Abstract
High mitochondrial DNA (mtDNA) amount has been reported to be beneficial for resistance and recovery of metabolic stress, while increased mtDNA synthesis activity can drive aging signs. The intriguing contrast of these two mtDNA boosting outcomes prompted us to jointly elevate mtDNA amount and frequency of replication in mice. We report that high activity of mtDNA synthesis inhibits perinatal metabolic maturation of the heart. The offspring of the asymptomatic parental lines are born healthy but manifest dilated cardiomyopathy and cardiac collapse during the first days of life. The pathogenesis, further enhanced by mtDNA mutagenesis, involves prenatal upregulation of mitochondrial integrated stress response and the ferroptosis-inducer MESH1, leading to cardiac fibrosis and cardiomyocyte death after birth. Our evidence indicates that the tight control of mtDNA replication is critical for early cardiac homeostasis. Importantly, ferroptosis sensitivity is a potential targetable mechanism for infantile-onset cardiomyopathy, a common manifestation of mitochondrial diseases.
Similar content being viewed by others
Introduction
The depletion of mitochondrial DNA (mtDNA) contributes to the pathogenesis of a wide range of severe human disorders1, while increased mtDNA amounts have been reported to promote health and recovery from disease2,3,4,5,6,7,8,9. Two nuclear-encoded proteins regulate the amount of the multicopy genome: Twinkle helicase by mtDNA replication licensing10,11 and mitochondrial transcription factor A (TFAM) by packaging and half-life increase11,12. Overexpression of either Twinkle or TFAM in mice (TwOE, TfamOE, respectively) leads to high mtDNA amount and enhance recovery from ischemic heart damage [TwOE2,3,4,5] or delay male infertility [TfamOE6,7,8]. The double-transgenic TwOE/TfamOE showed synergistic effects, further increasing mtDNA copy number and enlarging mtDNA nucleoid size. These animals remained healthy and had a normal lifespan, despite mild respiratory chain deficiency in the aged heart11. The evidence indicates that even a 6-fold increase in steady-state mtDNA amount, when boosted by replication licensing and packaging11, causes no major health effects.
Overexpression of DNA polymerase gamma (POLG), the mtDNA replicase, does not affect mtDNA copy number in mammals, emphasizing the rate-limiting role of the replicative helicase Twinkle in healthy animals. However, if the exonuclease function of POLG is inactivated (PolgD257A knock-in; mtDNA “mutator” mice), both mtDNA replication frequency and mutagenesis increase13,14,15. These lead to progeroid syndrome, progressively manifesting after 6 months of age, with anemia as the initial symptom, followed by osteoporosis, thin skin, and late-onset cardiac hypertrophy14,15. While these symptoms were attributed to mtDNA mutagenesis, recent data suggest an alternative explanation. The exonuclease-deficient POLG synthesizes mtDNA at an increased rate leading to imbalanced nucleotide pools, nuclear replication fork stalling, and chronic nuclear genome replication stress in stem cells13. Supporting these conclusions, TFAM-mediated increased mtDNA packaging in PolgD257A cells decreased genomic DNA breaks13 and delayed testicular degeneration in mutators7, likely by hindering replisome activity. These mechanisms highlight the importance of context-specific regulation of mtDNA replication for the maintenance of tissue homeostasis.
The data from the PolgD257A knock-in, TwOE, and TfamOE mice together suggest that each factor regulates mtDNA replication and amount via different mechanisms. Therefore, they offer exciting tools to test the roles of mtDNA amount and synthesis for health. Here, we simultaneously increased both mtDNA replication licensing and processivity (double-transgenic mice with TwOE and PolgD257A alleles). We hypothesized that if a high steady-state mtDNA amount indeed promoted health as an independent factor9, TwOE should alleviate the mutator phenotype, similar to what was reported for TfamOE/PolgD257A mice7,13. Meanwhile, if the metabolic consequences of PolgD257A-driven increased mtDNA replication are dominant in the progressive pathology, TwOE should aggravate the mutator phenotype. The molecular understanding of these mechanisms is essential for the efficacy and safety of therapeutic approaches targeting mtDNA amount and replication.
Here, we show that increased mtDNA replication activity can disrupt postnatal cardiac maturation, leading to fatal neonatal dilated cardiomyopathy. Our data indicate the mechanism is separate from mtDNA mutagenesis, and instead raises from the maladaptive interaction of metabolic stress responses and the ferroptotic inducer MESH1.
Results
High mtDNA replication in PolgD257A carriers elicits fatal neonatal cardiomyopathy
To increase mtDNA copy number by enhancing both mtDNA replication licensing and processivity, we crossed TwOE mice to the PolgD257A background (Fig. 1a). The parental TwOE line ubiquitously overexpresses wildtype murine Twinkle cDNA, resulting in a two-to-four fold increased mtDNA amount already at birth, and causing no robust phenotype even by two years of age10,11. The second parental line, PolgD257A homozygote “mutator” mice manifest with progressive anemia and progeric symptoms at six months of age (before which they remain asymptomatic), shortening their lifespan to maximally 14 months14,15. The PolgD257A x TwOE crossing represented all combinations of PolgD257A and TwOE genotypes in near-mendelian ratios, and the different genotypes were indistinguishable from each other at birth (Fig. 1b-c). The lines carrying the Twinkle transgene presented high Twinkle mRNA levels and a 2-3-fold increase in mtDNA amount in the heart, as well as WT-like weight at birth (Fig. 1c–f). However, the double transgenic PolgMutTwOE offspring developed a striking neonatal pathology and stalled growth apparent at postnatal day (P) 4 (Fig. 1c, d). Most of the pups died around P7.
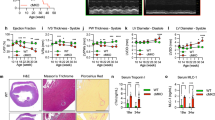
a Summary of genetic manipulations of mice and their effects on mtDNA. b Mouse genotype array and naming. c Representative WT and PolgMutTwOE mouse phenotypes, postnatal (P) days P1 and P7. d Weight of the P7 transgenic mice. e Relative Twnk mRNA expression, P1 heart. f Relative mtDNA amount to nuclear Rbm15; mtDNA probes: mt-Rnr2, mt-Co1, and mt-Nd4 genes, P7 heart. g Heart-to-body weight ratio, P7. h Representative P7 hearts; hematoxylin/eosin and Masson’s trichrome staining. Hemorrhagic, fibrotic lesions (black arrowhead), vacuolation (white arrowhead), and interstitial fibrosis (black arrowhead) (n = 3 WT, 2 PolgHet, 7 PolgMut, 7 TwOE, 7 PolgHetTwOE, 3 PolgMutTwOE). i Myofibrillar structure (black arrowhead), and mitochondrial ultrastructure (white arrowhead) in transmission electron-micrograph (n = 3 WT, 3 PolgMut, 3 TwOE, 1 PolgHetTwOE, 5 PolgMutTwOE). j, k Representative heart cardiomyocyte immunostaining; P7; laminin (j) and DAPI (k) (n = 3 WT, 1 PolgHet, 3 PolgMut, 5 TwOE, 2 PolgHetTwOE, 4 PolgMutTwOE). l, m PolgMutTwOE dilated cardiomyopathy and heart failure; echocardiography, quantified indicators: Ejection fraction (%), left-ventricular volume (µL), left-ventricular wall thickness (mm). mtDNA = mitochondrial DNA; POLG = DNA Polymerase Gamma; H&E = hematoxylin and eosin staining; Masson’s = Masson’s trichrome stain; EF = ejection fraction; LV = left ventricle. Bar plots represent mean ± SEM, boxplot representing median, Q1-Q3 as box bounds and minima-maxima as whiskers, p values from Kruskal-Wallis/Dunn’s test with Bonferroni correction, compared to WT (p < 0.05*, 0.01**, 0.001***).
