Abstract
Aberrant repair underlies the pathogenesis of pulmonary fibrosis while effective strategies to convert fibrosis to normal regeneration are scarce. Here, we found that thyroid hormone is decreased in multiple models of lung injury but is essential for lung regeneration. Moreover, thyroid hormone receptor α (TRα) promotes cell proliferation, while TRβ fuels cell maturation in lung regeneration. Using a specific TRβ agonist, sobetirome, we demonstrate that the anti-fibrotic effects of thyroid hormone mainly rely on TRβ in mice. Cellularly, TRβ activation enhances alveolar type-2 (AT2) cell differentiation into AT1 cell and constrains AT2 cell hyperplasia. Molecularly, TRβ activation directly regulates the expression of KLF2 and CEBPA, both of which further synergistically drive the differentiation program of AT1 cells and benefit regeneration and anti-fibrosis. Our findings elucidate the modulation function of the TRβ-KLF2/CEBPA axis on AT2 cell fate and provide a potential treatment strategy to facilitate lung regeneration and anti-fibrosis.
Similar content being viewed by others
Introduction
The lung is the primary organ targeted by a great variety of environmental and microbial insults, which result in lung diseases accounting for one-sixth of all deaths worldwide1,2,3. Functional lung regeneration is required for the restoration of normal lung homeostasis after injury, while aberrant repair represents the vital pathogenesis feature of many acute and chronic lung diseases4,5,6. Indeed, pulmonary fibrosis (PF) is caused by consistent lung remodeling and scarring7. However, what goes awry in abnormal lung repair at cellular and molecular levels remains elusive, and there are no effective therapeutic strategies to convert fibrosis to normal regeneration.
The distal lung has evolved an elegant architecture, the alveolar, lined by two types of epithelial cells, the AT2 cell and the AT1 cell, forming a large and tight barrier for gas exchange8. AT1 cells cover 95% of the alveolar surface with a flat and thin shape, while AT2 cells are cuboidal and play multifaceted roles in pulmonary surfactant generation and recycling and host defense9. Most importantly, AT2 cells serve as local progenitor cells for AT1 cells, facilitating alveolar epithelial turnover through precisely coordinated proliferation and differentiation5,10,11. Disruption of the balance between proliferation and differentiation leads to hyperplasia or metaplasia of AT2 cells, underlying many life-threatening lung diseases, such as idiopathic pulmonary fibrosis (IPF), acute lung injury (ALI), and lethal coronavirus disease 2019 (COVID-19)2,12,13,14,15,16,17. However, the molecular signals that drive activated AT2 cells differentiation into AT1 cells are poorly understood.
Decreased serum thyroid hormone (TH) concentrations in a range of severe diseases have been noted since the 1970s, known as nonthyroidal illness syndrome (NTIS). However, the functional significance and regulation mechanism of the low TH remain enigmatic18,19,20. The thyroid gland secretes predominantly prohormone thyroxine (T4) and ~ 20% of circulating T3 (the most bioactive form) under the regulation of the hypothalamic-pituitary-thyroid (HPT) axis. Strikingly, peripheral conversion from T4 to T3 by iodothyronine deiodinase (DIO) provides the remaining 80%, in which DIO1 contributes more circulating T3 and DIO2 determines the intracellular T3 on account of their unique subcellular localization21,22,23. We have reported that TH inhibited lung fibrosis by restoring mitochondrial function in AT2 cells24. Indeed, hypothyroidism is more prevalent in IPF, is an independent predictor of IPF mortality, and is the true causal determinant of IPF based on analysis of clinical data25,26,27. In addition to regulating metabolism, TH is indispensable for the biological processes of growth, development, and tissue homeostasis by orchestrating cell proliferation and differentiation28,29,30, but these functions are less studied in lung repair after injury. Furthermore, TH primarily depends on thyroid hormone receptors (TRs) to convert the repressed state to an active state of target genes’ transcription (canonical TH signaling pathway by genomic binding), but whether TRα and TRβ have different functions in lung regeneration is still unknown.
Here, we show that the various lung injury mouse models consistently present low T3 in serum, but T3 is essential for lung regeneration in local lung cells. Using overexpression and pharmacological activation of TRβ with sobetirome (a specific TRβ agonist, also known as GC-1)31,32,33,34, we identified the unique function of TRα for cell proliferation and TRβ for cell differentiation. Further, we evaluated the efficiency and safety of GC-1 in multiple PF models. Using multi-omics analysis and lineage-tracing mice, we ascertained the functions of TRβ activation in inhibiting AT2 cell hyperplasia and prompting terminal differentiation of AT1 cells. Mechanistically, we revealed a direct transcriptional control of TRβ on lung Kruppel-Like Factor 2 (KLF2) and CCAAT-enhancer binding protein alpha (CEBPA) and verified the impacts of these two transcriptional factors (TFs) on AT1 cell differentiation. The use of conditional knockout (CKO) mice and adeno-associated viruses (AAV) confirmed that KLF2 and CEBPA are required for TRβ to promote lung regeneration and anti-fibrosis.
