Abstract
Spatial patterns of genetic variation compared across species provide information about the predictability of genetic diversity in natural populations, and areas requiring conservation measures. Due to their remarkable fish diversity, rivers in Neotropical regions are ideal systems to confront theory with observations and would benefit greatly from such approaches given their increasing vulnerability to anthropogenic pressures. We used SNP data from 18 fish species with contrasting life-history traits, co-sampled across 12 sites in the Maroni– a major river system from the Guiana Shield –, to compare patterns of intraspecific genetic variation and identify their underlying drivers. Analyses of covariance revealed a decrease in genetic diversity as distance from the river outlet increased for 5 of the 18 species, illustrating a pattern commonly observed in riverscapes for species with low-to-medium dispersal abilities. However, the mean within-site genetic diversity was lowest in the two easternmost tributaries of the Upper Maroni and around an urbanized ___location downstream, indicating the need to address the potential influence of local pressures in these areas, such as gold mining or fishing. Finally, the relative influence of isolation by stream distance, isolation by discontinuous river flow, and isolation by spatial heterogeneity in effective size on pairwise genetic differentiation varied across species. Species with similar dispersal and reproductive guilds did not necessarily display shared patterns of population structure. Increasing the knowledge of specific life history traits and ecological requirements of fish species in these remote areas should help further understand factors that influence their current patterns of genetic variation.
Similar content being viewed by others
Introduction
Genetic diversity has long been acknowledged as an essential aspect of biodiversity (Pereira et al. 2013) and a critical support to the resilience of individuals (Dobzhansky 1965), populations (Frankham 1995), and communities (Crutsinger 2016). A loss of genetic diversity could have irreversible impacts on populations (Allendorf et al. 2008) and genetic monitoring is crucial for managing them. Despite analytical and technical issues challenging the integration of genetic information into conservation practices (Hoban et al. 2021), researchers are pursuing efforts to understand the evolution of molecular data for a variety of taxa. Collecting genetic information at several levels of organization (populations, species, communities) can help to understand evolutionary processes that occur in the wild (Vellend and Geber 2005). Multispecies databases are increasingly developed to georeference new molecular markers or samples collected at various spatio-temporal scales (Deck et al. 2017). Comparative population genetics is an emerging field with promising applications for conservation (Pauls et al. 2014; Leigh et al. 2021).
Comparing the evolutionary trajectories between species (comparative phylogeography) can reveal long-term processes that influenced their current patterns of genetic variation (Jenkins et al. 2018). Similarly, comparing patterns of genetic variation across species that live in sympatry (or share landscape features) has provided insights into the landscape-related factors that influence them (DeWoody and Avise 2000; Paz-Vinas et al. 2015; Selkoe et al. 2016). For instance, riverscape topography is known to influence patterns of genetic diversity strongly in fish populations (Prunier et al. 2017a). Indeed, river networks typically display a fractal, “dendritic” structure (dendritic ecological network DEN; Campbell Grant et al. 2007) where primary streams and confluences show higher connectivity within the network than peripheral areas like headwaters. This causes a heterogeneous spatial repartition of genetic diversity in relation to the network complexity because genetic diversity tends to accumulate within highly-connected areas at migration-drift equilibrium (Paz-Vinas and Blanchet 2015; Thomaz et al. 2016). Furthermore, empirical observations from strictly freshwater species frequently reported higher local genetic diversity in downstream, highly-connected areas than in upstream headwaters (Chaput-Bardy et al. 2009; Paz-Vinas et al. 2015).
Subsidiarily to the network properties per se, downstream-biased migration, greater downstream habitat availability, or upstream-directed colonization with successive founder effects were suggested as potential evolutionary processes generating such pattern of a downstream increase in genetic diversity (DIGD) (Paz-Vinas et al. 2015). Species with low-to-medium dispersal abilities are also more likely to be influenced by network properties and to show DIGD (Paz-Vinas and Blanchet 2015), since gene flow is restricted further by the topography of the river network.
However, many biological or environmental factors can also modify this general, expected pattern of genetic variability, leading to idiosyncratic patterns that challenge the predictability of spatial genetic variation in natural populations. Notably, particular non-equilibrium processes, specific colonization routes, or environmental constraints that decrease effective population size (Ne) locally can alter expected spatial patterns such as DIGD (Salisbury et al. 2016; Pilger et al. 2017; Richmond et al. 2018). Interactions among local (e.g., water quality) or regional (e.g., DEN properties) riverscape features, species’ life history traits and their evolutionary trajectories are complex (Pilger et al. 2017; Paz-Vinas et al. 2018). Their relative influence on patterns of genetic variation is not completely understood (Ruzzante et al. 2019; Blanchet et al. 2020), and it remains unclear how diverse these patterns might actually be across a range of co-distributed species belonging to the same clade.
Several meta-analysis studies have compared patterns of spatial genetic variation between species at large geographical scales, aggregating previously published datasets (e.g., DeWoody and Avise 2000; Paz-Vinas et al. 2015; Lino et al. 2018). These approaches provide a dense multi-taxa overview of drivers of genetic variability in the wild (Leigh et al. 2021). Another more-recently widening approach is based on the simultaneous sampling of several co-distributed species. This co-sampling can be used to directly compare species with a variety of ecological characteristics (Selkoe et al. 2014; Leigh et al. 2021) from a common environment. Overall, comparative approaches can identify common patterns of genetic variation shared by most species or, conversely, reveal species showing atypical patterns. Comparing patterns of genetic variation among co-distributed species can also reveal spatial hot- or coldspots of genetic diversity (Paz-Vinas et al. 2018) and provide information about species’ vulnerabilities to localized anthropogenic pressures (Blanchet et al. 2010), with direct applications for management policies. However, because sampling multiple species simultaneously is often logistically and technically burdensome, most co-sampling studies to-date have included a limited number of species (<10). The lack of available molecular markers also hinders collection of multispecies datasets (Horne 2014).
Neotropical freshwater habitats harbor a high specific and functional ichthyodiversity (Abell et al. 2008), with more than 5600 species described to-date. Coastal rivers in the Guiana Shield are smaller than in other Neotropical regions, yet contain similar habitats and exceptional ichthyodiversity. The Maroni is one of its most speciose basins, with 264 strictly freshwater fish species, of which 16.9% are considered endemic (Le Bail et al. 2012; see also Papa et al. 2021 suggesting 25 additional cryptic species using molecular data). This unique biodiversity heritage provides cultural and commercial resources for an expanding community of native people (Longin et al. 2021a;b). However, its intraspecific facet remains poorly documented (Hilsdorf and Hallerman 2017) and genetic diversity and conservation status of most species remain unknown, even though these ecosystems are experiencing increasing anthropogenic pressures (Reis et al. 2016; Longin et al. 2021b).
Here we used genotypic data obtained for 18 native, co-distributed fish species in the Upper Maroni (French Guiana) during a conservation program led by the Parc Amazonien de Guyane in collaboration with local fishermen. We aimed at examining and comparing spatial patterns of genetic variation across species in order to reveal areas of potential conservation interest (i.e., hot- or coldspots of genetic diversity) across the Upper Maroni, to evaluate if genetic structure could be explained by factors such as riverscape distance, to identify common or idiosyncratic patterns of spatial genetic variation across species in relationship with their known biological characteristics and, overall, to provide a reference snapshot for local conservation practitioners. We focused on species with a variety of biological characteristics and contrasting dispersal abilities, sampled across the main river network of the Upper Maroni. Firstly, we examined how local genetic diversity varied across our study zone. We tested whether a repeatable pattern of DIGD exists among the species sampled. We considered that a species displayed DIGD when local genetic diversity was higher (and local genetic isolation lower) in downstream areas than in headwaters, and we expected this pattern to be more distinct for brood hider or guarder species with little active or passive dispersal of eggs (both of which suggest low dispersal ability). We also examined how genetic diversity varied spatially on average, i.e., using species as independent observations within each site. Secondly, we examined the relative influence of isolation by stream distance, isolation by discontinuous river flow and isolation by spatial heterogeneity in effective size on genetic structure across species. We evaluated whether species with similar life history traits displayed comparable patterns of spatial genetic variation. We also unfolded a preliminary outline of evolutionary processes that may generate DIGD for the species concerned.
Methods
Notation and definitions for statistical indices used in this study are provided in Table 1.