The early postnatal fatality of the PolgMutTwOE genotype was caused by a dramatic rapidly progressing dilated cardiomyopathy leading to complete cardiac collapse within one week (Fig. 1g-h). At P1, the histological analysis of the hearts showed myocardial disarray, developing by P7 into interstitial fibrosis of the left ventricle wall, with cardiomyocytic lysis, vacuolation, and large hemorrhagic regions (Fig. 1h). Ultrastructural examination showed large cardiomyocytes with disrupted mitochondrial ultrastructure (Fig. 1i). The nuclei were enlarged and less numerous (Fig. 1j, k) suggesting defective polyploidization, typical for heart development within the first postnatal week16. Echocardiography at P7 confirmed heart failure of PolgMutTwOE: decreased ejection fraction and enlarged left ventricles with thin walls, characteristic of dilated cardiomyopathy (Fig.1l, m).
P7 PolgMutTwOE lungs and liver developed hepatic fibrosis and steatosis (Supplementary Fig. 1a) and disrupted alveolar structure (Supplementary Fig. 1b). While possible aggravating primary defects cannot be discarded, lungs and liver lacked the increase in mtDNA copy number at P7 (Supplementary Fig. 1c) and showed no histological sign of disease at P1 in stark contrast to the heart, suggesting a secondary organ failure likely caused by the failing heart. Blood hemoglobin and glucose were also decreased at P7 compared to WT (Supplementary Fig. 1d, e), while somatic stem cells demonstrated normal proliferation and DNA integrity (testes, skin epidermis, and hair follicles) (Supplementary Fig. 1 f). Despite its structural and functional similarities with the myocardium, skeletal muscle histology presented little-to-no changes in PolgHetTwOE (cross-sectional area or signs of degeneration), both in neonates and 3-week-old mice (Supplementary Fig. 1g), similar to the typical presentation of the late-onset parental lines.
Remarkably, PolgD257A heterozygotes (typically WT-like) also presented the early phenotype when combined with TwOE: The low body weight phenotype appears to be aggravated by the PolgD257A allele dose (Fig. 1d, g), and the penetrance of the cardiac phenotype was ~60% in PolgHetTwOE (in contrast to 100% in PolgMutTwOE).
These intriguing results indicate that licensing more mtDNA molecules to replication is highly deleterious for postnatal heart development in a background of even mild mtDNA stress. Paradoxically, the same TwOE line that is protected against genetic and ischemic cardiac damage2,3,4,5, is deleterious for the heart when combined with over-processive POLG and/or mtDNA defects in early life.
Boosted mtDNA replication exacerbates the PolgD257A mutagenic behavior
These exciting results prompted our search for the causes of the cardiac failure. As both POLG and Twinkle collaborate in the replisome, we first assessed mtDNA integrity. MtDNA nucleoid size was increased in cardiac tissue and cultured embryonic fibroblasts (MEFs) of TwOE groups (Fig. 2a, b; Supplementary Fig. 1h), as well as mtDNA replication frequency by EdU incorporation was elevated (Fig. 2c). The nucleoid enlargement was exclusive to replicating nucleoids (Fig. 2b), suggesting the delayed resolution of mtDNA daughter molecules. MtDNA deep-sequencing revealed an increased mutation load in the Mutator-TwOE-mice compared to non-TwOE controls, as expected from augmented mtDNA replication by mutagenic PolgD257A (Fig. 2d, e). The majority of misincorporations were T > A mutations (Fig. 2f), consistent with PolgD257A mutagenic behavior17. However, heteroplasmy levels were low (<10%) (Fig. 2e), and the asymptomatic PolgMut pups presented an over 3-fold higher load of mutations than the cardiomyopathic PolgHetTwOE –heterozygous for mutator allele–, excluding mtDNA mutations as causative for the phenotype (Fig. 2d–f). MtDNA topology analysis revealed a remarkable increase of abnormal mtDNA forms, with an accumulation of linear mtDNA fragments spanning the two mtDNA replication origins especially enriched in PolgMutTwOE (Fig. 2g; Supplementary Fig. 1i, j), consistent with the large nucleoids. Even these abnormal mtDNA topologies did not explain the disease manifestation pattern, as the symptomatic PolgHetTwOE did not show detectable amounts of the mtDNA rearrangements, agreeing with previous reports of cardioprotective effects of TwOE without prevention of large-scale mtDNA rearrangements3. These data suggest that the PolgD257A background –heterozygous or homozygous– can sensitize the animals to develop heart failure, without being sufficient to cause it alone.
a Replicating nucleoids in MEFs; dsDNA (nucleoids and nucleus, magenta), EdU (replicating mtDNA, green), DAPI (nucleus, blue). b Nucleoid radius distribution by genotype and replication status (classified by EdU positivity). c Percentage of replicating nucleoids over the total dsDNA foci, n = 5 + 4 + 2 + 5. Bar plots represent mean ± SEM, adjusted p values from an ANOVA/Tukey’s HSD test. d Heart mtDNA mutation rate (standardized to each base’s occurrence); P7, n = 4 for all groups; p values from two-tailed t-test, boxplot representing median, Q1–Q3 as box bounds and minima-maxima as whiskers. e Heteroplasmy percentage of each mtDNA point mutation detected over the total number of locus reads, along the full length mtDNA sequence. f Mean standardized mutation rate as in (d), by type of substitution, n = 4 for all groups; point-error plots represent mean ± SEM. g MtDNA topology; Southern hybridization analysis showing the different topological species of mtDNA present in the sample. mtDNA = mitochondrial DNA, dsDNA = double-stranded DNA, EdU = 5-ethynyl-2’-deoxyuridine, OH-OL = region between the origins of heavy and light strand of mtDNA.
Multi-omic analysis reveals disrupted cardiac metabolic maturation in double transgenics
Intrigued by the cardiac failure pathomechanism being independent of mtDNA mutations or rearrangements, we analyzed the proteome and metabolome of P1 hearts, to characterize the metabolic and signaling landscape at this critical neonatal stage. Altogether 2423 proteins were significantly changed (Qval<0.01) when compared to WT (Fig. 3a, b; Supplementary Fig. 2a), with a prominent mitochondrial integrated stress response (ISRmt) signature (Phgdh, Psat1, Asns, Mthfd2, Pycr1)18, 19 and decreased coenzyme Q biosynthesis. About 50% of the detected mito-proteome (MitoCarta3.020) was significantly changed in PolgMutTwOE, being mostly downregulated (Fig. 3c). Despite the increased mtDNA amount, mtDNA- and nuclear-encoded OXPHOS subunits were reduced (Fig. 3a and d). The response also included a robust induction of the cytosolic translation machinery (aminoacyl-tRNA synthetases, ribosomal subunits, and assembly factors) (Fig. 3a; Supplementary Fig. 2a), pointing at a cross-talk between mtDNA maintenance and cytoplasmic translation. Although mitochondrial aminoacyl-tRNA synthetases were mildly induced, the entire mito-ribosome machinery and mt-DNA encoded genes were downregulated, (Fig. 3a). Cardiomyopathy markers (e.g., Tgfb1, Cd36)) were induced, cytoskeleton/myofibrillar genes decreased (e.g., Actc1, Ctnna3, Myh6, Tnni1), and the essential cardiac metabolism coordinator ERRα is downregulated, together with many Pgc1-α targets (Supplementary Fig. 2a, b). Hopx, the transcription factor governing cardiac maturation21, was remarkably low in expression (Supplementary Fig. 2b)22,23. These data indicate that the cardiac damage signature is already present before the visible damage.