Results
Thyroid hormone is decreased in serum after lung injury but is required for lung regeneration
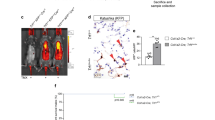
To test TH levels in serum after lung injury, we individually constructed multiple mouse models with lipopolysaccharide (LPS, 1 mg/kg), bleomycin (BLM, 1 U/kg), and silica (SiO2, 200 mg/kg) via lung intubation delivery to minimize the direct effect on other organs (Fig. 1a–c). The T3 and T4 levels significantly decreased in all models, with the T3 change being more dramatic (Fig. 1a–c). To unveil the causes underlying this phenomenon, we examined indicators of the HPT axis and the deiodinases of TH in peripheral tissues. The thyroid stimulating hormone (TSH) levels were not decreased (Fig. 1a–c). In contrast, the Tpo (thyroid peroxidase, an enzyme for TH production) mRNA expression in the thyroid gland was significantly decreased in the BLM model (Supplementary Fig. 1a). Furthermore, the circulating T3-regulated enzyme Dio1 was decreased in the lung, thyroid, and liver (Dio1 is highly expressed in thyroid and liver), demonstrating that the conversion from T4 to T3 and the diffusion to serum were reduced (Supplementary Fig. 1b). The intracellular T3-regulated enzyme Dio2 consistently increased in all injured lungs (Fig. 1d). In fact, DIO2 expression exhibited a consistent increase in ALI, IPF, chronic hypersensitivity pneumonitis (CHP), and lethal COVID-19 patients (Supplementary Fig. 1c). The increased Dio2 implied a requirement for T3 in the local cell to repair after injury.
a–c The levels of serum T3, T4, and TSH in mice after lung challenge with LPS (n = 6), BLM (n = 10, TSH n = 6), and SiO2 (n = 7, 9; 8, 8; 8, 8) at 24-hour, 14-day, and 56-day, individually. d Dio2 mRNA level of lung homogenates (n = 4). e, f Illustration of the experiment design and line graph show the right lung Dio2 mRNA dynamic change after left lung PNX (n = 4). g, h Representative gross pictures of the right lung and the internal volume at day 21 after PNX (n = 3). Hypothyroidism (Hypo) was induced by PTU in drinking water and TDX. i, j Representative H&E staining of lung sections and mean liner intercept of alveolar (n = 9, from 3 mouse lung). Arrows indicated the thickened alveolar walls. Scale bar, 100 μm. k–n AT2 lineage tracing mice treated with T3 after hypothyroidism and PNX construction (k), mRNA levels of proliferation and growth marker in lung homogenates (l, n = 4), and immunofluorescence images (m, the insets are shown on the right with individual fluorescence channels; tdT, tdTomato) and quantification (n, n = 5, from 3 mouse lung) of lung sections show the KI67-positive cells. The value of n indicates biologically independent samples (a–d, f, h, l). Data of different sections from three biologically independent mice (j, n). Similar results were repeated in two biologically independent experiments. Data are presented as mean ± SEM. P-values were obtained by two-tailed unpaired Student’s t test (a–d) and one-way ANOVA with Turkey’s multiple comparisons (h, j, l, and n).
After injuries, the repair process can be divided into two phases: cell activation (proliferation, growth, and migration) and re-quiescence (cell cycle arrest, maturation, and differentiation)35. To investigate the effects of TH on these sequential phases of lung regeneration precisely, we constructed a relatively normal regeneration model, the left pneumonectomy (PNX) (Fig. 1e), which exhibits maximum proliferation at day 5 and climaxed differentiation at day 14 in the remnant right lung36. Indeed, Dio2 maintained a high level in the regenerating lung during both the cell proliferation and differentiation phases (Fig. 1f). The enhancement of right lung size and internal volume after PNX was weakened at day 21 in the hypothyroidism group induced by propylthiouracil (PTU) combined with thyroidectomy (TDX) (Fig. 1g, h). Hypothyroidism thickened the alveolar walls and impeded the mean linear intercept increase after PNX, as shown by hematoxylin and eosin (H&E) staining; moreover, T3 administration rescued these phenotypes (Fig. 1i, j). In the proliferation phase, 5 days after PNX, T3 rescued the mRNA expression of proliferation markers Mki67, Pcna, Mcm6, and Aurkb, which was inhibited by hypothyroidism (Fig. 1k, l). The same pattern of KI67 expression was also detected at the protein level with immunofluorescence (IF) images (Fig. 1m, n).
