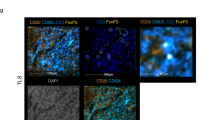
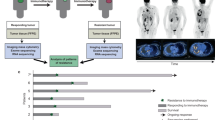
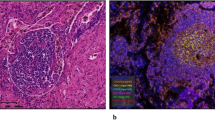
Abstract
Tumor-Infiltrating Lymphocytes (TILs) immunotherapy is a highly promising treatment for Non-small Cell Lung Cancer (NSCLC), which is responsible for 18% of all cancer-related deaths. The heterogeneity of TILs remains poorly understood. Here, we utilized combined single-cell RNA (scRNA)/T cell receptor sequencing (scTCR-seq) data from lung adenocarcinoma (LUAD) patients. Naïve CD4+ and effector memory CD8+ T cells were increased in tumor tissue compared with circulating blood samples. Activated signaling pathways were detected, and GZMA was identified as a potential novel diagnostic biomarker. During the transitional phase, macrophages (FTL) and dendritic (AIF1) cells transported the most CD3 TCR clones to T cells, while cytotoxicity CD8+ T (NKG7) cells transported to terminal exhausted CD8+ T cells. In both transition and expansion phases, T helper cells (CXCL13) are transported to regulatory T cells (Tregs). Additionally, we investigated the expression profiles of key cytokines, checkpoint receptors, and their ligands. Cytotoxicity CD8+ T cells (CCL5 and IFNG), T helper cells (FTL, TNFRSF4, and TIGIT), and regulatory T cells (CTLA4, TIGIT and FTL) exhibited functional roles in both primary and metastatic tumor stages. Taken together, our study provides a single-cell resolution of the TIL immune landscape and suggests potential treatment strategies to overcome drug resistance.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 digital issues and online access to articles
118,99 € per year
only 19,83 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
A comprehensive survey of the literature on next-generation sequencing (NGS) studies involving TILs in NSCLC identified three recent publications: 1. Gueguen et al., referred to as “scRNA-seq and scTCR-seq data of resident and circulating precursors to tumor-infiltrating CD8+ T cell populations in lung cancer” (GSE162498 and GSE162499) [18]. 2. Laughney et al., based on the “scRNA-seq transcriptional landscape of primary tumors and metastases human LUAD” (GSE123902) [39]. 3. Ganesan et al., referred to as “Analysis of purified populations of CD8 T cells (isolated from primary lung tumors and matched adjacent lung tissue of lung cancer patients) at the transcriptomic level by RNA sequencing” (GSE90728) [40].
References
Schabath MB, Cote ML. Cancer progress and priorities: lung cancer. Cancer Epidemiol Biomark Prev. 2019;28:1563–79.
Chen G, Hu J, Huang Z, Yang L, Chen M. MicroRNA-1976 functions as a tumor suppressor and serves as a prognostic indicator in non-small cell lung cancer by directly targeting PLCE1. Biochem Biophys Res Commun. 2016;473:1144–51.
Granhoj JS, Witness Praest, Jensen A, Presti M, Met O, Svane IM, et al. Tumor-infiltrating lymphocytes for adoptive cell therapy: recent advances, challenges, and future directions. Expert Opin Biol Ther. 2022;22:627–41.
Kasper HU, Drebber U, Stippel DL, Dienes HP, Gillessen A. Liver tumor infiltrating lymphocytes: comparison of hepatocellular and cholangiolar carcinoma. World J Gastroenterol. 2009;15:5053–7.
Chen J, He Q, Liu J, Xiao Y, Xiao C, Chen K, et al. CD8+ tumor-infiltrating lymphocytes as a novel prognostic biomarker in lung sarcomatoid carcinoma, a rare subtype of lung cancer. Cancer Manag Res. 2018;10:3505–11.
Lee SW, Choi HY, Lee GW, Kim T, Cho HJ, Oh IJ, et al. CD8(+) TILs in NSCLC differentiate into TEMRA via a bifurcated trajectory: deciphering immunogenicity of tumor antigens. J Immunother Cancer. 2021;9:1–14.
Altorki NK, Markowitz GJ, Gao D, Port JL, Saxena A, Stiles B, et al. The lung microenvironment: an important regulator of tumour growth and metastasis. Nat Rev Cancer. 2019;19:9–31.