a Proteomic volcano plots of top enriched pathways from P1 proteomic analysis, PolgMutTwOE vs WT (n = 6 + 5); areas above the Q < 0.01 significance threshold shaded according to positive (red) or negative (blue) log2 fold-change. b Enriched KEGG pathways in proteomic hits, fold enrichment, and log10 p-value in lollipop plot (FDR-corrected hypergeometric p-values, see methods). c MitoCarta3.0 classification of detected proteins based on mitochondrial localization, P1. d Respiratory chain subunits change from P1 proteomics, by respiratory complex (I-IV). e Metabolomics volcano plot of PolgMutTwOE vs WT (n = 7 + 6), P1. FDR-adjusted two-tailed t-test p < 0.01 significance threshold shaded as in (a). f Simplified cysteine metabolic pathway summarizing the direction of significant changes in P1 metabolomics by color. g Heatmap of log2 fold-change of significantly affected metabolites across all measured genotypes. P values from t-test vs WT (n = 6): (p < 0.05*, 0.01**, 0.001***). h Transcriptomics volcano plot of PolgMutTwOE vs WT (n = 4 + 4), E16.5; p < 0.01 threshold shaded as in (a), significantly changed hits categorized into key pathways. i Enriched KEGG pathways in the transcriptomic hits, log10 p-value in lollipop plot (as in b). j Representative examples of ISRmt gene expression landscapes across genotypes, bar plots represent mean ± SEM. See methods for details on each p-value derivation from the omic analyzes. 1C = one-carbon, Mito = Mitochondrial protein, OXPHOS = oxidative phosphorylation machinery, NS = not significantly changed, ECM = extracellular matrix, ISRmt = mitochondrial integrated stress response.
P1 heart metabolomics showed a remarkable decrease in coenzyme A (CoA), an essential cofactor and regulator of oxidative metabolism and lipid biosynthesis (Fig. 3e, f). Its synthesis requires cysteine and pantothenate, and while cysteine was not detected by our metabolomics analysis, cystathionine (cysteine precursor) and pantothenate accumulated. Alongside the decreased downstream pathway products (hypotaurine, taurine, and glutathione) (Fig. 3e), the data suggest that increased mtDNA replication results in cysteine derivative deficiency (Fig. 3f, g). N-acetyl-L-cysteine supplementation during pregnancy was, however, insufficient to rescue the P1 proteomic remodeling: despite a mild rescue of cytoskeletal proteins, it generally replicated the PolgMutTwOE landscape and even exacerbated the loss of OXPHOS machinery. This result indicates that local cysteine production might play an essential role that externally provided cysteine cannot fulfill (Supplementary Fig. 3a, b).
To further clarify the timeline of the pathology we investigated the transcriptomic profiles of E16.5 embryos, which revealed 174 significantly changed transcripts between PolgMutTwOE and WT (Fig. 3h). The modest number of changes (in a three-fold larger gene set) suggested an earlier disease stage, characterized by the induction of ISRmt and cytosolic aminoacyl-tRNA synthetases, and Atf5 as the strongly upregulated likely effector (Fig. 3h–j). Most downregulated genes in PolgMutTwOE hearts participate in heart contraction and extracellular matrix maintenance, indicating that heart changes initiate already during embryogenesis.
Our molecular and histological data indicate that tight regulation of mtDNA replication is essential for the maturation of the postnatal heart. PolgMutTwOE hearts showed defective OXPHOS induction, fetal-like 1C-metabolism, disrupted cysteine metabolism, abnormal nuclear morphology, and disrupted myocardial structure. The first week of postnatal life is known to be crucial for heart development, entailing a gradual loss of regenerative capacity and a rapid shift from fetal glycolytic metabolism to mitochondrial-dependent ketone and lipid oxidation; eventually resulting in mature polyploid cardiomyocytes24,25. The shift involves the repression of mitochondrial 1 C metabolism25, while cysteine metabolism and CoA-biosynthesis are among the most enriched pathways during this period24. The loss of these essential pathways for cardiac homeostasis in PolgMutTwOE appears to fatally halt cardiac maturation shortly after exposure to the oxygen-rich postnatal environment.
Ferroptosis sensitivity mediated by mtDNA stress and MESH1 induction
Our multi-omics data analysis highlighted a notable enrichment of the ferroptotic pathway, with 20/38 and 11/24 of ferroptosis markers from curated ferroptosis databases26,27 altered in the P1 proteomic analysis, typically upregulated (Fig. 4a, b). Of ferroptosis suppressors, 15/30 were changed, most of them downregulated. The complete loss of OXPHOS function and coenzyme Q biosynthesis [critical factors in anti-ferroptotic lipid peroxidation maintenance28], alongside the canonical ferroptotic responses mediated by Gpx4, Chac1, Hmox1, Alas1, and other Nrf2 target genes strongly indicate the large-scale disruption of cellular homeostasis via ferroptotic cell death (Fig. 4a, Supplementary Fig. 2b), further supported by the visible rupture of cardiac mitochondrial membranes (Fig. 1i). This finding is in line with the simultaneous report of ferroptotic cell death in a model with a heart-specific deletion of the OXPHOS factor Cox10 combined with Oma1 or Dele1 ablation29. Interestingly, the data from Ahola et al. suggest a protective role for ISRmt against lipid peroxidation and ferroptosis, which begs the question of why PolgMutTwOE exhibits such a severe and acute ferroptotic signature despite the high and severity-correlated induction of ISRmt.
a Illustrative ferroptosis-related pathways colored by their proteomic (normal font) and metabolomic (italics) changes in P1 hearts. b Pie charts of ferroptosis involvement (FerrDb marker/suppressor or KEGG pathway) in PolgMutTwOE vs WT (n = 6 + 5), by significant change (Qval<0.01) in P1 heart proteomics, colors as in (a). c Time-course cell death assay for RSL3 sensitivity titration, point-error plot indicating mean and standard error of each time point (n = 3 + 3 cell lines derived from separate mice). d Ferroptosis sensitivity assay showing differential response to RSL3 and rescue by Fer-1, data from t = 6 h. e Venn diagram of significantly changed genes (FDR-corrected p value > 0.01) per genotype against WT (n = 4) in E16.5 transcriptomics, with detailed highlighted overlaps. f Scatterplot of PolgMut vs PolgMutTwOE p-values (against WT) from E16.5 transcriptomic analysis, highlighting Hddc3 in red. P values derived from multiple testing-corrected Wald test, as described in the methods. g, h Relative heart Hddc3 mRNA (g, E16.5) and MESH1 protein (h, P1, n = 5 + 1 + 6) amounts from omics data. i Relative Hddc3 mRNA amount by RT-qPCR from control and siRNA-treated MEF cells (n = 3 for all groups). j Ferroptosis sensitivity and rescue assay in control and siRNA-silenced cells, data from t = 6 h, n = 24 (untreated controls) + 18 (all other groups). Individual data points overlaid on summarized mean and standard error bars, p values from two-sided t-test unless otherwise stated: (p < 0.05*, 0.01**, 0.001***).
To functionally test the role of ferroptosis in PolgMutTwOE cells, we pharmacologically modulated ferroptosis in MEFs and studied their sensitivity. Treating the cells with different concentrations of the Gpx4-inhibitor and ferroptosis inducer drug RSL330 revealed increased cell death in PolgMutTwOE at concentrations between 10 and 100 nM (Fig. 4c, d & Supplementary Fig. 3c, d), as measured by the relative change of SYTOX-positive cells (cell death marker). The synthetic ferroptosis suppressor ferrostatin-1 (Fer-1) was sufficient to rescue the RSL3-driven cell death (Fig. 4d & Supplementary Fig. 3c–f), supporting ferroptosis as the pathway driving PolgMutTwOE cell death. In our experimental cohort, ISRmt induction was unique to TwOE genotypes and correlated with PolgD257A allele number and disease severity (Figs. 1g, 3j). We thus sought to identify factors that uniquely and dose-responsively reacted to the mutator genotype, reasoning that such a candidate would sensitize the heart, and maladaptively synergize with TwOE/ISRmt to drive the pathogenesis instead of protecting the heart.