In summary, these results suggested that decreased synthesis and conversion in the thyroid and peripheral tissues cause low serum T3 after lung injury. More importantly, T3 is essential for regeneration in the local lung.
TRα promotes cell proliferation, while TRβ facilitates AT2 cells differentiation into AT1 cells in lung regeneration
As a ligand, TH mainly exerts its function through the nuclear receptors TRα (THRA) and TRβ (THRB). THRA is widely expressed, with a higher level in smooth muscle cells and fibroblasts than others, while THRB is mainly expressed in alveolar epithelial cells, as analyzed using the Human Lung Cell Atlas (HLCA)37 and the Mouse Cell Atlas (MCA)38 (Fig. 2a). There being no TRα-specific agonist, in order to investigate the different effects between TRα and TRβ activation on cell proliferation (first phase of lung repair), we overexpressed THRA and THRB in MRC5 (human fetal lung fibroblasts), followed by T3 treatment. In contrast to TRβ activation, which scarcely affected proliferation, TRα activation significantly promoted the cell proliferation and growth tested by the EdU assay and markers (KI67, AURKB, and EIF4E) at mRNA level (Fig. 2b and Supplementary Fig. 2a). Intriguingly, THRA facilitated the expression of THRB, whereas THRB inhibited the expression of THRA (Supplementary Fig. 2b). Further, a well-studied TRβ agonist GC-131,32,33,34 was chosen to verify the effects of TR activation. GC-1 demonstrated about 14-fold specificity to TRβ comparable to that of T3, consistent with the literature (Supplementary Fig. 2c)31. Under these treatments, only T3 fueled the proliferation and migration of MRC5 compared with GC-1 at an equal molar concentration in the EdU and wound healing assays (Fig. 2c and Supplementary Fig. 2d). These results, combined with Fig. 1, indicated that TRα activation mainly contributes to the proliferation and growth of lung cells (consistent with the recent finding in brown adipose tissue39) and implied that TRβ functions for epithelial cell differentiation (Supplementary Fig. 2e).
a The heatmap of indicated gene expression in the HLCA and MCA data. b EdU assay in MRC5 after THRA and THRB overexpression for 24 h with 10 nM T3 for another 24 h (n = 3). Scale bar, 100 μm. c EdU assay in MRC5 with or without 10 nM T3 or GC-1 treatment (n = 5). Scale bar, 100 μm. d Workflow of the intervention with hypothyroidism and GC-1 in the differentiation phase of the PNX model. e qPCR analysis of AT1 cell markers (Ager, Hopx, and Pdpn) in lung homogenates (n = 3, 3, 4, 4). f, g Representative IF images and quantification show AT1 cells differentiated from AT2 cells (indicated by arrows) (n = 3). Scale bars, 50 μm. h–k Experimental workflow for 3D-organoid culture and GC-1 treatment. Cont., continued GC-1 from day 0. Late, GC-1 for the last 4 days (h). Representative images of colonies in wells, H&E staining, and IF. Scale bars, top 2 mm, middle and bottom, 50 μm (i). Colony average area and colony-forming efficiency of 3D-organoids (n = 4) (j). Quantification of AT1 cell markers by qPCR (n = 4) (k). The value of n indicates biologically independent samples. Similar results were repeated in two biologically independent experiments. Data are presented as mean ± SEM. Statistical significance among groups was determined using one-way ANOVA.
To investigate TRβ activation on cell differentiation in the late phase of PNX, hypothyroidism was induced starting at day 6 (Fig. 2d). The increase of AT1 cell markers was dampened by hypothyroidism and rescued by GC-1 treatment on day 14 (Fig. 2e). The IF images and quantification confirmed this effect of TRβ activation on AT1 cell differentiation (Fig. 2f, g). Because primary AT2 cells spontaneously differentiate into AT1 cells in plastic culture vessels, we cultured primary lineage AT2 cells with an air-liquid interface 3D-organoid system to assess the effects of TRβ activation on differentiation in vitro over 16 days (Fig. 2h). After GC-1 treatment in the late 4 days, the structure of the cell spheroid became hollower and larger with AGER-marked AT1 cells differentiation toward the lumen (Fig. 2i). The colony area and AT1 cell markers in mRNA level were significantly increased in the late-GC-1 group, although colony-forming efficiency did not significantly change (Fig. 2j, k). Based on this pro-differentiation benefit, we hypothesize that TRβ activation exerts mainly anti-fibrotic effects in the late phase of lung repair. Therefore, BLM-induced fibrotic mice were individually treated with an equal molar dosage of GC-1, MGL-3196 (another TRβ-specific agonist40) and T3 from day 11 to day 20 to compare the effects (Supplementary Fig. 2f). Hydroxyproline levels and Masson’s trichrome staining exhibited the same patterns in treatment groups, suggesting that the anti-fibrotic effect of TH mainly depends on TRβ (Supplementary Fig. 2g, h).