Study zone and model species
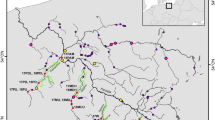
The Maroni is the largest river in French Guiana (>68,700 km²). With the river Mana sharing the same outlet, this biogeographical subunit of the Guianas ecoregion harbors a particular ichthyofauna which marks a transition between an extended Proto-Berbice ecoregion (Lujan and Armbruster 2011), with strong influence from the Amazon, in the west, and other coastal rivers in French Guiana in the east (Lemopoulos and Covain 2019). Our study zone comprises a portion of the main channel (Lawa) flowing above the Abattis Cottica, and extends upstream towards four main tributaries, from west to east: the Litani, Marouini, Tampok, and Waki (Fig. 1). Sampling sites (Fig. 1, Table 2) were located in large and open river channels with Strahler index > 4. Five sites were located near the Lawa and the urbanized area of Maripasoula (La.0, La.1a, La.1b, Li.1, and Ta.1), while seven sites were located in more remote regions along secondary streams (Li.2, Li.3, Ma.2, Ma.3, Ta.2, Ta.3, and Wa.3). Although sites La.1b and Li.1 lay close to each other in Euclidian distance, they are relatively isolated from each other by steep rapids. Nevertheless, there exist no natural nor artificial barriers across our study zone and flowing is considered continuous, even though all river sections may be subject to seasonal variations in depth and width.
Each sampling site is inner-colored according to the mean (i.e., across species) of internal diversity (ID, which increases as local genetic diversity decreases) and outer-colored according to the mean of the standardized local genetic isolation (STFST.P). The blue-to-red gradient indicates low to high values of ID and STFST.P, i.e., decreasing local genetic diversity and increasing local genetic isolation.
The 18 focal fish species are ubiquitous across the study zone and were selected because they could potentially be found across all sites. They have different life history traits according to their reproductive guild and their migratory behavior as adults (Table S1.1 from Appendix 1). Six species are broadcast spawners and long-distance migrants; they release a large number of eggs into the water column; they can move and reproduce across the entire study zone during the rainy season. Seven species are brood hiders and short-distance migrants; they fertilize and lay their eggs on the substrate; they usually only migrate along secondary streams to reproduce, mostly in the early rainy season. Finally, five species are brood guarders that provide parental care to their progeny; they are usually linked to particular and restricted habitats and display very limited dispersal compared to the other species. All species have overlapping generations and a generation time longer than 1 year. Beyond these categories, five of the 18 species are endemic to the Maroni River and its two nearest adjacent basins (Le Bail et al. 2012).
Genetic data and analyses
We used the genotypic data of Delord et al. (2018) for all 18 species. Here, we summarize essential information about field sampling and the development of molecular markers. Additional details are provided in the Supplementary note associated to Table S1.1 and Fig. S1.1. Fish were sampled from late 2013 to early 2016 during a program led by the Parc Amazonien de Guyane, using mainly trammel and cast nets and mostly during the dry season (July–December). SNP markers were developed for each species independently, using the protocol described by Delord et al. (2018): for each species, a subsample of ∼20 individuals was selected across all sampling sites to build a SNP-discovery set. Their DNA extracts were mixed into a single library for pooled-RAD sequencing. Next, sequencing reads were analyzed using a custom bioinformatics pipeline (RAD4SNPs) designed to select polymorphic sites without individual nor population information, but mainly on the basis of depth coverage and the quality of flanking sequences to enable the design of amplification primers for future individual genotyping. Finally, 111 to 157 neutral, biallelic, nuclear SNP markers were scored for each species (full marker data available from the Dryad repository: https://doi.org/10.5061/dryad.2b6b43k). For one species (Cynodon meionactis), the dataset was supplemented with 31 SNPs available from the same protocol. During sample collection, the ideal sample size was set at 20 specimens per species and per site, which was sometimes missed due to a variety of field constraints. Ultimately, 145–283 individuals were retained per species, with each species sampled from at least 9 sites (Table S1.1), and each site represented by at least 12 species in the final dataset (Table 2). Sample sizes per species and site in this study are provided in Table 3.
Describing DIGD and riverscape patterns of genetic variation across species
We based our statistical analyses on the twelve sampling sites with species representing independent replicates of these sites, irrespective of their overall genetic structure. For each species and each site, we first computed several indices of local genetic diversity: the polymorphism rate (P95) (proportion of loci with minor allele frequency ≥ 0.05) using a modified rarefactor function from the R package DIVERSITY (Keenan et al. 2013) with R version 4.0.4 (Core Team 2021); expected (HS) and observed (HO) heterozygosity using the R package HIERFSTAT (Goudet and Jombart 2017). For each species, we assessed the overall genetic structure using the fixation index FST (Weir and Cockerham 1984) and its significance using the G-test in HIERFSTAT. We also calculated pairwise FST values between sites and computed their confidence intervals using 999 bootstraps. Then, we calculated a site-specific FST (FST.P) for each sampling site by averaging its pairwise FST values with all other sampling sites to assess local genetic isolation, or “uniqueness” (Pflüger and Balkenhol 2014; Paz-Vinas et al. 2015). Because none of the species displayed locally private alleles (i.e., present at only one site), indices developed for them were not considered.
To observe the upstream reduction in local genetic diversity expected for species under the DIGD pattern, we calculated (for each species) the Pearson coefficient of correlation between the ___location of each site along the downstream-to-upstream gradient (upstream distance) and each of the three site-specific genetic indices computed previously (P95, HS, and FST.P). Those coefficients will be hereafter referred as CORP, CORHS, and CORFST.P, respectively. For species that display DIGD, we expected that P95 and HS (local genetic diversity) would decrease (leading to negative coefficients CORP and CORHS), but FST.P (local genetic isolation) would increase (leading to positive coefficient CORFST.P), as upstream distance increased. We defined the upstream distance of each site as the distance to the town of Grand Santi (in km; Fig. 1, Table 2), which lays at the downstream edge of our study zone. All riverscape distances were obtained from the national GIS database IGN BD TOPO® Hydrographic Network, version 2.2. We also performed three linear regression models with P95, HS, and FST.P as response variables, respectively, to see how the relationship between local genetic diversity (or isolation) and upstream distance varied across species and to test for generalization of the DIGD pattern. To this end, we set species as a categorical fixed-effect factor and upstream distance as a continuous fixed-effect factor in each linear regression model (analyses of covariance performed using the R function lm). From each model, we extracted the estimated marginal mean (EMM) per species using the R package EMMEANS (Lenth 2018). EMM, along with their confidence intervals, enable to compare the trend (and associated uncertainty) of the relationship between the response variable (P95, HS, or FST.P) and upstream distance between species.
To provide an overall, multispecies vision of genetic variation across the study zone, we calculated two additional genetic diversity indices for each species at each site. Our objective was to assess whether certain sites displayed remarkable patterns of diversity or isolation and to reveal hot- and coldspots of diversity. First, for each species, we calculated the local contribution of each site to the species overall genetic variation across the study zone. For that, we used an estimate of local “internal diversity” (hereafter referred as ID, Caballero and Toro 2002) calculated using MOLKIN software, version 2.0 (Gutiérrez et al. 2005). ID, calculated for a given site, illustrates the direction and magnitude of the change in overall genetic diversity if this site was excluded from the full species dataset. If negative, ID indicates that overall diversity would decrease if this site was absent from the full species dataset, implying it has a high contribution to overall diversity. Conversely, if positive, ID indicates that overall diversity would increase if this site was absent from the full species dataset, implying it has a low contribution to overall genetic diversity. In addition, the absolute value of ID indicates its magnitude. Expressed as percentages, ID values were used to compare each site’s contribution to overall diversity, across species. Second, for each species, we standardized all FST.P values (hereafter referred as STFST.P) between 0 (lowest FST.P) and 1 (highest FST.P) to identify sites frequently associated to species lowest or highest value of local genetic isolation.
Comparing patterns of spatial genetic variation across species
To better understand drivers of spatial structure across species, we assessed the relative influence of stream distance (in km), connectivity by river flow, and spatial heterogeneity in effective size on pairwise genetic differentiation (linearized pairwise FST = FST/(1-FST)) using multiple linear regression on distance matrices (MRM) (Paz-Vinas et al. 2015; Pilger et al. 2017).
For each species, the response matrix contained linearized pairwise FST values, while three independent matrices were used as explanatory factors. The first matrix contained riverscape distances (in km) between each pair of sites to test for isolation by stream distance. The second matrix described whether each pair of sites was connected by continuous, unidirectional water flowing (value of 0) or not (value of 1) (Paz-Vinas et al. 2015) to test for isolation by discontinuous river flow.