Hddc3/MESH1 was the 5th most significantly changed transcript in E16.5 PolgMutTwOE heart, and the only one lacking an ontological classification (Fig. 3h). It was also significantly changed in all groups carrying PolgD257A alleles, in a unique dose-responsive manner and seemingly independent of TwOE or ISRmt status (Fig. 4f, g), as well as strongly upregulated as a protein in P1 hearts (Fig. 4h). PolgMutTwOE MEFs presented an approximately 10-fold increase in Hddc3 expression compared to WT MEFs (Supplementary Fig. 3g). MESH1 is the mammalian orthologue of the stringent-response-regulator SpoT, displaying both NADPH phosphatase31 and ppGpp alarmone degradation activities32. MESH1 was reported to facilitate ferroptosis, as its overexpression or downregulation renders cancer cells respectively sensitive or resistant to ferroptosis31,33. Interestingly, the integrated stress response has been suggested to mediate the protective effects of MESH1 knockdown, as ATF4 depletion seems to re-sensitize MESH1-depleted cells to ferroptosis33. Why cardiomyocytes seem vulnerable to ferroptosis is one of the outstanding questions in the field34. Heart muscle is consistently within the 5th and 6th highest expressing tissues for MESH1 in expression databases35, and there is a heart-specific genomic regulatory system that maintains MESH1 levels low to maintain cardiac homeostasis36. To dissect the putative involvement of MESH1 in PolgMutTwOE ferroptosis sensitivity, we silenced MESH1 in MEFs, and repeated the RSL3/Fer-1 pharmacological assay. Both siRNAs utilized were successful in downregulating Hddc3/MESH1 mRNA to wildtype levels (Fig. 4i), and both were able to rescue the RSL3-induced cell death phenotype similarly to Fer-1 (Fig. 4j). This strongly supports a causal role for MESH1 in sensitizing PolgMutTwOE cells to ferroptotic cell death.
While the exact mechanism behind MESH1 sensing of mtDNA replication stress remains to be elucidated, the identification of MESH1 as a putative mitochondrial DNA stress/replication rate signal and a sensitizing factor to ferroptosis in the heart in a full-body transgenic mouse brings forth exciting new possibilities for targeted therapy.
Discussion
Our evidence indicates that perinatally increased mtDNA replication activity, especially in the context of mtDNA mutations, sensitizes the mammalian heart to dilated cardiomyopathy and causes early postnatal cardiac failure. This is a noteworthy finding, as mtDNA-enhancing therapeutic approaches have been suggested for diseases caused by mtDNA mutations8,9. We boosted the replisome function by introducing an overactive mtDNA replicase together with increased licensing of mtDNA synthesis by helicase Twinkle. Despite being innocuous or even protective on their own in early life10,11,14,15, they together become acutely maladaptive for neonatal heart maturation and prevent the metabolic shift to postnatal oxidative metabolism. The simultaneous inability to activate postnatal energy metabolism (coenzyme A -dependent lipid oxidation) and promotion of growth (induction of cytoplasmic translation machinery) causes a metabolic crisis and cellular death.
Our multi-omic analysis reveals a major remodeling of 1C-metabolism with embryonic ISRmt induction, followed by the inability to undergo most of the components of typical neonatal cardiac maturation: oxidative metabolism shift, lipid catabolism, CoA biosynthesis, and nuclear polyploidization24. These components essential for cardiac homeostasis and growth render the system unable to adapt to the oxidative metabolism required for postnatal life. Altogether, our results demonstrate the importance of tight regulation of mtDNA amount and replication for healthy perinatal heart development. Indeed, mtDNA amount is known to be also low in the human liver at birth, increasing slowly during the first year37.
While the postnatal hearts present a clear ferroptotic signature, such a signal is not yet present during fetal life, except for the recently characterized inducer MESH131. The uniquely proportional increase of MESH1 along with PolgD257A allele count proposes a sensing role for MESH1 in the heart, sensitizing the tissue to ferroptosis upon non-sustainable metabolic stress. Indeed, the silencing of MESH1 in vitro could mimic the effects of the ferroptosis inhibitor Fer-1, and protect the cells from the Gpx4 inhibitor RSL3, mechanistically supporting the causal role of MESH1 in ferroptosis sensitization.
Our study introduces a mechanistically novel neonatal mitochondrial cardiomyopathy model, boosting, instead of inactivating, the function of the mitochondrial DNA replisome. Mitochondrial childhood cardiomyopathies are typically lethal human diseases, commonly caused by mtDNA expression defects38. Murine models available for cardiomypathy studies involve postnatal heart-specific knock-outs39. To our knowledge, the PolgMutTwOE is the first model of infantile mitochondrial cardiomyopathy arising from the ubiquitous transgenic expression of mitochondrial proteins, boosting mtDNA replication in backgrounds of severe or mild mtDNA stress. This model provides an invaluable tool for research on the pathogenesis of heart failure during fetal and perinatal times, as well as the interaction and involvement of other organ systems. The data highlight the importance of the mtDNA replisome as a signaler and sensor of metabolic state and as a regulator of tissue homeostasis. Furthermore, the evidence emphasizes the importance of controlled mtDNA synthesis for the perinatal metabolic shift in the heart. Our evidence also demands caution in the development of mtDNA replication-inducing approaches to treat mitochondrial dysfunction and disease, especially in early life, even pre- or asymptomatically.
Finally, we show that metabolic sensitizing factors such as MESH1 can underlie the widely unexplained acute mitochondrial disease manifestations especially in the case of mitochondrial cardiomyopathies of infants. These diseases are often triggered by external metabolic challenges such as fever or viral infection. Importantly, our data also raise the exciting possibility for the testing of ferroptosis inhibitors in the treatment of childhood mitochondrial cardiomyopathies.
Methods
Mouse models and cell culture
The mouse strains used were the Mutator mice (a kind gift from Thomas Prolla), with a homozygous knock-in D257A mutation in DNA polymerase gamma14, and the Twinkle overexpressor mice, carrying a Twnk cDNA transgene10, all in a C57BL/6 J background. The experimental group consists of the different genotype combinations arising from the crossing between those two strains (Fig. 1b), obtained by first crossing Polgwt/D257A males with TwOE females, and then crossing the resulting Polgwt/D257ATwOE to one another. The Mutator PolgD257A allele was maintained through the paternal line to avoid the inheritance of mtDNA mutations, and the Mutator-allele was present in females only in the last crossing, to result in homozygote Polg D257A/D257A offspring.
The mice were housed in controlled rooms under a 12h-12h light-dark cycle and 22 °C, with ad libitum access to water and food (Altromin 1324 standard diet). Adult mice were euthanized by gradual inhalation of excess CO2 and younger mice (<P7) by cervical dislocation. Tissues were weighed and collected immediately, snap-frozen in liquid nitrogen and stored at −80 °C or histologically processed as detailed below.
Mouse embryonic fibroblasts (MEFs) were extracted from E16.5 embryos using standard procedures: dissociation of the embryo body (without head, heart and liver) using a scalpel and followed by incubation in trypsin:EDTA, plating in gelatine-coated plates and growth in standard MEF media.
The animal work was performed according to regulations and approval of The Ethical Review Board of Finland (permit: ESAVI/3686/2021).