Taken together, TRα promotes cell activation in the early phase of lung repair, while TRβ facilitates epithelial cell differentiation in the late phase of regeneration. Further taking into account the side effects of TRα activation on tachycardia and bone loss41, TRβ activation with a specific agonist would be a superior option to T3 in pulmonary fibrosis treatment.
TRβ activation is an efficient strategy to treat lung fibrosis in two mouse models
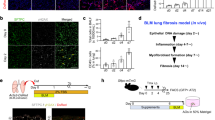
Next, we choose GC-1 to evaluate the efficiency and safety of TRβ agonist in lung fibrosis because GC-1 is well-studied and commonly used31,32,33,34 and because of the similarity of TRβ agonists (Supplementary Fig. 2f–h). The dose-response relationship exhibited that GC-1 impeded hydroxyproline with a maximum effect at 20–40 μg/kg (Fig. 3a, b). 30 μg/kg was chosen in all the following experiments in vivo. This dose of GC-1 is comparable to 8-fold of the physiological replacement by T3 on TRβ activation and presumably will maintain its high specificity on TRβ (Supplementary Fig. 2c)31,32,34,42. Systemic administration of GC-1 resulted in a significant decrease in hydroxyproline (Fig. 3c). The lung internal volume under 25 cm H2O pressure, which indicates static compliance, was markedly raised (Fig. 3d). GC-1 reduced total lung weight, white blood cell (WBC) counts and protein content in bronchoalveolar lavage fluid (BALF) (Fig. 3e–g). After GC-1 treatment, mice presented a rescued body weight compared to the BLM group (Fig. 3h). A higher dose of BLM (2 U/kg) was used to investigate the effects of GC-1 on survival (treatment advanced to day 7). In contrast to pirfenidone, GC-1 significantly improved overall survival (Fig. 3i). Analyses of the protein and mRNA levels of extracellular matrix (ECM) markers (COL1A1, TN-C, COL3A1), myofibroblast and mesenchymal cell markers (α-SMA, VIM, N-Cad), the epithelial cell marker (E-Cad), and pro-fibrogenic cytokines (CTGF, SPP1) from lung homogenates confirmed the antifibrotic and pro-epithelialization effects of GC-1 (Fig. 3j, k). Furthermore, X-ray micro-computed tomography (micro-CT) renditions showed that opacifications were distinctly mitigated in the therapy group. Histological staining demonstrated diminished cystic airspaces, traction bronchiectasis/bronchiolectasis, immune cell infiltration, and ECM deposition, suggesting the restriction of lung pathological remodeling with GC-1 (Fig. 3l).
a Diagram of GC-1 delivery starting on day 11 after the bleomycin challenge. b Right lung hydroxyproline contents with gradient GC-1 treatment from 5 to 80 μg/kg (n = 5). c Right lung hydroxyproline contents (n = 8, 8, 8, 7) with or without GC-1 30 μg/kg. d Total lung internal volume under static pressure of 25 cm H2O for 1 h (n = 4). e Total lung wet weight (n = 10, 10, 9, 10). f, g Protein content and white blood cell count in BALF (f, n = 11, 12, 8, 8; g, n = 8, 9, 8, 8). h Percentage of body weight change (n = 12, 11, 10, 12). i Percent survival of mice treated as indicated from d7 after 2U/kg BLM challenge (n = 10). j Immunoblotting of ECM proteins (COL1A1, FN1), mesenchymal cell markers (N-Cad, α-SMA, VIM), and the epithelial cell marker (E-Cad) in lung homogenates. k qPCR analysis of indicated genes in the lungs (n = 3). l Representative micro-CT images, H&E, and trichrome-stained lung sections at d21 after BLM. Scale bar of whole sections, 3 mm; Scale bar of 10 × images, 100 μm. The value of n indicates biologically independent samples. Similar results were repeated in three biologically independent experiments. Data are presented as mean ± SEM. P-values were obtained by one-way ANOVA with Turkey’s multiple comparison and log-rank (i), respectively.