The third matrix contained the difference in local effective population size (Ne) between each pair of sites. Spatial heterogeneity in effective size was identified as a potential driver of between-site genetic structure even in the presence of gene flow (Prunier et al. 2017b). Thus, for each species, we calculated the matrix of pairwise Ne values as the harmonic mean of local Ne point estimates for all pairs of sites (Eq. (3) of Prunier et al. 2017b). The harmonic mean weights smaller values more heavily, and highlights the contribution of local genetic drift on pairwise genetic variation that is expected when one or both local Ne estimates are small. We estimated current local Ne for each species at each site independently, using the linkage-disequilibrium method implemented within the NEESTIMATOR software version 2.1 (Do et al. 2014), setting the minimum allele frequency to 0.05. Estimates from this method need to be interpreted with caution when it is applied on samples drawn from populations with overlapping generations or potential admixture with immigrant individuals over recent generations, as both could influence patterns of linkage-disequilibrium (Waples and England 2011; Waples et al. 2014). Biases may also occur when sample size is low, in which case estimates may be arbitrarily high, infinite and associated to wide confidence intervals. Thus, we did not focus on the quantitative estimates of Ne except when these were strikingly small and associated to finite confidence intervals (see Results section). Instead, we were interested in whether pairwise genetic variation could be explained by steepness of Ne variations between sites. When actual Ne point estimates were higher than 500 (or infinite), we arbitrarily set them to 500 before building our matrix of pairwise Ne values for the MRM analyses. 500 was chosen as a fairly typical value, commonly considered as a relatively large effective size (Franklin 1980). This enabled to avoid statistical artifacts introduced by punctually very high and possibly unreliable Ne estimates, and rather focus the MRM on differences between smaller Ne estimates. We also ran MRM analyses using the lower bound of the confidence intervals of Ne instead of point estimates, and we also ran MRM analyses while keeping all “outlier” (i.e., larger than 500) Ne estimates and replacing infinite estimates by the mean Ne calculated across all sites for the species.
From each of the 18 MRM analyses, we extracted standardized beta-coefficients, which described the effect of each explanatory factor on pairwise genetic differentiation: isolation by stream distance (CORIBD), isolation by discontinuous river flow (CORIBF), and isolation by spatial heterogeneity in effective size (CORNE).
We performed a multivariate approach, including all 18 species and the previously computed indices of spatial genetic variation, to visualize potential clusters of species displaying similar patterns and evaluate how species with similar life history traits would position one from each other (Selkoe et al. 2014). We used estimates of overall genetic structure (FST), descriptors of DIGD (CORP, CORHS, and CORFSTP) and factors explaining pairwise genetic variation (CORIBD, CORIBF, and CORNE) as continuous variables within a principal component analysis (PCA) with species as statistical individuals, using the R package FACTOMINER (Lê et al. 2008).
Inferring evolutionary processes that influence DIGD across species
For species that displayed DIGD (negative CORP and/or CORHS, positive CORFST.P and relatively high overall FST), we used the simulation-based results obtained by Paz-Vinas et al. (2015) to obtain preliminary information about which of several evolutionary processes (or combination of processes) might generate it. Paz-Vinas et al. (2015) simulated genotypic data for subpopulations evenly distributed across a dendritic ecological network (DEN). These data were generated under seven evolutionary scenarios all likely to generate DIGD, but with distinct genetic signatures as provided by a small set of easily computable summary statistics. Those seven scenarios are asymmetrical, downstream-biased migration (ASYM), upstream-biased colonization (COLON), higher downstream habitat availability (DIFFNE) or any combinations of two or all processes. Paz-Vinas et al. (2015) also generated data under a null model (NULL) in which none of the three processes applied, indicating that global genetic variation results only from the dendritic structure of the river network itself. Data were simulated under a wide range of local effective sizes (50 to 10,000), migration rates (0.001 to 0.30) and other demographic parameters for each scenario. The summary statistics obtained from those simulations could be used under an ABC model-choice procedure (Approximate bayesian computation, Csilléry et al. 2010), in order to determine which evolutionary scenario would most likely explain the DIGD pattern observed from an empirical dataset. Paz-Vinas et al. (2015) applied this framework to several previously published datasets in order to outline which processes might most frequently underlie DIGD in riverine species. This approach has to be led very cautiously, as it relies on a generic set of genotype data simulated under a DEN with “generic” properties (symmetric fixed branching, fixed number of sampled subpopulations) and compared to empirical datasets, which do not necessarily fully match those properties. Nevertheless, this framework provides a valuable baseline to understand the various processes that may exist in riverscapes along with their specific genetic signatures (Blanchet et al. 2020). More importantly, it may be of particular interest when applied on a set of co-sampled and co-distributed species, where direct comparisons might be even more informative because of the common sampling scheme.
Thus, we applied the approach developed by Paz-Vinas et al. (2015) to the five species that displayed DIGD (see Results), using the ABCRF method (Pudlo et al. 2016) from the eponymous R package, which performs model selection based on a random forest of classification and regression trees (Breiman 2001) instead of classical ABC algorithms. As a reference table, we used values of eight summary statistics, provided from the generic simulated data from Paz-Vinas et al. (2015) under all evolutionary processes (https://doi.org/10.5061/dryad.38c1g). Those summary statistics were overall fixation indices FIS, FST, and FIT; CORAR (analogous to our CORP), CORHS, CORFST.P, CORIBD, and CORIBF. Thus, for the species that displayed DIGD, we used overall FIS, FST, and FIT, CORP, CORHS, CORFST.P, CORIBD-PAZ, and CORIBF-PAZ computed from our own data as observed summary statistics (Table S1.4) for model selection using the ABCRF method. CORIBD-PAZ and CORIBF-PAZ were calculated in the same way as CORIBD and CORIBF from our original MRM analysis, but this time without including our matrix of spatial heterogeneity in effective size, since Paz-Vinas et al. (2015) did not consider this factor or pairwise genetic differentiation within their model selection framework.
Using their full set of 10 summary statistics, Paz-Vinas et al. (2015) obtained an “out-of-bag” error rate (percentage of sets of simulated summary statistics that were not correctly assigned to the evolutionary scenario under which they were generated) of 11.9%. In our case, we obtained an “out-of-bag” error rate of 14.3% using eight summary statistics and generating 500 classification trees. Albeit this value may seem fairly high, it remains comparable to figures reported from other ABCRF applications (e.g., Rougemont et al. 2016; Fraimout et al. 2017) and so was considered acceptable to apply the method, albeit with caution. The ABCRF model-choice procedure enabled us to identify which evolutionary scenario might best fit the set of summary statistics observed for each species that displayed DIGD, with the associated posterior probabilities. Additional information about the ABCRF framework, its application and potential limitations to discriminate between processes underlying DIGD in our case is provided in the Supplementary note associated to Table S1.4.
Results
All genetic indices calculated for each species and sampling site are available in Appendix 2. P95 and HS varied among sites and species, from 0.697 to 0.999 and from 0.257 to 0.436, respectively. Local FST.P ranged from −0.001 to 0.072. Overall FST, calculated for each species, ranged from 0.000 to 0.038; they were significant for eleven species (Table 4). Pairwise FST ranged from 0.000 to 0.099 across all 18 species (Tables S1.2.1–S1.2.18). Significant values often revealed stronger genetic differentiation between the eastern (Tampok and Waki rivers) and the rest of our study zone, converging with results from individual-based genetic clustering methods such as DAPC (Jombart et al. 2010) or STRUCTURE (Pritchard et al. 2000) (not shown). Additionally, significant values often (but not always) involved sites located in upstream reaches of the Upper Maroni.
DIGD and spatial contrasts in local genetic diversity
CORP, calculated for each species, ranged from −0.447 to 0.584 (Table 4). CORHS ranged from −0.485 to 0.557, while CORFST.P had more and greater positive values that ranged from −0.326 to 0.752, indicating that local genetic isolation (FST.P) showed a stronger relationship with upstream distance across species. Analyses of covariance showed large and significant differences in local values of P95, HS, and FST.P across species. For P95 and HS, the interaction between upstream distance and species was non-significant, and species explained 88 and 96% of overall variation, respectively (Table S1.3). Upstream distance had no significant effect overall on P95 or HS across the 18 species. In contrast, for FST.P, the interaction between upstream distance and species was significant and explained 5.0% of overall variation in FST.P (Table S1.3). EMM values (mean and confidence intervals) illustrated the contrasting covariation of upstream distance with P95, HS, and FST.P values across species (Fig. 2). For four species (Tometes lebaili, Serrasalmus eigenmanni, Pseudancistrus barbatus and Geophagus harreri) P95 and HS both tended to decrease, and FST.P to increase as upstream distance increased, with substantial genetic structure (overall FST > 0.010). In addition, Myloplus rhomboidalis showed no trend with upstream distance for HS, but a decreasing P95, increasing FST.P and overall FST = 0.008. We considered that these five species displayed DIGD.