DNA isolation
DNA was extracted from frozen samples by standard phenol-chloroform extraction. For extraction of DNA from paraffinized samples, 6-7 slices of paraffin were cut from the block. Paraffin was removed by washing the slides twice with xylene and the samples were rehydrated in a descending ethanol series (100%, 90%, 75%). The samples were dried and the extraction was continued according to the standard protocol, adding a 20-minute incubation at 90 °C to de-crosslink DNA at the end of the proteinase K incubation.
RNA isolation and RT-qPCR
RNA was extracted using Trizol reagent (Invitrogen) and purified through Qiagen RNeasy minicolumns (Qiagen), following the manufacturer’s instructions.
For RT-qPCR, cDNA was synthesized with Maxima First Strand cDNA Synthesis Kit (Thermo Scientific) according to the manufacturer’s instructions. 100 ng of RNA was used in the cDNA synthesis and the cDNA was diluted 1:4 before its use in RT-qPCR. RT-qPCR was performed with SensiFAST™ SYBR No-ROX Kit (BIO-98020, Bioline) and the relative gene expression levels were normalized against β-actin expression. The primers used are listed in the supplementary material (Supplementary Table 1).
mtDNA quantitation
MtDNA copy number was quantified by quantitative PCR (qPCR) using the SensiFAST™ SYBR No-ROX Kit (BIO-98020, Bioline), by amplification of three mtDNA fragments (12S-rRNA, COX1 and ND4) against nuclear-DNA-encoded RBM15. 25 ng of DNA was used per PCR reaction, all qPCR assays were run in triplicates. The primers used are listed in the supplementary material (Supplementary Table 1).
Histology & immunofluorescence
Formalin-fixed paraffin-embedded tissues or frozen sections were used for histological stainings and histochemical activity analyzes. For immunohistology, standard protocols were used with slight modifications together with manufacturer instructions. Antigen retrieval from paraffin-derived samples was done in 10 mM citric acid buffer pH 6,0. The antibodies used are found in the supplementary material. Frozen sections were used for the laminin immunostaining and the OilRedO staining.
For immunofluorescence, coverslips with cultured cells were fixed in 4% PFA at room temperature for 15 min and washed with PBS. Slides were blocked in blocking buffer (1% BSA, 0.1% TritonX and 10% horse serum in PBS), followed by overnight primary antibody incubation in antibody buffer (1% BSA, 0.1% TritonX and 1% horse serum in PBS). Following PBS washing, samples were incubated for 1 hour at room temperature in secondary antibodies conjugated with Alexa Fluor fluorescent probes (Thermo Fisher Scientific) diluted 1:400 in 1% PBS. Finally, the coverslips were mounted onto slides with DAPI-containing medium (Vectashield #H-1200- 10).
EdU mtDNA replication assay
EdU incorporation into nucleoids was performed using the Click-iT™ EdU Alexa Fluor™ 488 Imaging Kit (Fisher Scientific), following the manufacturer’s instructions with minor deviations (non-confluent cells incubated for 1 h in 50 μM EdU-containing media, and two sequential Click reactions to ensure detection). The resulting fixed cells were blocked and immunostained with a dsDNA antibody (details in supplementary material) and DAPI, to identify nucleus and mtDNA nuclei as above.
Microscopy and image analysis
For transmission electron microscopy, fresh heart samples were fixed in 2.5% glutaraldehyde (EM-grade) in 0.1 M sodium phosphate buffer for 2 h, RT, and transferred into 0.1 M sodium phosphate buffer (pH 7.4) after fixation. The samples were post-fixed with 1% reduced osmium tetroxide in 0.1 M sodium phosphate buffer for 1 h, on ice. Samples were again washed with phosphate buffer and then dehydrated through a series of ethanol and acetone prior to gradual infiltration into Epon (TAAB 812, Aldermaston, UK). The resin was polymerized at 60 °C for a minimum of 16 hours. A pyramid was trimmed on the tissue and 60-70 nm sections were cut with an ultramicrotome and placed on Pioloform-coated copper grids. Sections were post-stained with uranyl acetate and lead citrate. TEM micrographs were acquired using a JEM-1400 transmission electron microscope (Jeol Ltd., Tokyo, Japan) running at 80 kV, with a bottom-mounted CCD camera (Orius SC 1000B, AMETEK-Gatan Inc., Pleasanton, CA), with images of 4008 × 2672 pixels.
Fluorescent images were acquired using the Andor Dragonfly spinning disk confocal microscope, and image quantification was performed with the CellProfiler software, using its standard segmentation and classification modules.
Echocardiography
To analyze cardiac function and dimensions of the left ventricular chamber, P7 pups were anesthetized by inhalation with 2,5% isoflurane mixed with 0.5 L/min 100% oxygen (Vevo Compact Dual Anesthesia System). The mice were placed on a warm stage (Vevo Imaging Station) in supine position and 2,5% isoflurane mixed with 0.5 L/min 100% oxygen was supplied continuously via a nose cone to maintain the mice under sedation. Pre-warmed ultrasound gel was applied to the thoracic area and two-dimensional ECG images were acquired using MS550D 22-50 MHz linear array solid-state transducer (Vevo 2100 Ultrasound, FUJIFILM VisualSonics). Long-axis ECG images in B-mode were used to confirm the anatomic boundaries of the ventricular chambers. M-mode images along the parasternal short axis were used to measure left ventricular internal diameter, left ventricular posterior wall thickness, and interventricular septum thickness at end-systole and end-diastole. These parameters were used to calculate left ventricular mass, volume, ejection fraction, and fractional shortening (Vevo Vasc Analysis software).
MtDNA sequencing
mtDNA was enriched from whole P7 heart DNA by long-range PCR amplification of three fragments; processed and analyzed as in ref. 40, using the Phusion hi-fidelity enzyme to ensure a negligible impact of PCR mutagenesis to the expected mutation rates (one-to-two orders of magnitude lower). DNA concentrations were measured with Qubit and quality was assessed using Genomic DNA ScreenTape Assay (Agilent TapeStation 4200). 100 ng of pooled amplicon DNA (DIN > 6.8) was converted into sequencing libraries using NEBNext Ultra II FS DNA Library Prep Kit for Illumina. Completed libraries were pooled and then sequenced with a NextSeq Mid Output 300 cycle flow cell on the NextSeq 500 to produce 2 × 150 bp reads.
Several approaches were taken to account for potential PCR mutation and nuclear mitochondrial DNA segments (NUMTs). On the former, the LoFREQ* detection pipeline accounts for non-strand-matched SNPs, further minimizing the impact of amplification on the detection. The lack of NUMT contamination was corroborated by two approaches: by probing for the appearance of two or more of the defined SNPs in a thoroughly characterized NUMT (spanning mtDNA regions 4950–5940)41, and by ensuring an unaffected mutation load in regions with a known high-identity NUMT (mtDNA 6390 – 11043 region inserted in chromosome 1)40.
MtDNA topology
The analysis of mtDNA topology was performed as in ref. 42, 1 μg of total DNA was separated over a 0,4 % agarose gel (UltraPure Agarose, #16500, Life Technologies) without ethidium bromide in 1x TBE buffer overnight at 32 V. The gel was treated for 2 × 15 min with 0,25 M HCl and 2 × 20 min with 0,5 M NaOH, 1,5 M NaCl, transferred by capillary blot onto Hybond XL-nylon membrane and hybridized in Church’s buffer with a cytochrome b probe (nucleotides 14783–15333 of mouse mtDNA). Uncropped gel in Supplementary Fig. 4.
RNA sequencing
The sequencing services were provided by the Biomedicum Functional Genomics Unit at the Helsinki Institute of Life Science and Biocenter Finland at the University of Helsinki.