Next, the chronic and irreversible PF model induced by SiO2 was used to further test the anti-fibrotic effect of GC-1 (Supplementary Fig. 3a). GC-1-treated mice showed a significant decline in lung hydroxyproline and soluble collagen (Supplementary Fig. 3b, c). The whole lung internal volume, inflammatory cells protein content in BALF, and fibrosis markers in protein and mRNA levels manifested remarkably reversed effects of GC-1 (Supplementary Fig. 3d–h). GC-1 suppressed the progress of fibrotic tissues surrounding the silica nodules, albeit not enough to eliminate the nodules (Supplementary Fig. 3i).
To assess the safety of GC-1, the serum of mice was collected to assay the liver and kidney functions. The levels of alanine aminotransferase (ALT), aspartate aminotransferase (AST), and creatinine (CREA) did not change significantly after GC-1 administration (Supplementary Fig. 4a–c). Although serum T3 didn’t change in disease groups after GC-1 treatment, it declined in health groups (Supplementary Fig. 4d), demonstrating a critical role of TRβ in TH negative feedback22. Aerosol delivery would be an alternative route to strengthen organ specificity and minimize the systemic effects. Using the nose-only inhalation tower, serum T3 did not significantly decline with GC-1 inhalation, but the anti-fibrotic effects were retained, as verified by hydroxyproline, WBC in BALF, and histology (Supplementary Fig. 4e–i).
Activation of TRβ constrains the hyperplasia and metaplasia of Maladaptive-AT2 cells in lung fibrosis
To decipher the influences of TRβ activation on biological processes in fibrosis, we performed bulk RNA-seq and scRNA-seq in the lung after BLM challenge with or without GC-1 treatment. As predicted, the Thrb regulon (co-expression and significant motif enrichment of the correct upstream regulator analysis) was activated primarily in epithelial cells in scRNA-seq data (Fig. 4a, b). In bulk RNA-seq, GC-1 down-regulated the processes of epithelium proliferation and morphogenesis and up-regulated epithelium development (Fig. 4c). Focused on the epithelium, AT2 cells present several aberrant states in PF identified by scRNA-seq recently, such as basal-like state (basaloid, KRT5-/KRT17-)43,44,45 and AT2-to-AT1 transitional cell state (Krt8+/Cldn4+)2,12,13,46. We reanalyzed these data and found that they are extremally similar subpopulations, exhibiting the ostensibly contradictory characteristics of senescence, apoptosis, and proliferation (Supplementary Fig. 5a, b). To unify, we termed these dysregulated AT2 cells Maladaptive AT2 (M-AT2) cells with markers Krt8, Cldn4, etc. TRβ activation compromised the expression of M-AT2 cell markers (Supplementary Fig. 5c). In immunohistochemistry (IHC), SFTPC-marked AT2 cells were sporadically scattered in the corner of alveoli with a small and cuboidal shape in healthy mice; in stark contrast, aggregated AT2 cells were located adjacent to thickened-interstitial regions with a swollen or stretched shape in the BLM group, which returned to normal after GC-1 treatment (Fig. 4d). GC-1 also attenuated the increase of proliferation markers in fibrotic lung in RNA-seq data (Supplementary Fig. 5d). Based on these findings, we hypothesized that TRβ activation exerts its pro-regeneration and anti-fibrotic effects by modulating the cell fate of M-AT2 cells.
a The uniform manifold approximation and projection (UMAP) plot displays cells colored by cell type identity in scRNA-seq. b UMAP visualization of Thrb regulon activity. c GO enrichment analysis of GC-1-regulated genes in differential expression between BLM and PBS groups in mouse lung bulk RNA-seq (n = 3). d Immunochemistry staining for SFTPC and KRT8 in lung sections of mice, same as Fig. 3. Arrows indicate normal AT2 cells, and arrowheads indicate elongated or hypertrophic AT2 cells. Scale bar, 100 μm. e Protein analysis in lung homogenates for M-AT2 cell marker CLDN4. f, g Workflow, representative IF images, and quantification of Ki67-positive AT2 cells in lineage-labeled mice lungs with or without GC-1 after BLM challenge. Arrowheads indicate basaloid cells from lineage-labeled cells (indicated by arrows) (n = 3). Scale bar, 50 μm. h Representative pictures and quantification of EdU assay (n = 12, images from four experiments) in A549. GC-1 15 nM for 36 h. Scale bar, 100 μm. i–l AT2 cells isolated from different groups were used to stimulate normal primary fibroblasts for 48 h. The activation of lung fibroblasts was assayed by the 3D-collagen gel contraction (j, n = 3), expression of a-SMA (k, Scale bar, 20 μm), COL1A1, and FN1 (l). The value of n indicates biologically independent samples (c, g, j). Data of different views from four biologically independent samples (h). Similar results were repeated in two biologically independent experiments. Significant differences were assessed using two-tailed unpaired Student’s t test (h) and one-way ANOVA with Tukey test (g, j). Results are presented as mean ± SEM.