Significant EMM (i.e., CIs not including zero) are shown in black. All values on the x-axis were multiplied by 103 to increase intelligibility. Species labels indicate their class of life history traits with broadcast spawners, long-distance migrants in white, brood hiders, short-distance migrants in gray and guarders, sedentary species in black.
ID, calculated for each sampling site for each species, ranged from −0.690 to 0.750 (Fig. S1.2 from Appendix 1). Mean ID and STFST.P values (i.e., averaged across species for each sampling site) revealed a distinct difference between the eastern and western areas of the study zone (Fig. 1). Sites in the Tampok and the Waki tributaries (Ta.1, Ta.2, Ta.3, and Wa.3) in the eastern area displayed higher mean ID and STFST.P (lower local genetic diversity and higher local genetic isolation) compared with sites in the Litani and Marouini tributaries (Li.1, Li.2, Li.3, Ma.2, and Ma.3) in the western area (Fig. 1 and S1.2). In both eastern and western areas, however, mean ID and STFST.P were lower (higher local genetic diversity and lower local genetic isolation) near confluences with the Lawa than within upstream sites. On the Lawa, two sites (La.1a and La.1b) with intermediate position had the lowest mean ID and STFST.P while the most downstream site of our study zone (La.0) had a higher mean ID and lower mean STFST.P.
Shared and idiosyncratic patterns of genetic structure across species
Estimates of Ne for each species and sampling site ranged from 7 to infinite (Table S1.5). Ninety-eight Ne estimates (of 185) were set to 500 for the MRM analyses (Appendix 2), comprising 41 Ne estimates higher than 500 and 57 infinite estimates. Expectedly, MRM analyses revealed no significant isolation by stream distance, discontinuous river flow (i.e., positive CORIBF) or spatial heterogeneity in effective size for any broadcast spawner, long-distance migrant species. By contrast, seven short-distance migrant or sedentary species showed significant isolation by stream distance with CORIBD ranging from 0.345 to 0.826 and seven short-distance migrant or sedentary species showed significant isolation by discontinuous river flow with CORIBF ranging from 0.236 to 0.545 (Table 4). Only two species (M. rhomboidalis and T. lebaili) displayed both significant CORIBD and CORIBF. S. eigenmanni displayed the highest CORIBF (=0.545) in accordance with a particularly strong genetic break (according to pairwise FST values, Table S1.2.11) between the eastern tributaries (Tampok and Waki) and the rest of the study zone. Finally, spatial heterogeneity in effective size had a significant influence on pairwise genetic differentiation for four species, along with isolation by stream distance (Hoplias aimara; CORNE = 0.616), isolation by discontinuous river flow (G. harreri and Serrasalmus rhombeus; CORNE = 0.398 and 0.365, respectively), or both (T. lebaili; CORNE = 0.378). MRM results for alternative handling of outlier and infinite Ne estimates are provided in Table S1.6 and did not show any striking difference with the current analyses (See also Fig. S1.4).
The first two components of the PCA explained more than 66% of the between-species variability (Fig. 3). The first PCA axis opposed all broadcast spawners, long-distance migrant (but also a few short-distance migrant and one sedentary) species with lower overall FST and higher CORP and CORHS (left half of the PCA biplot) to other short-distance migrant and sedentary species displaying higher overall FST, higher CORIBF, and lower (including negative) CORP and CORHS (right half of the biplot), such as the five species that displayed DIGD. The second PCA axis opposed species with higher CORIBD to species with higher CORNE.
Variables are global FST values, the strength of isolation by stream distance (CORIBD), by discontinuous river flow (CORIBF), by spatial heterogeneity in effective size (CORNE) extracted from multiple linear regression on distance matrices (MRM) analyses, and values of CORP, CORHS, and CORFST.P which correspond to Pearson correlation coefficients between upstream distance and local polymorphism rate (P95), expected heterozygosity (HS), and local genetic isolation (FST.P), respectively.
Outline of the evolutionary processes influencing DIGD
For four of the five species that displayed DIGD, COLONDIFFNE (i.e., a combination of upstream-biased colonization and higher downstream habitat availability) was the most frequently assigned evolutionary scenario according to the ABCRF model-choice procedure (Table 5), indicating that DIGD may result from a combination of higher downstream habitat availability (DIFFNE) and upstream-directed colonization (COLON). For one species (P. barbatus), COLON was the most frequently assigned process, indicating that DIGD may result from upstream-directed colonization only. However, posterior probabilities associated with each selected model were low, ranging from 0.554 to 0.779 (Table 5). Indeed, for all five species, a non-negligible number of regression trees assigned the empirical statistics to other evolutionary scenarios, usually COLON or DIFFNE alone, or a combination of both for P. barbatus. For one species (S. eigenmanni), the selected model (COLONDIFFNE) was closely followed by one that combined all three evolutionary processes: DIFFNE, COLON, and ASYM (asymmetrical, downstream-biased migration).
Discussion
Comparative approaches are increasingly contributing to the field of conservation genetics. Studying a large number of species should be possible through close collaborations with biodiversity managers in the field and the development of efficient collecting and genotyping tools (Hoban et al. 2013), including the use of high-throughput-sequencing to develop molecular resources for non-model species (Lepais et al. 2020). Here, we used SNP data from 18 co-sampled Neotropical fish species at 12 sites in the Upper Maroni to describe riverscape genetic variability across species in this remote (but increasingly inhabited by people) area of high ichthyodiversity. Currently, few studies focus on comparative multispecies population genetics, especially in Neotropical areas (Leigh et al. 2021) and, to our knowledge, few comparative studies have included as many species and samples collected in the same area (but see Selkoe et al. 2016 for an example of combining meta-analysis and co-sampling; and Zbinden et al. 2022 for a recent, extensive co-sampling application).
We observed DIGD for a few species displaying moderate or low dispersal abilities. This pattern, previously documented in temperate freshwater habitats (Blanchet et al. 2020), may result from increasing habitat availability downstream and upstream-directed colonization, according to the ABCRF analysis. Nonetheless, spatial comparison of local genetic indices (per species and averaged across all species) also highlighted contrasts among sites, some of which were genetic hot- or coldspots regardless of their upstream/downstream position along the main channel or tributaries of the Upper Maroni. Moreover, riverscape patterns of genetic variation and the influencing factors varied across species. Some species had contrasting patterns of genetic structure despite having similar ecological characteristics and life history traits. This coexistence of shared and idiosyncratic patterns of genetic variation will constitute the basis of the discussion.
Occurrences of DIGD in the Upper Maroni and their underlying processes
For all six species documented as long-distance migrants and broadcast spawners, we were not able to reject panmixia based on our dataset nor to detect any presence of DIGD. In contrast, we identified significant genetic structure and DIGD in five species, which were all short-distance migrants or sedentary species with medium-to-low fecundity. This supports previous observations that the dendritic structure of river networks has more influence on genetic variation for species with low dispersal ability and/or small Ne (Chaput-Bardy et al. 2009; Paz-Vinas and Blanchet 2015; Zbinden et al. 2022). Nevertheless, the other seven short-distance migrant, sedentary, brood-hider, or guarder species did not display this particular riverscape pattern, which ended up underrepresented in our dataset (i.e., DIGD being detected in only 5 of 18 species). Based on our results, DIGD thus did not seem to be a prevalent pattern of genetic variation in the Upper Maroni -although we must recall that, in spite of the consequent number of species studied here, it still represents an infinitesimal proportion of the significant ichthyodiversity of the region. Instead, there seem to exist constrated patterns of genetic variation across species, as discussed later.