3’UTR RNA sequencing “Bulkseq” was performed as in ref. 43 using whole E16.5 heart RNA (isolated as described above). Shortly, Bulkseq involves mRNA priming with an oligo dT primer which also contains a 12 bp sample barcode and an 8 bp UMI sequence, which can be used to remove PCR duplicates during data analysis. Single-stranded cDNA is then converted to double-stranded cDNA using the template switch effect and the double-stranded cDNA product is then amplified with PCR and another set of primers (SMART PCR primer). Samples are then pooled together, and PCR sequencing pools are made with Nextera i7 primers and the Dropseq P5 primer. Sequencing is then performed on the NextSeq High Output 75 cycle flow cell on the NextSeq 500 to produce (Read 1: 20 bp, Index 1 (i7): 6 bp and Read 2 (61 bp)).
For data analysis, bcl2fastq2 was used to convert BCL files to FASTQ file format and demultiplex the samples. Sequenced reads were trimmed for adapter sequence and reads shorter than 20 nt were also removed using the next-generation sequencing preprocessing tool Trimmomatic (v0.40). Read processing was performed using drop-seq tools (version 2.4.0); In short, reads were additionally filtered to remove polyA tails of length 6 or greater and then trimmed reads were aligned to GRCm38 reference genome (GENCODE Mouse Release 25) using STAR aligner (2.7.6a). Gene expression was quantified with drop-seq tools after removing PCR duplicates using unique molecular identifier -sequences (UMIs).
Differential expression analysis was done in the DESeq2 software in the R environment. The count values were normalized between samples using a geometric mean. Sample-wise factors were estimated to correct for library size variability and estimation of dispersion (i.e. variance, scatter) of gene-wise values between the conditions. A negative binomial linear model and Wald test were used to produce p-values. Low-expression outliers were removed using Cook’s distance to optimize the p-value adjustment and finally, multiple testing adjustment of p-values was done with the Benjamini-Hochberg procedure. For volcano plot visualization, genes directly affected by the TwOE transgene (Twnk, Tmprss11a, Ugt2b37) were cropped.
Targeted metabolomic profiling analysis
Metabolites were extracted from 6–10 mg P1 mouse heart tissue using a 2 mL Precellys homogenization tube (Bertin Technologies, Montigny-le-Bretonneux, France) with 2.8 mm ceramic (zirconium oxide) beads with 400 µL of cold extraction solvent (Acetonitrile:Methanol:Milli-Q Water; 40:40:20). Subsequently, samples were homogenized using a tissue homogenizer (Bertin Technologies, France) for 3 cycles (30 sec at 3 000 × g with 60 sec pause at 4 °C). Followed by centrifugation at 20 000 × g at 4 °C for 5 minutes. The supernatant was loaded into a Phenomenex, Phree Phospholipid removal 96 well plate 30 mg (Part No. 8E-S133-TGB) and passed through using a robotic vacuum. The filtrate was transferred into polypropylene tubes and placed into a nitrogen gas evaporator to dry the solvent completely. Dried samples were suspended with 40 µL of mobile phase solvent (Acetonitrile: 20 mm Ammonium Hydrogen Carbonate Buffer, pH 9.4; 80:20) and vortex for 2 minutes and transfer into HPLC glass autosampler vials. 2 µL of sample injected with Thermo Vanquish UHPLC coupled with Q-Exactive Orbitrap quadrupole mass spectrometer equipped with a heated electrospray ionization (H-ESI) source probe (Thermo Fischer Scientific). A SeQuant ZIC-pHILIC (2.1 × 100 mm, 5 μm particle) column (Merck) used for chromatographic separation. Gradient elution was carried out with a flow rate of 0.100 mL/minutes using 20 mM ammonium hydrogen carbonate, adjusted to pH 9.4 with ammonium solution (25%) as mobile phase A and acetonitrile as mobile phase B. The gradient elution was initiated from 20% mobile phase A and 80% of mobile phase B and maintained till 2 min. After that, 20% mobile phase A gradually increase up to 80% till 17 min, then 80% to 20% Mobile phase A decreased in 17.1 min and maintained up to 24 minutes. The column oven and auto-sampler temperatures were set to 40 ± 3 °C and 5 ± 3 °C, respectively. MS equipped with a heated electrospray ionization (H-ESI) source using polarity switching and the following setting: resolution of 35,000, the spray voltages: 4250 V for positive and 3250 V for negative mode, the sheath gas: 25 arbitrary units (AU), and the auxiliary gas: 15 AU, sweep gas flow 0, Capillary temperature: 275 °C, S-lens RF level: 50.0. Instrument control operated with the Xcalibur 4.1.31.9 software (M/s Thermo Fischer Scientific, Waltham, MA, USA).
In the data processing, the final peak integration was done with the TraceFinder 4.1 software (Thermo Fischer Scientific) using confirmed retention times of 462 metabolites in-house library developed using library kit MSMLS-1EA (Merck). For further data analysis, the peak area data were exported as an excel file. Data quality was monitored throughout the run using a pooled sample as Quality Control (QC) prepared by pooling 5 µL from each suspended sample and interspersed throughout the run as every 10th sample. After integration of QC data with TraceFinfer 4.1 detected metabolites were checked for peak, % RSD were calculated, and acceptance limit was set ≤20%. Blank samples for carryover were injected after every fifth randomized sample to monitor the metabolites’ carryover effect and calculated against the mean QC area and the acceptance limit was set ≤ 20% for each metabolite. Background % noise was calculated with respect to the first blank against the mean QC area and the acceptance limit was set ≤20% for each metabolite.
The obtained data were analyzed and explored using the MetaboAnalyst software following the recommended path. The data was normalized by the sum of the sample, Generalized log transformation (glog2) and Autoscaling (mean-centered and divided by the standard deviation of each variable) settings were used.
Proteomic analysis
The mass spectrometry analyzes proteomic were performed at the Turku Proteomics Facility supported by Biocenter Finland, following their pipeline. Shortly, proteins were isolated from approximately five (5) mg of snap-frozen hearts pulverized with polypropylene grinding rods in liquid nitrogen, extracted with lysis buffer (6 M Urea, 100 mM TEAB and 1 x Halt protease inhibitor cocktail) and sonication (3 × 5 min cycles of 30 s on/off; Diagenode Bioruptor sonication device). Extracted proteins were digested to peptides with in-solution digestion performed at the Turku Proteomics Facility, desalted with a Sep-Pak C18 96-well plate (Waters) and evaporated to dryness and stored at −20 °C.
Digested peptide samples were dissolved in 0.1% formic acid and peptide concentration was determined with a NanoDrop device. For DIA analysis 800 ng peptides was injected and analyzed in random order. Wash runs were submitted between each sample to reduce the potential carry-over of peptides. The LC-ESI-MS/MS analysis was performed on a nanoflow HPLC system (Easy-nLC1200, Thermo Fisher Scientific) coupled to the Orbitrap Lumos Fusion mass spectrometer (Thermo Fisher Scientific) equipped with FAIMS interface and a nano-electrospray ionization source. Peptides were first loaded on a trapping column and subsequently separated inline on a 15 cm C18 column (75 μm x 15 cm, ReproSil-Pur 3 μm 120 Å C18-AQ, Dr. Maisch HPLC GmbH, Ammerbuch-Entringen, Germany). The mobile phase consisted of water with 0.1% formic acid (solvent A) or acetonitrile/water (80:20 (v/v)) with 0.1% formic acid (solvent B). A 110 min gradient was used to elute peptides (70 min from 5% to 21% solvent B followed by 40 min from 21 % to 36 min solvent B). Samples were analyzed by a data-independent acquisition (DIA) LC-MS/MS method. MS data were acquired automatically by using Thermo Xcalibur 4.1 software (Thermo Fisher Scientific). In a FAIMS-DIA method, a duty cycle contained two compensation voltages (-50V and -70V) with one full scan (400-1000 m/z) and 30 DIA MS/MS scans covering the mass range 400–1000 with variable width isolation windows in each of the compensation voltages. Data analysis consisted of protein identifications and label-free quantifications of protein abundances. Data were analyzed by Spectronaut software (Biognosys; version 15.0.2). DirectDIA approach was used to identify proteins and label-free quantifications were performed with MaxLFQ. Main data analysis parameters in Spectronaut included - Enzyme: Trypsin/P, Missed cleavages: 2, Fixed modifications: Carbamidomethyl, Variable modifications: Acetyl (protein N-term) and oxidation (M), Protein database: Swiss-Prot 2021_02, Mus musculus, Normalization: Global median normalization, Differential abundance testing: Unpaired t-test and multiple-testing correction of the p-values with Benjamini-Hochberg method (Q-values). P-values for KEGG pathway enrichment were obtained from false discovery rate-corrected hypergeometric p-values as calculated by g:profiler44.