KRT8+ epithelial cells were only found in the airways of the control group but presented in fibrotic alveoli with hyperplasia and malformation morphology, which was reversed by GC-1 (Fig. 4d). The protein level of CLDN4 in lung homogenates suggested the increase of M-AT2 cells and the rescue effects of GC-1 (Fig. 4e). Based on the hyperplastic property of M-AT2 cells in fibrosis (Supplementary Fig. 5b)14,47,48,49, we stained the Ki67 in AT2-lineage mice and found that the proliferation cell predominantly presented in the fibrotic region with airway-like morphology, which was significantly restricted by GC-1 at day 21 (Fig. 4f, g). In A549 and MLE12 cell lines, which all possess a hyperplastic characteristic, EdU assay, colony formation assay, CCK8, and cell cycle detection thoroughly confirmed the anti-hyperplastic effect of TRβ activation by GC-1 (Fig. 4h and Supplementary Fig. 5e–g).
M-AT2 cells exhibited damage-associated molecular patterns (DAMPs) and a senescence-associated secretory phenotype (SASP) (Supplementary Fig. 5b)2,13. To investigate whether AT2 cells stimulate fibroblasts by paracrine in PF, we individually isolated the AT2 cells from the BLM and GC-1 groups to co-culture with normal primary fibroblasts (Fig. 4i). AT2 cells from fibrotic mice profoundly stimulated fibroblast contractility in the 3D collagen-gel contraction assay and the expression of α-SMA, COL1A1, and FN1, which was diminished with AT2 cells from GC-1-treated mice (Fig. 4j–l).
In summary, TRβ activation hampered the metaplasia and hyperplasia of M-AT2 cells; as a result, it curbed the stimulation of fibroblasts by M-AT2 cells in pathological remodeling.
TRβ activation promotes M-AT2 cells differentiation into AT1 cells in the fibrotic lung
AT1 cell differentiation and maturation are required to restore lung function after injury. We performed pseudotime analysis with the progression of epithelial differentiation in scRNA-seq. The RNA velocity analysis demonstrated that the differentiation of AT1 cells from AT2 cells was blocked in Krt8+ cells, and GC-1 administration restored the trajectory close to normal (Fig. 5a). AT1 cell markers in bulk RNAseq demonstrated the impeded AT1 cell differentiation in fibrosis and the rescue effects of GC-1, which were also verified by immunoblotting and qPCR (Fig. 5b–d).
a RNA-velocity analysis of AT2 cells, Krt8+ cells, and AT1 cells in scRNA-seq data. The arrows indicate the predicted lineage trajectories. b Heatmap showing the average FPKM value of AT1 cell genes per group in mouse lung bulk RNA-seq (same as Fig. 3c, n = 3). c, d Protein and mRNA levels of AT1 cell markers in lung homogenates (n = 3), same as Fig. 3. e AT2-lineage tracing mice were used to detect AT2 differentiation with a time-lapse. f Representative IF images from lung cryosections stained with antibodies against KRT8 and PDPN. Arrows point to KRT8+ cells, arrowheads indicate AT1 from lineage-labeled AT2 differentiation, and unfilled arrows point to KRT8+ lineage-labeled cells with basaloid morphology (n = 3). Scale bars, 50 μm. g Quantification of indicated cells in (f) (left, n = 15; right, BLM n = 10, BLM + GC-1 n = 11). h The percentage of KT8+tdT+ cells were tested by flow cytometry at d14 after lung digestion (n = 3). i, j Immunoblotting and qPCR analysis of AT1 marker genes after GC-1 treatment with dose gradient in A549 (j, n = 3) and MLE12. The value of n indicates biologically independent samples (d, h, j). Data of different sections from three biologically independent mice (g). Similar results were repeated in two biologically independent experiments. Significant differences were obtained using one-way ANOVA with Turkey’s multiple comparison tests. Error bars, SEM.