The ABCRF approach usually identified a combination of higher downstream habitat availability (DIFFNE) and upstream-directed colonization (COLON) as the processes most likely to influence the DIGD for the species concerned, albeit with low posterior probability. This could have been because the approach cannot distinguish between these processes in our case, or because they both influenced genetic diversity of the species in the study zone. In their review of riverscape genetics, Blanchet et al. (2020) highlighted that upstream-directed colonization was the process that most frequently influenced DIGD in rivers. In the Maroni though, diversification and past colonization routes of most fish taxa were documented as resulting from the formation of a Pleistocene refuge and relatively ancient stream-capture events between headwaters of the Maroni and neighboring watersheds (e.g., between the Litani and the river Jari in the Tucumaque mountains, Fisch-Muller et al. 2018) and less likely from exchanges of fish with other coastal rivers through estuaries (Lemopoulos and Covain 2019). In addition, current patterns of landscape genetic variation are considered to result mainly from environmental heterogeneity (Borstein et al. 2022) and species life history traits (Papa et al. 2021), although specific reconstructions of past demography and evolutionary trajectories are missing. Thus, upstream-directed colonization from the river outlet to the headwaters seems less likely to have occurred in our study zone, unless it relies on contemporary non-equilibrium processes such as extinction-recolonization of upstream reaches due to local environment constraints. This would support the parallel implication of higher downstream habitat availability to explain DIGD there, as suggested by the ABCRF results. However, low posterior probabilities and the relatively high out-of-bag score of the ABCRF in our case (14.3%) hampers a greater understanding of the relative influence of these two processes at this stage. The power of the ABCRF analysis to further distinguish between them may be limited by our small number of sites, the use of biallelic SNP markers (See supplementary note from Table S1.4) or by relatively important gene flow or local effective size. Ultimately, we tested only three evolutionary processes (and combinations thereof), although other processes may potentially generate DIGD in the wild (Blanchet et al. 2020).
Hot- and coldspots of local genetic diversity in the Upper Maroni
The DIGD pattern, when it existed, was often weak. CORP, CORHS, and CORFST.P never exceeded 0.584, 0.557, and 0.752, respectively, and the EMM from the analyses of covariance had large confidence intervals that often included zero. Most DIGD studies of fish rarely consider more than ∼15 sampling sites (e.g., Salisbury et al. 2016; Pilger et al. 2017; Richmond et al. 2018); however, the Upper Maroni is a dense watershed, and our 12 sampling sites may lack the resolution required to reveal stronger DIGD patterns (but see Alther et al. 2021, who also highlighted complex patterns of DIGD in Gammarus fossarum even though 95 sampling sites were sampled). In particular, a sampling design biased against the most upstream sites (which are usually more isolated, especially within a network with complex branching, Thomaz et al. 2016) may underestimate the strength of DIGD. A weak DIGD pattern may also result from analyzing too few molecular markers. However, we used an average ∼150 SNP per species, which, for most applications, is consistent with the genetic information provided by 10–15 microsatellites (Morin et al. 2004), a fairly common design used in other DIGD studies (Pilger et al. 2017; Ruzzante et al. 2019; Alther et al. 2021). Moreover, these SNP markers were developed from a discovery panel drawn from the same place and time as the current study, in order to capture the main patterns of spatial genetic variation in the Upper Maroni (Delord et al. 2018).
The weak DIGD pattern for the five species could ultimately have resulted from the contrasting local genetic diversity between the eastern and western areas of the study zone, as revealed by local ID and FST.P values. We observed lower mean genetic diversity in the Tampok and the Waki tributaries than in the Marouini and Litani, regardless of the upstream distance. This trend was also observed for certain species with low genetic structure, such as C. meionactis and S. rhombeus (Figs. S1.3.2, S1.3.12). This diverging trend suggests that the dendritic structure of the river network is not the only feature that influences spatial patterns of genetic diversity along the upstream gradient. At a larger geographical scale, Papa et al. (2021) identified significant global genetic divergence between western (i.e., Suriname) and eastern (i.e., our study zone) regions of the Maroni River using multispecies genetic landscapes based on the mitochondrial gene COI. This may result from current expansion of population ranges from the Maroni toward adjacent watersheds. They also highlighted divergence between the Marouini and the Tampok to a lesser extent, a finding strengthened by our results, and suggested to arise mainly from species dispersal abilities or intra- and interspecific competition. Here, we further hypothesize that lower habitat availability, accessibility, or quality may explain the lower local genetic diversity detected in association with higher local genetic isolation in the eastern area of our study zone. The Waki is narrower than the other tributaries sampled, which implies lower habitat availability. The Tampok is a narrow valley, with large variations in seasonal water levels but little river overflow, which may decrease access to certain habitats. In addition, local practitioners have observed that the Tampok and Waki were more influenced by local, small-scale gold mining activities in the past decade than the Marouini or Litani, which may have polluted the water, increased water turbidity, or degraded riverbanks, thus reducing habitat quality for fish. In conclusion, local genetic diversity was generally lower, and the DIGD more pronounced, in the eastern area (including the Tampok and Waki) than in the western area (including the Litani, Marouini, and sites La.1a and La.1b of the Upper Lawa).
In the main channel (Lawa), local genetic diversity was slightly lower (and local genetic isolation slightly higher) at site La.0 than at La.1a or La.1b for several species (Fig. 1 and S1.2). La.0 harbors the highest human density in the study zone, surrounding the downstream village of Papaïchton and the city of Maripasoula, and where fishing pressure is higher than that at other locations (Longin et al. 2021b). For instance, H. aimara, one of the species most targeted by fishermen in this ___location, had a low polymorphism rate, high local genetic isolation, and small Ne (Table S1.5), similar to other species such as Leporinus friderici and Brycon falcatus. This may indicate that their populations experience substantial fishing pressure in these areas (Allendorf et al. 2008; Longin et al. 2021b).
In conclusion, the expected overall DIGD pattern in the Upper Maroni is likely to be influenced by local habitat characteristics along a west-to-east gradient of decreasing habitat quality or availability, in addition to the dendritic structure and upstream distance. Moreover, anthropogenic pressures at certain sites, such as gold mining or fishing, may influence patterns of local genetic variation in areas with increasing human presence. Collecting local physico-chemical data at high spatial resolution across the study zone (e.g., turbidity, dissolved oxygen or mercury concentration, diversity and availability of fish habitats) may enable to better understand the influence of environmental features on spatial genetic variation in the Upper Maroni, e.g., through the application of complementary riverscape genetic frameworks (Davis et al. 2018). Anthropogenic influences on spatial genetic variation may be much less predictable than the influence of the river network, and perhaps weaker (Prunier et al. 2017a). However, it could be a non-negligible predictor of riverscape genetic variation, and quantifying it will require additional studies to address multicollinearity among environmental and anthropogenic variables (Blanchet et al. 2020).
Contrasting patterns and factors affecting genetic structure across species
The infrequent occurrence of DIGD in our results, and its interference with potentially independent local constraints also lead to consider the various patterns of genetic structure observed across the 18 focal species. As mentioned earlier, all species documented as long-distance migrants and broadcast spawners displayed seemingly negligible genetic structure. However, the other species displayed contrasting patterns, as revealed by MRM and multivariate analyses where species sharing similar life history traits were not necessarily grouped together. Endemic species did not associate to any particular structuring pattern either. All of them were evenly distributed along both axes of the PCA biplot, with, for instance, a distinct contrast between species displaying high isolation by stream distance, as revealed by CORIBD (e.g., H. gymnorhynchus), and species whose genetic structure depended more on riverscape topography or spatial heterogeneity in effective size, as revealed by CORIBF and CORNE (e.g., G. harreri). The six Serrasalmidae species, all of which are documented as brood-hiders and short-distance migrants (Table 4 and Supplementary Table S1.1), had highly contrasting patterns of genetic variation. Conversely, H. aimara, a species considered to be sedentary and a guarder, had surprisingly low genetic structure (Table 4, Fig. 3), except at site La.0, as mentioned. Such idiosyncratic patterns were identified in other studies as well (Paz-Vinas et al. 2018), whereas other work suggest more consistent patterns with a strongly predominant influence of stream hierarchy across a number of co-distributed species sharing similar traits (Zbinden et al. 2022). Several hypotheses can be discussed to explain the idiosyncratic patterns observed in our case.