Ferroptosis sensitivity in vitro assays
To assess ferroptosis sensitivity, cells were plated in multi-well plates (96- or 24-well plates) and media was replaced for SYTOX Green Dead Cell Stain-containing media 20 minutes before the treatment, according to manufacturer’s instructions (Invitrogen, S34860). Depending on the treatment, SYTOX-containing MEF media was supplemented with: RSL3 (Sigma, SML2234) in concentrations ranging between 0.1–100 nM, and Ferrostatin-1 (MedChem, HY-100579), at a concentration of 10 μM. Cells were imaged every hour in an IncuCyte S3 Live-Cell Analysis Platform (Essence BioScience, Sartorius). The segmentation and quantification of the images was performed using the IncuCyte software. For the manual replication of the experiment, SYTOX images were obtained using an EVOS FL Digital Inverted Microscope (Invitrogen), and cells were manually counted using a hemocytometer.
Hddc3/MESH1 silencing
To knock-down Hddc3 expression, two Silencer™ siRNA against Hddc3 were used (siRNA1 ID# s206122, siRNA2 ID# s86962) along with the Silencer™ Select Negative Control (Invitrogen, 4390846). 24 hours after seeding, the cells were treated with Lipofectamine® RNAiMAX Transfection Reagent (Thermo, 13778030), Opti-MEM™ I Reduced Serum Medium, (Gibco, 31985062) and the specific siRNA following manufacturer’s instructions (1 pmol of siRNA per well of a 96-well plate). The transfection medium was replaced by fresh MEF media after 24-hours, and 24-hours later the ferroptosis sensitivity assay was performed as described above. The efficacy of the siRNA treatment was assessed by RT-qPCR.
Statistics and reproducibility
The experimental cohorts were generated as detailed in the section mouse models and cell culture. Each mouse was considered an independent biological replicate (n), and the experimentation largely consisted in comparing different genotypes and their littermate controls. Due to the limited amount of biological material (e.g. minute neonatal hearts), many methods required the entirety of the sample and thus the n number varies across measurements, ensuring the statistical power and optimizing the use of the material.
Mouse sampling and data collection was performed blinded to genotype, and data analysis (including image analysis and omics data) was performed in blinded bulk pipelines. No data were excluded from the analyzes. Sampling occurred perinatally, all mice were sampled and grouping was performed by genotype, thus all sampling and analysis was blinded to and included both sexes.
By nature of the experimental design, most data arise from several independent cohorts from different times, parental combinations and litters, while always containing litter-mate controls. The reported phenotype was replicated in different litters from different parental lines across three years and a large number of crossings. Statistically strong groups and strict multiple comparison adjustments were used to ensure the robustness of the molecular results, detailed in each method section or in the figure legends.
For in vitro experiments cell lines derived from 5 WT, 4 TwOE, 2 PolgMut and 5 PolgMutTwOE embryos were used, with the ferroptosis sensitivity experiments using 3 + 3 WT and PolgMutTwOE lines, independently treated (siRNA and drug treatments performed independently) across multiple experiments (3 and 2 independent experiments for baseline sensitivity assay and siRNA modulation respectively). Technical replicates were used to obtain experimental means, and only independently treated repetitions were considered biological replicates and used for data visualization and statistical analysis.
Unless otherwise stated, all data points are clearly displayed in the data visualization and represent biological replicates, frequently overlayed on boxplots [representing the median as the center, the two quartiles (Q1 and Q3) as box bounds, and the maxima and minima as whiskers (outliers defined as beyond 1.5 times the inter-quartile range IQR)], or point/bar plots with error bars (representing the mean and SEM). Statistical tests were performed on biological replicates and are specified in the legends or method section for each analysis. Unless otherwise specified, all statistical tests were two-sided, and the exact p-values are provided in the figure legends.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data generated or analyzed during this study are included in this published article, its supplementary files, source data, or available in the following repositories: Sequencing data have been deposited at the NCBI SRA database; accession: PRJNA830670. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD047603. Other supplementary data including the detailed results of the omics analyzes and representative videos of echocardiograms can be found in the Mendeley Data repository with the identifier https://doi.org/10.17632/wm2dhwpghm.145. Source data are provided with this paper.
References
Gorman, G. S. et al. Mitochondrial diseases. Nat. Rev. Dis. Prim. 2, 16080 (2016).
Ikeda, M. et al. Overexpression of TFAM or twinkle increases mtDNA copy number and facilitates cardioprotection associated with limited mitochondrial oxidative stress. PLoS ONE 10, e0119687 (2015).
Pohjoismäki, J. L. et al. Overexpression of Twinkle-helicase protects cardiomyocytes from genotoxic stress caused by reactive oxygen species. Proc. Natl Acad. Sci. USA. 110, 19408–19413 (2013).
Tanaka, A. et al. The overexpression of twinkle helicase ameliorates the progression of cardiac fibrosis and heart failure in pressure overload model in mice. PLoS ONE 8, e67642 (2013).
Inoue, T. et al. Twinkle overexpression prevents cardiac rupture after myocardial infarction by alleviating impaired mitochondrial biogenesis. Am. J. Physiol. - Heart Circ. Physiol. 311, 509–519 (2016).
Filograna, R. et al. Modulation of mtDNA copy number ameliorates the pathological consequences of a heteroplasmic mtDNA mutation in the mouse. Sci. Adv. 5, eaav9824 (2019).
Jiang, M. et al. Increased total mtdna copy number cures male infertility despite unaltered mtdna mutation load. Cell Metab. 26, 429–436.e4 (2017).
Bianco, A. et al. High mitochondrial DNA copy number is a protective factor from vision loss in heteroplasmic leber’s hereditary optic neuropathy (LHON). Invest. Ophthalmol. Vis. Sci. 58, 2193 (2017).
Filograna, R., Mennuni, M., Alsina, D. & Larsson, N. G. Mitochondrial DNA copy number in human disease: the more the better? FEBS Lett. 595, 976–1002 (2021).
Tyynismaa, H. et al. Twinkle helicase is essential for mtDNA maintenance and regulates mtDNA copy number. Hum. Mol. Genet. 13, 3219–3227 (2004).
Ylikallio, E., Tyynismaa, H., Tsutsui, H., Ide, T. & Suomalainen, A. High mitochondrial DNA copy number has detrimental effects in mice. Hum. Mol. Genet. 19, 2695–2705 (2010).
Ekstrand, M. I. et al. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum. Mol. Genet. 13, 935–944 (2004).
Hämäläinen, R. H. et al. Defects in mtDNA replication challenge nuclear genome stability through nucleotide depletion and provide a unifying mechanism for mouse progerias. Nat. Metab. 1, 958–965 (2019).