To precisely investigate the effects of TRβ activation by GC-1 on the differentiation of alveolar epithelial cells, we added more time points of analysis and extended the duration of GC-1 in AT2-lineage mice after the BLM challenge (Fig. 5e). In the control group, lineage-labeled AT2 cells were relatively quiescent, with none expressing KRT8. In contrast, M-AT2 cells (tdT+ KRT8+) emerged at all detection points and exhibited airway-like morphological features at day 28 in the BLM group (Fig. 5f). Indeed, GC-1 minimized the accumulation of M-AT2 cells and facilitated their differentiation into AT1 cells, as scored by the KRT8+ and PDPN+ proportions and further verified by flow cytometry (Fig. 5g, h). In cell lines, GC-1 showed the best ability to increase PDPN and HOPX expression at 10-20 nM in protein and mRNA levels (Fig. 5i, j). Taken together, TRβ activation with GC-1 facilitates M-AT2 cell differentiation into AT1 cells in the fibrotic lung.
TRβ orchestrates KLF2 and CEBPA to facilitate AT1 cell differentiation
Given the decisive role of TFs in cell fate and the fact that TRβ is a member of the steroid receptor-coregulator transcriptional complex, we computed the principal components (PCs) of TFs in bulk RNA-seq. The top five PCs explained 95% of the RNA variance, in which PC-2 and PC-5 exhibited the therapeutic effects of GC-1 (Fig. 6a). Furthermore, we found that Id2, Cebpa, Cenpb, Klf2, and Atf4 are the top five genes in terms of high quality and contribution to the PC-2 and PC-5 (Fig. 6b). However, only Klf2 and Cebpa were retained with the regulon analysis in public AT2-scRNA-Seq (Supplementary Fig. 6a). In fact, CEBPA mainly expressed in AT2 cells2,12 and KLF2 has a high expression level in endothelial cells50 and AT2 cells. KLF2 and CEBPA decreased in all ALI, IPF, and CHP whole lung data (Supplementary Fig. 6b), while KLF2 was slightly increased in AT2 cells in IPF scRNA-seq data, presumably indicating a stress response (Supplementary Fig. 6c). IHC staining in IPF lung sections verified these data (Supplementary Fig. 6d).
a Principal component analysis (PCA) of TFs in bulk RNA-Seq of mouse lung (same as Fig. 3c, n = 3). The lower and upper bounds of the boxplot correspond to the first and third quartiles (the 25th and 75th percentiles); whiskers represent minima/maxima or 1.5*IQR. b Histogram showing the contributions of TFs in PC.2 and PC.5. c, d Protein and mRNA levels (n = 3) of KLF2 and CEBPA in the mouse lung, as shown in Fig. 3a. e qPCR analysis of KLF2 and CEBPA expression in isolated AT2 cells from mice (n = 3). f, g Immunoblotting and qPCR analysis (n = 3) of KLF2 and CEBPA in A549 and MLE12 with a GC-1 gradient. h The mRNA level of Klf2 and Cebpa in primary AT2 cells after 10 nM GC-1 treatment for 6 h (n = 3). i–l Protein (i, k) and mRNA (j, l) tests of AT1 cell markers, KLF2, and CEBPA expression in A549 after KLF2 (i, j) and CEBPA (k, l) overexpression (n = 3). m Luciferase activity of KLF2, CEBPA, and AT1-marker promoters cloned in pGL3.0 after THRB transfection for 36 h with or without GC-1 10 nM for 12 h (n = 3). Values were normalized to the transfection vector. n, o Luciferase activity of indicated promoters after KLF2 (n) and CEBPA (o) transfection with 600 ng plasmid for 36 h (n = 3). p–r TRβ (p), KLF2 (q), and CEBPA (r) bind to the promoter regions (pro) of KLF2, CEBPA, and AT1 cell genes in ChIP q-PCR assays (n = 3). KRT5 promoter as negative control. Values were normalized to IgG. s Luciferase activity of AT1-maker promoters with KLF2 (300 ng) and CEBPA (300 ng) co-transfection for 36 h, as shown in (n, o) (n = 3). t IF image of KLF2 and CEBPA after co-transfection in A549. Scale bar, 10 μm. u CoIP of KLF2 and CEBPA overexpressed in A549. v Illustration of the regulatory processes of GC-1 on AT1 cell markers. The value of n indicates biologically independent samples. Similar results were repeated in two biologically independent experiments. The statistical tests used were one-way ANOVA (d, e, and g), two-tailed unpaired Student’s t test (h), and two-way ANOVA (j, l, m–s). Data are mean ± SEM.
