated gene expression, proliferation and immune surveillance in lung adenocarcinoma. Oncogene. 2016;35:3209–16.
Therneau T. A package for survival analysis in S. R package version, 2015. 2.
Kassambara, A, Kosinski M, Biecek P, Fabian S. Survminer: Drawing Survival Curves Using Ggplot2. 2021. https://CRAN.R-project.org/package=survminer. R package version 0.4, 2021. 9.
Sturm G, Finotello F, List M. Immunedeconv: An R Package for Unified Access to Computational Methods for Estimating Immune Cell Fractions from Bulk RNA-Sequencing Data. Methods Mol Biol. 2020;2120:223–32.
Yoshihara K, Kim H, Verhaak R. estimate: Estimate of Stromal and Immune Cells in Malignant Tumor Tissues from Expression Data. R package version, 2016. 1: p. r21.
Fu J, Li K, Zhang W, Wan C, Zhang J, Jiang P, et al. Large-scale public data reuse to model immunotherapy response and resistance. Genome Med. 2020;12:21.
Jiang P, Gu S, Pan D, Fu J, Sahu A, Hu X, et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med. 2018;24:1550–8.
Wong KY, Cheung AH, Chen B, Chan WN, Yu J, Lo KW, et al. Cancer-associated fibroblasts in nonsmall cell lung cancer: From molecular mechanisms to clinical implications. Int J Cancer. 2022;151:1195–215.
Xiang H, Ramil CP, Hai J, Zhang C, Wang H, Watkins AA, et al. Cancer-Associated Fibroblasts Promote Immunosuppression by Inducing ROS-Generating Monocytic MDSCs in Lung Squamous Cell Carcinoma. Cancer Immunol Res. 2020;8:436–50.
Wang L, Cao L, Wang H, Liu B, Zhang Q, Meng Z, et al. Cancer-associated fibroblasts enhance metastatic potential of lung cancer cells through IL-6/STAT3 signaling pathway. Oncotarget. 2017;8:76116–28.
Scholl C, Frohling S, Dunn IF, Schinzel AC, Barbie DA, Kim SY, et al. Synthetic Lethal Interaction between Oncogenic KRAS Dependency and STK33 Suppression in Human Cancer Cells. Cell. 2009;137:821–34.
The Human Protein Atlas. 2022; Available from: http://www.proteinatlas.org.
Thul PJ, Akesson L, Wiking M, Mahdessian D, Geladaki A, Ait Blal H, et al. A subcellular map of the human proteome. Science. 2017;356:eaal3321.
Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419.
Wang T, Hao D, Yang S, Ma J, Yang W, Zhu Y, et al. miR-211 facilitates platinum chemosensitivity by blocking the DNA damage response (DDR) in ovarian cancer. Cell Death Dis. 2019;10:495.
Zhao DD, Zhao X, Li WT. Identification of differentially expressed metastatic genes and their signatures to predict the overall survival of uveal melanoma patients by bioinformatics analysis. Int J Ophthalmol. 2020;13:1046–53.
Zhang D, Park D, Zhong Y, Lu Y, Rycaj K, Gong S, et al. Stem cell and neurogenic gene-expression profiles link prostate basal cells to aggressive prostate cancer. Nat Commun. 2016;7:10798.
Wang YX, Marino-Enriquez A, Bennett RR, Zhu MJ, Shen YP, Eilers G, et al. Dystrophin is a tumor suppressor in human cancers with myogenic programs. Nat Genet. 2014;46:601–6.
Ding Z, Zu S, Gu J. Evaluating the molecule-based prediction of clinical drug responses in cancer. Bioinformatics. 2016;32:2891–5.
Acknowledgements
This work was funded by the National Key R&D Program of China (2022YFA1303200 to XS), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB29030104 to TJ), the National Natural Science Foundation of China (Grant 82272301 to TJ), and the Fundamental Research Funds for the Central Universities (WK9100000001 to TJ).
Author information
Authors and Affiliations
Contributions
Qingyu Cheng: Conceptualization, Data curation, Investigation, Visualization, Writing - original draft. Weidong Zhao: Writing - review & editing. Xiaoyuan Song: Project administration, Writing - review & editing. Tengchuan Jin: Conceptualization, Project administration, Supervision, Writing - review & editing.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Cheng, Q., Zhao, W., Song, X. et al. Machine-learning and scRNA-Seq-based diagnostic and prognostic models illustrating survival and therapy response of lung adenocarcinoma. Genes Immun 25, 356–366 (2024). https://doi.org/10.1038/s41435-024-00289-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41435-024-00289-0
This article is cited by
-
Single-cell expression and immune infiltration analysis of polyamine metabolism in breast cancer
Discover Oncology (2024)