Kujoth, G. C. et al. Medicine: mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science 309, 481–484 (2005).
Trifunovic, A. et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature 429, 417–423 (2004).
Soonpaa, M. H., Kim, K. K., Pajak, L., Franklin, M. & Field, L. J. Cardiomyocyte DNA synthesis and binucleation during murine development. Am. J. Physiol. - Heart Circ. Physiol. 271, 2183–2189 (1996).
Johnson, A. A. & Johnson, K. A. Fidelity of nucleotide incorporation by human mitochondrial DNA Polymerase. J. Biol. Chem. 276, 38090–38096 (2001).
Kühl, I., et al. Transcriptomic and proteomic landscape of mitochondrial dysfunction reveals secondary coenzyme Q deficiency in mammals. eLife 6, e30952 (2017).
Nikkanen, J. et al. Mitochondrial DNA replication defects disturb cellular dNTP pools and remodel one-carbon metabolism. Cell Metab. 23, 635–648 (2016).
Rath, S. et al. MitoCarta3.0: An updated mitochondrial proteome now with sub-organelle localization and pathway annotations. Nucleic Acids Res. 49, D1541–D1547 (2021).
Friedman, C. E. et al. HOPX-associated molecular programs control cardiomyocyte cell states underpinning cardiac structure and function. Dev. Cell 59, 91–107.e6 (2024).
Huss, J. M. et al. The nuclear receptor ERRα is required for the bioenergetic and functional adaptation to cardiac pressure overload. Cell Metab. 6, 25–37 (2007).
Wang, T. et al. Estrogen-related receptor α (ERRα) and ERRγ are essential coordinators of cardiac metabolism and function. Mol. Cell. Biol. 35, 1281–1298 (2015).
Lalowski, M. M. et al. Characterizing the key metabolic pathways of the neonatal mouse heart using a quantitative combinatorial omics approach. Front. Physiol. 9, 365 (2018).
Pohjoismäki, J. L. et al. Postnatal cardiomyocyte growth and mitochondrial reorganization cause multiple changes in the proteome of human cardiomyocytes. Mol. Biosyst. 9, 1210–1219 (2013).
Zhou, N. & Bao, J. FerrDb: A manually curated resource for regulators and markers of ferroptosis and ferroptosis-disease associations. Database 2020, baaa021 (2020).
Kanehisa, M., Sato, Y., Kawashima, M., Furumichi, M. & Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457–D462 (2016).
Deshwal, S. et al. Mitochondria regulate intracellular coenzyme Q transport and ferroptotic resistance via STARD7. Nat. Cell Biol. 25, 246–257 (2023).
Ahola, S. et al. OMA1-mediated integrated stress response protects against ferroptosis in mitochondrial cardiomyopathy. Cell Metab. 34, 1875–1891 (2022).
Dixon, S. J. et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell 149, 1060–1072 (2012).
Ding, C. C. et al. MESH1 is a cytosolic NADPH phosphatase that regulates ferroptosis. Nat. Metab. 2, 270–277 (2020).
Sun, D. et al. A metazoan ortholog of SpoT hydrolyzes ppGpp and functions in starvation responses. Nat. Struct. Mol. Biol. 17, 1188–1194 (2010).
Lin, C.-C. et al. The regulation of ferroptosis by MESH1 through the activation of the integrative stress response. Cell Death Dis. 12, 1–10 (2021).
Ahola, S. & Langer, T. Ferroptosis in mitochondrial cardiomyopathy. Trends Cell Biol. 34, 150–160 (2023).
Uhlen, M. et al. Towards a knowledge-based human protein atlas. Nat. Biotechnol. 28, 1248–1250 (2010).
Huang, R. et al. A long non-coding RNA LncSync regulates mouse cardiomyocyte homeostasis and cardiac hypertrophy through coordination of miRNA actions. Protein Cell 14, 153–157 (2023).
Morten, K. J. et al. Liver mtDNA content increases during development: A comparison of methods and the importance of age- and tissue-specific controls for the diagnosis of mtDNA depletion. Mitochondrion 7, 386–395 (2007).
Vasilescu, C. et al. Genetic basis of severe childhood-onset cardiomyopathies. J. Am. Coll. Cardiol. 72, 2324–2338 (2018).
Ruzzenente, B., Rötig, A. & Metodiev, M. D. Mouse models for mitochondrial diseases. Hum. Mol. Genet. 25, R115–R122 (2016).
Isokallio, M. A. & Stewart, J. B. High-throughput detection of mtDNA mutations leading to tRNA processing errors. Methods Mol. Biol. 2192, 117–132 (2021).
Hagström, E., Freyer, C., Battersby, B. J., Stewart, J. B. & Larsson, N.-G. No recombination of mtDNA after heteroplasmy for 50 generations in the mouse maternal germline. Nucleic Acids Res. 42, 1111–1116 (2014).
Goffart, S. et al. Twinkle mutations associated with autosomal dominant progressive external ophthalmoplegia lead to impaired helicase function and in vivo mtDNA replication stalling. Hum. Mol. Genet. 18, 328–340 (2009).
Freitag, T. L. et al. Gliadin nanoparticles induce immune tolerance to gliadin in mouse models of celiac disease. Gastroenterology 158, 1667–1681.e12 (2020).
Raudvere, U. et al. G:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 47, W191–W198 (2019).
Landoni, J. C. et al. Overactive mitochondrial DNA replisome causes neonatal heart failure via ferroptosis - Supplemental data. 1, 10.17632/wm2dhwpghm.2 (2022).
Acknowledgements
The authors wish to thank Markus Innilä, Tuula Manninen, Sonja Jansson, Babette Hollmann, Satu Malinen, and Maria Arraño de Kivikko for their technical contributions and expertize; Amy Platt, Aurora Hämälainen and the team of animal caretakers are thanked for their excellent handling and flexibility. We want to thank Howard Jacobs for the enriching scientific discussion, and Olesia Ignatenko, Gulayse Ince Dunn, Rocio Maldonado, Swagat Pradhan, Saara Forsström, Nahid Khan, and Anastasiia Marmyleva for their support and technical contributions. We thank the core facilities that enabled the performance of this work: the Electron Microscopy Unit at the Institute of Biotechnology, the Biomedicum Functional Genomics Unit, the FIMM Metabolomics Unit (funded by Biocenter Finland and HiLIFE), the Turku Proteomics Facility at Biocenter Finland, and Franziska Metge of the Bioinformatics Core facility at MPI-AGE, Cologne, Germany. This work was supported by the following funders (recipient initials in parentheses): Sigrid Jusélius Foundation (AS, RK), Research Council of Finland (AS [361873, 345248], RK [297245]), Helsinki University Hospital (AS), Max Planck Society (JS), Finnish Foundation for Cardiovascular Research (RK), University of Helsinki Funds (JCL), Maud Kuistila Foundation (JCL), Päivikki ja Sakari Sohlberg Foundation (JCL), and the Biomedicum Helsinki Foundation (JCL).
Author information
Authors and Affiliations
Contributions
Conceptualization: J.C.L., A.S. Investigation: J.C.L., S.E., T.L., S.G., J.S., R.K., K.S., A.N., S.W. Funding acquisition: A.S., J.C.L. Supervision: A.S. Writing – original draft: J.C.L., A.S. Writing – review& editing: J.C.L., S.E., J.S., T.L., S.G., R.K., K.S., A.N., S.W., A.S.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Rita Horvath, Gary Lopaschuk and Douglas Wallace for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Landoni, J.C., Erkul, S., Laalo, T. et al. Overactive mitochondrial DNA replication disrupts perinatal cardiac maturation. Nat Commun 15, 8066 (2024). https://doi.org/10.1038/s41467-024-52164-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-52164-1