Body size, although usually considered to be positively associated with higher dispersal abilities (Radinger and Wolter 2014), did not complement the differences observed across species. For instance, small-bodied herbivorous Serrasalmidae species had lower genetic structure than larger-bodied herbivorous Serrasalmidae (e.g., Myloplus rubripinnis, with non-significant overall FST = 0.001, compared with larger-bodied M. rhomboidalis and T. lebaili, with FST ≥ 0.008). A more restricted ecological niche or reproduction timing might better explain these observations. For instance, T. lebaili and M. rhomboidalis depend completely on flowing water of large streams, including during the breeding period (Planquette et al. 1996; Jégu and Keith 2005), which may limit gene flow at larger spatial scales. T. lebaili also has considerable foraging specialization (Planquette et al. 1996; Jégu and Keith 2005), since it feeds almost exclusively on Podostemaceae plants that bloom in shallow rapids. S. eigenmanni, another Serrasalmidae species with high genetic structure, is found only in densely vegetated riverbanks (fisherman Janakale Makiloewala, pers. comm.). Results for H. aimara could be explained by its high fecundity (Planquette et al. 1996), if early dispersal compensates for adult territoriality.
Neotropical rivers host high species richness, and tremendous efforts were deployed to inventory it. However, information about the ecological optima, reproductive cycles, vital rates, and vulnerability of these species to environmental constraints is still lacking. In such cases, comparative population genetic approaches help by providing a holistic overview of intraspecific patterns, but biological interpretation of spatial genetic signatures appear limited by current knowledge. For instance, it would be useful to assess whether the higher genetic structure observed for large-bodied herbivorous Serrasalmidae results solely from a narrower spatial ecological niche that limits genetic connectivity, or also from naturally low abundance or local pressures (e.g., habitat degradation, fishing) resulting in smaller reproductive output and smaller Ne (which is more difficult to assess).
Similarly, collecting additional information about life history traits and the ecology of the focal species, in parallel to considering physico-chemical parameters from the environment, may enable us to better understand in which case we might observe a particular pattern of genetic structure across riverine species -and in which way it may, or not, be associated to DIGD. Subsidiarily, we may recall that all species studied here remain relatively ubiquitous across the study zone. It is possible that studying more sedentary, guarder species with narrower niche would provide additional illustrations of DIGD in the Upper Maroni.
An initial snapshot of genetic variation in fish in the Upper Maroni: conclusions and perspectives
Our study provides useful information for identifying geographic areas and species in the Upper Maroni that could be of conservation interest. Additional environmental data could ease our understanding of the patterns observed, especially with regard to habitat quality. The information provided at the intraspecific level could be combined with traditional biodiversity surveys at the interspecific level by local professionals and managers, to propose relevant actions that maximize the conservation of both aspects of biodiversity. Our comparative approach could also serve as a baseline for future similar studies, for instance, with higher sampling coverage and/or high-density genomic datasets for species of particular interest in the Upper Maroni. The genetic approach is a useful conservation tool in the Neotropics, where data collection to apply conventional tools for population monitoring (such as mark-recapture, catch-per-unit-effort standardization, or telemetry) is infeasible or technically challenging in such diverse and remote areas (Meunier et al. 1994).
Our genetic approach highlighted species that may be more vulnerable to certain local anthropogenic pressures. For instance, T. lebaili and M. rhomboidalis have high cultural and commercial importance to local human communities in the Upper Maroni. We observed significant spatial genetic variation for both species. In T. lebaili, linearized pairwise FST values were significantly influenced by stream distance, discontinuous river flow and spatial heterogeneity in effective size. This indicates potentially higher susceptibility to local pressures such as habitat degradation or fishing, which could lead to local losses of genetic diversity. Similarly, the significant genetic isolation and lower polymorphism of the economically important H. aimara around an area of high fishing pressure should be considered in future management policies. Once available, integrating more detailed information about species’ life history traits and ecological requirements in a comparative population genetics framework, in addition to riverscape features, will be crucial to further identify the relative influence (and potential predictability) of riverscape and biological factors on genetic diversity in fish species in the Upper Maroni and at a larger scale.
The increasing anthropogenic pressures on Upper Maroni fish populations (e.g., water pollution, habitat degradation, fishing) raises questions about their vulnerability and threatens the prosperity of subsistence fishing in southern French Guiana. Their potential influence on life history traits (e.g., mean body size; Longin et al. 2021b) may also influence patterns of genetic variation and community dynamics at a larger scale (Evangelista et al. 2021). Although we observed no direct evidence of local anthropogenic genetic erosion in the study zone, these pressures can occur even if they are currently compensated by large Ne or gene flow. For instance, local anthropogenic genetic drift in downstream areas might be compensated by DIGD, thus making it difficult to identify these pervasive impacts. Thus, long-term patterns of genetic variation in the riverscape depend on the spatial variation in Ne, the intensity of anthropogenic pressure, and the strength and direction of inter-site gene flow. Methods to finely quantify the strength and direction of gene flow had too low power to provide reliable results with our low-density data (results not shown) and we were unable to identify any of the sampling sites as a source or sink of genetic variation. A less descriptive, more mechanistic study of processes that influence patterns of genetic variation in fishes in the Upper Maroni (e.g., combining high-density genomic tools with past and current demography inference) could provide important insights into the potential influence of anthropogenic pressures on fish populations. To develop sustainable management policies for fish stocks, it could help to use a simulation-based framework that explicitly incorporates all spatial features of the Upper Maroni and focuses specifically on distinguishing between natural evolutionary scenarios that generate DIGD, and scenarios that also include the influence of anthropogenic factors, like harvesting. This was considered beyond the scope of the current study, but identified as a priority focus for future work to illustrate long-term patterns of riverscape genetic variation under anthropogenic pressures in the Upper Maroni. In addition, long-term genetic monitoring could be a promising approach to follow patterns of genetic variation over time, identify a gradual loss of local or global riverscape genetic diversity (Mathieu-Bégné et al. 2018), and determine the influence of local pressures on intraspecific dynamics of an entire metapopulation (Palstra and Ruzzante 2011).
Data availability