Sanmamed MF, Nie X, Desai SS, Villaroel-Espindola F, Badri T, Zhao D, et al. A burned-out CD8(+) T-cell subset expands in the tumor microenvironment and curbs cancer immunotherapy. Cancer Discov. 2021;11:1700–15.
Hsu CL, Ou DL, Bai LY, Chen CW, Lin L, Huang SF, et al. Exploring markers of exhausted CD8 T cells to predict response to immune checkpoint inhibitor therapy for hepatocellular carcinoma. Liver Cancer. 2021;10:346–59.
Liu Y, Liang L, Ji L, Zhang F, Chen D, Duan S, et al. Potentiated lung adenocarcinoma (LUAD) cell growth, migration and invasion by lncRNA DARS-AS1 via miR-188-5p/ KLF12 axis. Aging. 2021;13:23376–92.
Gohil SH, Iorgulescu JB, Braun DA, Keskin DB, Livak KJ. Applying high-dimensional single-cell technologies to the analysis of cancer immunotherapy. Nat Rev Clin Oncol. 2021;18:244–56.
Yang HQ, Wang YS, Zhai K, Tong ZH. Single-cell TCR sequencing reveals the dynamics of T cell repertoire profiling during pneumocystis infection. Front Microbiol. 2021;12:637500.
Philip M, Schietinger A. CD8(+) T cell differentiation and dysfunction in cancer. Nat Rev Immunol. 2022;22:209–23.
Gulati GS, Sikandar SS, Wesche DJ, Manjunath A, Bharadwaj A, Berger MJ, et al. Single-cell transcriptional diversity is a hallmark of developmental potential. Science. 2020;367:405–11.
Kubli SP, Berger T, Araujo DV, Siu LL, Mak TW. Beyond immune checkpoint blockade: emerging immunological strategies. Nat Rev Drug Discov. 2021;20:899–919.
Long M, Lu S, Sun G. Kinetics of receptor-ligand interactions in immune responses. Cell Mol Immunol. 2006;3:79–86.
Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48:W509–W514.
Gueguen P, Metoikidou C, Dupic T, Lawand M, Goudot C, Baulande S. et al. Contribution of resident and circulating precursors to tumor-infiltrating CD8(+) T cell populations in lung cancer. Sci Immunol. 2021;6:1–15.
Essogmo FE, Zhilenkova A.V, Tchawe YSN, Owoicho AM, Rusanov A.S, Boroda A. et al. Cytokine profile in lung cancer patients: anti-tumor and oncogenic cytokines. Cancers. 2023;15:1–17.
Yeo AT, Rawal S, Delcuze B, Christofides A, Atayde A, Strauss L, et al. Single-cell RNA sequencing reveals evolution of immune landscape during glioblastoma progression. Nat Immunol. 2022;23:971–84.
Saito S, Nakashima A, Shima T, Ito M. Th1/Th2/Th17 and regulatory T-cell paradigm in pregnancy. Am J Reprod Immunol. 2010;63:601–10.
Kaiko GE, Horvat JC, Beagley KW, Hansbro PM. Immunological decision-making: how does the immune system decide to mount a helper T-cell response? Immunology. 2008;123:326–38.
Metkar SS, Menaa C, Pardo J, Wang B, Wallich R, Freudenberg M, et al. Human and mouse granzyme A induce a proinflammatory cytokine response. Immunity. 2008;29:720–33.
van den Boogaard FE, van Gisbergen KP, Vernooy JH, Medema JP, Roelofs JJ, van Zoelen MA, et al. Granzyme A impairs host defense during Streptococcus pneumoniae pneumonia. Am J Physiol Lung Cell Mol Physiol. 2016;311:L507–16.
Elizondo DM, Brandy NZ, da Silva RL, de Moura TR, Lipscomb MW. Allograft inflammatory factor-1 in myeloid cells drives autoimmunity in type 1 diabetes. JCI Insight. 2020;5:1–12.
Hu ZW, Chen L, Ma RQ, Wei FQ, Wen YH, Zeng XL, et al. Comprehensive analysis of ferritin subunits expression and positive correlations with tumor-associated macrophages and T regulatory cells infiltration in most solid tumors. Aging. 2021;13:11491–506.