Supplementary notes, figures, and tables for this article are provided in MARONI_Appendix1.pdf. Individual genotypes and genetic diversity data used in the current study are accessible through the Dryad repository https://doi.org/10.5061/dryad.dbrv15f5n, as well as sequence information about the 31 additional SNPs used for species C. meionactis. The R command lines used to analyze those data are provided and commented in MARONI_Appendix2.pdf. There are no financial benefits to report.
References
Abell R, Thieme ML, Revenga C, Bryer M, Kottelat M, Bogutskaya N et al. (2008) Freshwater ecoregions of the world: a new map of biogeographic units for freshwater biodiversity conservation. BioScience 58(5):403–414. https://doi.org/10.1641/B580507
Allendorf FW, England P, Luikart G, Ritchie P, Ryman N (2008) Genetic effects of harvest on wild animal populations. Trends Ecol Evolut 23(6):327–337. https://doi.org/10.1016/j.tree.2008.02.008
Alther R, Fronhofer E, Altermatt F (2021) Dispersal behaviour and riverine network connectivity shape the genetic diversity of freshwater amphipod metapopulations. Mol Ecol 00:1–15. https://doi.org/10.1111/mec.16201
Blanchet S, Rey O, Etienne R, Lek S, Loot G (2010) Species-specific responses to landscape fragmentation: implications for management strategies. Evolut Appl 3(3):291–304. https://doi.org/10.1111/j.1752-4571.2009.00110.x
Blanchet S, Prunier JG, Paz-Vinas I, Saint-Pé K, Rey O, Raffard A et al (2020) A river runs through it: the causes, consequences, and management of intraspecific diversity in river networks. Evolut Appl 1–19. https://doi.org/10.1111/eva.12941
Borstein S, Lucanus O, Gajapersad K, Singer R, Mol JHA and López-Fernández H (2022). Fish Diversity of the Upper Tapanahony River, Suriname. Museum of zoology, University of Michigan. https://doi.org/10.7302/4816
Breiman L (2001) Random forests. Mach Learn 45:5–32
Caballero A, Toro MA (2002) Analysis of genetic diversity for the management of conserved subdivided populations. Conserv Genet 3:289–299
Campbell Grant EH, Lowe WH, Fagan WF (2007) Living in the branches: population dynamics and ecological processes in dendritic networks. Ecol Lett 10(2):165–175. https://doi.org/10.1111/j.1461-0248.2006.01007.x
Chaput-Bardy A, Fleurant C, Lemaire C, Secondi J (2009) Modelling the effect of in-stream and overland dispersal on gene flow in river networks. Ecol Model 220(24):3589–3598. https://doi.org/10.1016/j.ecolmodel.2009.06.027
Crutsinger GM (2016) A community genetics perspective: opportunities for the coming decade. N. Phytol 210(1):65–70. https://doi.org/10.1111/nph.13537
Csilléry K, Blum MGB, Gaggiotti OE, François O (2010) Approximate Bayesian Computation (ABC) in practice. Trends Ecol Evolut 25(7):410–418. https://doi.org/10.1016/j.tree.2010.04.001
Davis CD, Epps CW, Flitcroft RL, Banks MA (2018) Refining and defining riverscape genetics: How rivers influence population genetic structure. WIREs Water 5:e1269. https://doi.org/10.1002/wat2.1269
Deck J, Gaither MR, Ewing R, Bird CE, Davies N, Meyer C et al. (2017) The Genomic Observatories Metadatabase (GeOMe): A new repository for field and sampling event metadata associated with genetic samples. PLOS Biol 15(8):e2002925
Delord C, Lassalle G, Oger A, Barloy D, Coutellec M-A, Delcamp A et al. (2018) A cost-and-time effective procedure to develop SNP markers for multiple species: A support for community genetics. Methods Ecol Evolut 9(9):1959–1974. https://doi.org/10.1111/2041-210X.13034
DeWoody J, Avise JC (2000) Microsatellite variation in marine, freshwater and anadromous fishes compared with other animals. J Fish Biol 56(3):461–473. https://doi.org/10.1006/jfbi.1999.1210
Do C, Waples RS, Peel D, Macbeth GM, Tillett BJ, Ovenden JR (2014) NeEstimator v2: re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Mol Ecol Resour 14(1):209–214. https://doi.org/10.1111/1755-0998.12157
Dobzhansky T (1965) Genetic diversity and fitness. Genet Today 3:541–552
Evangelista C, Dupeu J, Sandkjenn J, Pauli BD, Herland A, Meriguet J et al. (2021) Ecological ramifications of adaptation to size-selective mortality. R Soc Open Sci 8:210842. https://doi.org/10.1098/rsos.210842
Fisch-Muller S, Mol JHA, Covain R (2018) An integrative framework to reevaluate the Neotropical catfish genus Guyanancistrus (Siluriformes: Loricariidae) with particular emphasis on the Guyanancistrus brevispinis complex. PLoS One 13:e0189789
Fraimout A, Debat V, Fellous S, Hufbauer RA, Foucaud J, Pudlo P et al. (2017) Deciphering the routes of invasion of Drosophila suzukii by means of ABC random forest. Mol Biol Evolut 34:980–996
Frankham R (1995) Conservation genetics. Annu Rev Genet 29:305–327
Franklin IR (1980) Evolutionary change in small populations. In: Soulé M, Wilcox B (eds) Conservation biology: an evolutionary‐ecological perspective. Sinauer Associates, Sunderland, MA, p 135–149
Goudet J and Jombart T (2017) hierfstat: Estimation and Tests of Hierarchical F-Statistics. R package version 0.04-22. http://www.r-project.org, http://github.com/jgx65/hierfstat
Gutiérrez JP, Royo LJ, Álvarez I, Goyache F (2005) MolKin v2.0: a computer program for genetic analysis of populations using molecular coancestry information. J Heredity 96(6):718–721
Hilsdorf AWS, Hallerman EM (2017) Genetic Resources of Neotropical Fishes. Springer International Publishing, Cham, 10.1007/978-3-319-55838-7
Hoban S, Bruford MW, Funk C, Galbusera P, Griffith MP, Grueber CE et al (2021) Global commitments to conserving and monitoring genetic diversity are now necessary and feasible. BioSciences, biab054. https://doi.org/10.1111/eva.12197
Hoban SM, Hauffe HC, Pérez-Espona S, Arntzen JW, Bertorelle G, Bryja J et al. (2013) Bringing genetic diversity to the forefront of conservation policy and management. Conserv Genet Resour 5(2):593–598
Horne JB (2014) Emerging patterns and emerging challenges of comparative phylogeography. Front Biogeogr 6(4):166–168
Jégu M, Keith P (2005) Threatened fishes of the world: Tometes lebaili (Jégu, Keith and Belmont-Jégu 2002) (Characidae: Serrasalminae). Environ Biol Fishes 72(4):378–378. https://doi.org/10.1007/s10641-004-4126-4
Jenkins TL, Castilho R, Stevens JR (2018) Meta-analysis of northeast Atlantic marine taxa shows contrasting phylogeographic patterns following post-LGM expansions. PeerJ 6:e5684. https://doi.org/10.7717/peerj.5684
Jombart T, Devillard S, Balloux F (2010) Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet 11(1):1–15
Keenan K, McGinnity P, Cross TF, Crozier WW, Prodöhl PA (2013) diveRsity: An R package for the estimation and exploration of population genetics parameters and their associated errors. Methods Ecol Evolut 4(8):782–788. https://doi.org/10.1111/2041-210X.12067
Lê S, Josse J, Husson F (2008) FactoMineR: An R package for multivariate analysis. J Stat Softw 25(1):1–18
Le Bail P-Y, Covain R, Jégu M, Fisch-Muller S, Vigouroux R, Keith P (2012) Updated checklist of the freshwater and estuarine fishes of French Guiana. Cybium 36(1):293–319
Leigh DM, van Rees CB, Millette KL, Breed MF, Schmidt C, Bertola L et al (2021) Opportunities and challenges of macrogenetic studies. Nat Rev Genet. https://doi.org/10.1038/s41576-021-00394-0
Lemopoulos A, Covain R (2019) Biogeography of the freshwater fishes of the Guianas using a partitioned parsimony analysis of endemicity with reappraisal of ecoregional boundaries. Cladistics 35(1):106–124. https://doi.org/10.1111/cla.12341
Lenth R (2018) emmeans: Estimated Marginal Means, aka Least-Squares Means. R package version 1.2.2. https://CRAN.R-project.org/package=emmeans
Lepais O, Chancerel E, Boury C, Salin F, Manicki A, Taillebois L et al. (2020) Fast sequence-based microsatellite genotyping development workflow. PeerJ 8:e9085. https://doi.org/10.7717/peerj.9085
Lino A, Fonseca C, Rojas D, Fischer E, Ramos Pereira MJ (2018) A meta-analysis of the effects of habitat loss and fragmentation on genetic diversity in mammals. Mamm Biol 94:69–76. https://doi.org/10.1016/j.mambio.2018.09.006
Longin G, Bonneau de Beaufort L, Fontenelle G, Rinaldo R, Roussel J-M, Le Bail P-Y (2021a) Fisher’s perceptions of river resources: case study of French Guiana native populations using contextual cognitive mapping. Cybium 2021 45(1):5–20. https://doi.org/10.26028/cybium/2021-451-001.
Longin G, Fontenelle G, Bonneau de Beaufort L, Delord C, Launey S, Rinaldo R et al (2021b) When subsistence fishing meets conservation issues: survey of a small fishery in a neotropical river with high biodiversity value. Fisheries Res 241. https://doi.org/10.1016/j.fishres.2021.105995
Lujan NK, Armbruster JW (2011) The Guiana Shield. In: Albert JS, Reis RE (eds) Historical biogeography of neotropical freshwater fishes. University of California Press, Berkeley, p 211–224
Mathieu-Bégné E, Loot G, Chevalier M, Paz-Vinas I, Blanchet S (2018) Demographic and genetic collapses in spatially structured populations: insights from a long-term survey in wild fish metapopulations. Oikos 128:196–207. https://doi.org/10.1111/oik.05511
Meunier FJ, Rojas-Beltran R, Boujard T, Lecomte F (1994) Rythmes saisonniers de la croissance chez quelques Téléostéens de Guyane française. Rev d’hydrobiologie Tropicale 27(4):423–440
Morin PA, Luikart G, Wayne RK, the SNP workshop group (2004) SNPs in ecology, evolution and conservation. Trends Ecol Evolut 19(4):208–216. https://doi.org/10.1016/j.tree.2004.01.009
Palstra FP, Ruzzante DE (2011) Demographic and genetic factors shaping contemporary metapopulation effective size and its empirical estimation in salmonid fish. Heredity 107(5):444–455. https://doi.org/10.1038/hdy.2011.31
Papa Y, Le Bail P-Y, Covain R (2021) Genetic landscape clustering of a large DNA barcoding dataset reveals shared patterns of genetic divergence among freshwater fishes of the Maroni Basin. Mol Ecol Resour 21:2109–2124. https://doi.org/10.1111/1755-0998.13402
Pauls SU, Alp M, Bálint M, Bernabò P, Čiampor F, Čiamporová-Zaťovičová Z et al. (2014) Integrating molecular tools into freshwater ecology: developments and opportunities. Freshw Biol 59(8):1559–1576. https://doi.org/10.1111/fwb.12381
Paz-Vinas I, Blanchet S (2015) Dendritic connectivity shapes spatial patterns of genetic diversity: a simulation-based study. J Evolut Biol 28(4):986–994. https://doi.org/10.1111/jeb.12626
Paz-Vinas I, Loot G, Hermoso V, Veyssiere C, Poulet N, Grenouillet G et al. (2018) Systematic conservation planning for intraspecific genetic diversity. Proc R Soc B Biol Sci 285:20172746. 10
Paz-Vinas I, Loot G, Stevens VM, Blanchet S (2015) Evolutionary processes driving spatial patterns of intraspecific genetic diversity in river ecosystems. Mol Ecol 24(18):4586–4604. https://doi.org/10.1111/mec.13345
Pereira HM, Ferrier S, Walters M, Geller GN, Jongman RHG, Scholes RJ et al. (2013) Essential biodiversity variables. Science 339(6117):277–278. https://doi.org/10.1126/science.1229931
Pflüger FJ, Balkenhol N (2014) A plea for simultaneously considering matrix quality and local environmental conditions when analysing landscape impacts on effective dispersal. Mol Ecol 23(9):2146–2156. https://doi.org/10.1111/mec.12712
Pilger TJ, Gido KB, Propst DL, Whitney JE, Turner TF (2017) River network architecture, genetic effective size and distributional patterns predict differences in genetic structure across species in a dryland stream fish community. Mol Ecol 26(10):2687–2697. https://doi.org/10.1111/mec.14079
Planquette P, Keith P, Le Bail P-Y (1996) Atlas des poissons d’eau douce de Guyane (tome 1). In: Collection du Patrimoine Naturel (ed), vol 22. IEGB-M.N.H.N., INRA, CSP, Min. Env., Paris, p 429
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155(2):945–959
Prunier JG, Dubut V, Chikhi L, Blanchet S (2017b) Contribution of spatial heterogeneity in effective population sizes to the variance in pairwise measures of genetic differentiation. Methods Ecol Evolut 8:1866–1877. https://doi.org/10.1101/031633
Prunier JG, Dubut V, Loot G, Tudesque L, Blanchet S (2017a) The relative contribution of river network structure and anthropogenic stressors to spatial patterns of genetic diversity in two freshwater fishes: a multiple-stressors approach. Freshw Biol 63:6–21. https://doi.org/10.1111/2041-210X.12820
Pudlo P, Marin J-M, Estoup A, Cornuet J-M, Gautier M, Robert CP (2016) Reliable ABC model choice via random forests. Bioinformatics 32(6):859–866
Radinger J, Wolter C (2014) Patterns and predictors of fish dispersal in rivers. Fish Fish 15(3):456–473. https://doi.org/10.1111/faf.12028
Reis RE, Albert JS, Di Dario F, Mincarone MM, Petry P, Rocha LA (2016) Fish biodiversity and conservation in South America. J Fish Biol 89(1):12–47. https://doi.org/10.1111/jfb.13016
Richmond JQ, Backlin AR, Galst-Cavalcante C, O’Brien JW, Fisher RN (2018) Loss of dendritic connectivity in southern California’s urban riverscape facilitates decline of an endemic freshwater fish. Mol Ecol 27(2):369–386. https://doi.org/10.1111/mec.14445
Rougemont Q, Roux C, Neuenschwander S, Goudet J, Launey S, Evanno G (2016) Reconstructing the demographic history of divergence between European river and brook lampreys using approximate Bayesian computations. PeerJ 4:e1910. https://doi.org/10.7717/peerj.1910
Ruzzante DE, McCracken GR, Salisbury SJ, Brewis HT, Keefe D, Gaggiotti OE et al. (2019) Landscape, colonization, and life history: Their effects on genetic diversity in four sympatric species inhabiting a dendritic system. Can J Fish Aquat Sci 76(12):2288–2302. https://doi.org/10.1139/cjfas-2018-0416
Salisbury SJ, McCracken GR, Keefe D, Perry R, Ruzzante DE (2016) A portrait of a sucker using landscape genetics: how colonization and life history undermine the idealized dendritic metapopulation. Mol Ecol 25(17):4126–4145. https://doi.org/10.1111/mec.13757
Selkoe KA, Gaggiotti OE, ToBo Laboratory, Bowen BW, Toonen RJ (2014) Emergent patterns of population genetic structure for a coral reef community. Mol Ecol 23(12):3064–3079. https://doi.org/10.1111/mec.12804
Selkoe KA, Gaggiotti OE, Treml EA, Wren JLK, Donovan MK, Hawai’i Reef Connectivity Consortium. et al. (2016) The DNA of coral reef biodiversity: predicting and protecting genetic diversity of reef assemblages. Proc R Soc B: Biol Sci 283(1829):20160354. https://doi.org/10.1098/rspb.2016.0354
Thomaz AT, Christie MR, Knowles LL (2016) The architecture of river networks can drive the evolutionary dynamics of aquatic populations: Brief Communication. Evolution 70(3):731–739. https://doi.org/10.1111/evo.12883
Vellend M, Geber MA (2005) Connections between species diversity and genetic diversity. Ecol Lett 8(7):767–781. https://doi.org/10.1111/j.1461-0248.2005.00775.x
Waples RS, England PR (2011) Estimating contemporary effective population size on the basis of linkage disequilibrium in the face of migration. Genetics 189:633–644. https://doi.org/10.1534/genetics.111.132233
Waples RS, Antao T, Luikart G (2014) Effects of overlapping generations on linkage disequilibrium estimates of effective population size. Genetics 197(2):769–780. https://doi.org/10.1534/genetics.114.164822
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38(6):1358. https://doi.org/10.2307/2408641
Zbinden ZD, Douglas MR, Chafin TK, Douglas ME (2022) Riverscape community genomics: a comparative analytical approach to identify common drivers of spatial structure. Mol Ecol 00:1–23. https://doi.org/10.1111/mec.16806
Acknowledgements
We thank all residents and fishermen of villages in the Upper Maroni for their hospitality and support, including local mediators and fishermen who participated in sample collection: Pitoma Mekouanali, Janakale Makiloewala, Kutaka Aïtalé, Kuliwaïkë Menali, Moloko Atiwaïke, Palanaïwa Alikana, Sébastien Amaïpetit, Etulano Yamo, Mones Tokotoko, Marc Pinson, Hervé Tolinga, Kalou, and Alfred Djaba, with a particular thought for Raymond Essimon†. We thank all those involved in the fieldwork: Philippe Cerdan†, Sébastien Le Reun, Brian Senechal and Régis Vigouroux (Hydreco Guyane), Raphael Covain (MNHG), Jean-Luc Baglinière, Damien Fourcy, Catherine Le Penven and Marie Nevoux (UMR DECOD, INRAE). We also thank Anne-Laure Besnard and Thibaut Jousseaume for their help with DNA extraction, quantification, and dilution and we thank Raphael Covain for helping to identify a few ambiguous samples through COI barcoding. We are grateful to Oscar E. Gaggiotti (Scottish Ocean Institute), Olivier Lepais (UMR Biogeco, INRAE), and Ivan Paz-Vinas (EDB, Paul Sabatier University) for their greatly helpful advice throughout this study. Finally, we thank Daniel Ruzzante and two anonymous reviewers for their very constructive feedback about earlier versions of this manuscript.
Funding
This study was funded by the Parc Amazonien de Guyane under contract R&D-2003-06 to Pierre-Yves Le Bail and Jean-Marc Roussel, with financial support from the DREAL and the Office de l’Eau de Guyane. The genetic resources studied here were collected in accordance with ethical considerations defined in the convention APA-973-7 in relation with the Nagoya protocol.
Author information
Authors and Affiliations
Contributions
CD contributed to specimen and molecular data collection, designed and ran the analyses, and wrote the manuscript. EJP and SB contributed to the design of analyses and reviewed the manuscript. GL, RR, and RV organized fieldwork, contributed to specimen collection and reviewed the manuscript. J-MR, P-YLB, and SL conceptualized and supervised the research, contributed to specimen collection, contributed to the design of analyses and reviewed the manuscript. All authors contributed critically to the drafts and gave final approval for publication.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Associate editor: Jane Hughes.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Delord, C., Petit, E.J., Blanchet, S. et al. Contrasts in riverscape patterns of intraspecific genetic variation in a diverse Neotropical fish community of high conservation value. Heredity 131, 1–14 (2023). https://doi.org/10.1038/s41437-023-00616-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41437-023-00616-7