Gu-Trantien C, Migliori E, Buisseret L, de Wind A, Brohee S, Garaud S. et al. CXCL13-producing TFH cells link immune suppression and adaptive memory in human breast cancer. JCI Insight. 2017;2:1–17.
Tang P, Shen X, Gao J, Zhang J, Feng Y, Zhang J, et al. Distinct characteristics of BTLA/HVEM axis expression on Tregs and its impact on the expansion and attributes of Tregs in patients with active pulmonary tuberculosis. Front Cell Infect Microbiol. 2024;14:1437207.
Wen T, Barham W, Li Y, Zhang H, Gicobi JK, Hirdler JB, et al. NKG7 is a T-cell-intrinsic therapeutic target for improving antitumor cytotoxicity and cancer immunotherapy. Cancer Immunol Res. 2022;10:162–81.
Zheng C, Zheng L, Yoo JK, Guo H, Zhang Y, Guo X, et al. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell. 2017;169:1342.1356.e16.
Sathe A, Ayala C, Bai X, Grimes SM, Lee B, Kin C, et al. GITR and TIGIT immunotherapy provokes divergent multicellular responses in the tumor microenvironment of gastrointestinal cancers. Genome Med. 2023;15:100.
Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102.
Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat Protoc. 2020;15:1484–506.
Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, et al. Comprehensive integration of single-cell data. Cell. 2019;177:1888.1902.e21.
Zhang X, Lan Y, Xu J, Quan F, Zhao E, Deng C, et al. CellMarker: a manually curated resource of cell markers in human and mouse. Nucleic Acids Res. 2019;47:D721–28.
Zhang Z, Wang ZX, Chen YX, Wu HX, Yin L, Zhao Q, et al. Integrated analysis of single-cell and bulk RNA sequencing data reveals a pan-cancer stemness signature predicting immunotherapy response. Genome Med. 2022;14:45.
Cao J, Spielmann M, Qiu X, Huang X, Ibrahim DM, Hill AJ, et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019;566:496–502.
Borcherding N, Bormann NL, Kraus G. scRepertoire: an R-based toolkit for single-cell immune receptor analysis. F1000Res. 2020;9:47
Laughney AM, Hu J, Campbell NR, Bakhoum SF, Setty M, Lavallee VP, et al. Regenerative lineages and immune-mediated pruning in lung cancer metastasis. Nat Med. 2020;26:259–69.
Ganesan AP, Clarke J, Wood O, Garrido-Martin EM, Chee SJ, Mellows T, et al. Tissue-resident memory features are linked to the magnitude of cytotoxic T cell responses in human lung cancer. Nat Immunol. 2017;18:940–50.
Acknowledgements
We thank everyone who contributed to this study and the GEO database for the analytical data.
Author information
Authors and Affiliations
Contributions
JGL analyzed scRNA-seq data. LY analyzed scTCR-seq data. XLG performed statistical analysis. XMH performed ligand–receptor analysis. ML conducted trajectory analysis. RYG validated data. BHZ performed Kaplan–Meier analysis. QYL generated GraphPad plots. WJZ prepared figures. PX annotated cell types. XHG analyzed correlations. YAC prepared T cell figures. XLY drafted the manuscript. YS approved the final version. ZHG provided immunological expertise. JHM edited the language. YXH improved image resolution. LML assisted cancer immunology. JH supported funding. HZ conceived the project. YL supervised the study. All authors reviewed and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
This study did not require ethical approval as it did not involve human participants, human data, or animal subjects. All analyses were based on publicly available, anonymized datasets (e.g., UALCAN, GEO, GEPIA, Kaplan–Meier) that had obtained appropriate ethical clearance prior to release and are openly accessible to the research community. Therefore, no institutional ethics review was necessary.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
41435_2025_330_MOESM1_ESM.pdf
Supplementary Material for Characterizing the Immune Landscape of Tumor-Infiltrating Lymphocytes in Non-small Cell Lung Cancer
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, JG., Yu, L., Guo, XL. et al. Characterizing the immune landscape of tumor-infiltrating lymphocytes in non-small cell lung cancer. Genes Immun 26, 229–241 (2025). https://doi.org/10.1038/s41435-025-00330-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41435-025-00330-w