To empirically test the effects of TRβ activation on KLF2 and CEBPA, we performed molecular detection in vivo and manipulation in vitro. GC-1 increased the expressions of KLF2 and CEBPA after BLM challenge at protein and mRNA levels in lung homogenates and isolated AT2 cells, further confirmed by IHC (Fig. 6c–e and Supplementary Fig. 6e). CEBPA ___location and expression in AT2 cells were also verified by IF images with or without GC-1 treatment (Supplementary Fig. 6f). Consistent with AT1 cell markers, GC-1 maximized KLF2 and CEBPA at concentrations of 10–20 nM in A549 and MLE12 cell lines (Fig. 6f, g). This impact was likewise demonstrated in primary AT2 cells (Fig. 6h). KLF2 and CEBPA both increased the expression of AT1 cell marker genes at the protein and mRNA levels when overexpressed in A549 (Fig. 6i–l). Moreover, KLF2 demonstrated the most potent effect on CAV1 and PDPN, while CEBPA exhibited the most potent effect on AGER (Fig. 6i–l). Interestingly, KLF2 promoted the expression of CEBPA but not vice versa (Fig. 6i–l).
We noticed that the promoter regions of CEBPA and KLF2 contain the classical direct repeat-4 (DR4) of TH response elements (TRE). To determine whether they are positive TRE, we cloned the promoters into pGL3 vectors to perform dual-luciferase reporter assays. Transfection of TRβ increased the luciferase activity of KLF2 and CEBPA promoters, and GC-1 further increased the KLF2 promoter activity (Fig. 6m). TRβ with GC-1 activation exhibited relatively weak effects on cross-species conserved promoters of PDPN, CAV1, HOPX, and AGER (Fig. 6m); therefore, the effects of TRβ on AT1 differentiation are mainly indirect. The binding of TRβ on KLF2 and CEBPA promoters was confirmed by chromatin immunoprecipitation qPCR (ChIP-qPCR) (see below). By the same methodology, KLF2 and CEBPA activated all the tested promoters of AT1 cell marker genes, demonstrating the preference of KLF2 on CAV1, PDPN, and HOPX and CEBPA on the AGER (Fig. 6n–r). Because of their similar functions, we examined whether KLF2 and CEBPA interacted synergistically or not. When co-transfected with a half dose of KLF2 (300 ng) and CEBPA (300 ng), the activity of the AT1 marker gene promoters did not obviously decline compared to the total dosage of each single one (600 ng), suggesting a synergistic effect (Fig. 6s, n, o). Moreover, we observed a clear co-___location of KLF2 and CEBPA on confocal images (Fig. 6t). To identify whether the interaction depends on the DNA chain, we assayed co-immunoprecipitation (Co-IP) with or without DNase, and the results suggested that their interaction mainly relied on the direct interaction independent of the DNA chain (Fig. 6u).
In summary, TRβ activation by GC-1 directly induced KLF2 and CEBPA expression, and KLF2/CEBPA synergistically drove the program of AT1 cell differentiation (Fig. 6v).
The benefits of TRβ activation are dependent on KLF2 and CEBPA in vivo
To determine whether the pro-regeneration and anti-fibrotic benefits of TRβ activation were mediated through KLF2, we generated Sftpc-CreERT2; Klf2flox/flox mice (AT2-KLF2-KO, Fig. 7a and Supplementary Fig. 7a). AT2-KLF2-KO mice exhibited significantly higher hydroxyproline, WBC and protein content in BALF, and COL1A1 expression than control littermates with or without bleomycin challenge. Albeit GC-1 slightly suppressed fibrosis in AT2-KLF2-KO mice, the therapeutic efficacy was significantly diminished (Fig. 7b–f and Supplementary Fig. 7c, d). AT2-KLF2-KO mice weakened the AT1 cell markers at protein and mRNA levels and were not rescued by GC-1 (Fig. 7e and Supplementary Fig. 7c). KRT8+ cells strikingly accumulated in AT2-KLF2-KO mice after BLM challenge, as detected by IF (Fig. 7g, h). In the PNX model, AT2-KLF2-KO blunted the elevation of AT1 cell markers in lung homogenates when tamoxifen was injected in the differentiation phase (Fig. 7i, j). On the other hand, AT2-KLF2-KO promoted Ki67-positive cell counts during the proliferation phase (Supplementary Fig. 7e, f).
a Schematic of the Klf2 Cre-loxp knockout site in AT2 cells and experimental design. b Hydroxyproline content in the right lung (n = 6, 6, 9, 6, 9, 6). c, d Protein concentration (n = 8, 7, 9, 7, 9, 7) and WBC numbers (n = 6, 6, 8, 7, 8, 7) in BALF. e Immunoblotting for COL1A1 and AT1 cell markers in lung homogenates. f Representative images of trichrome staining. Scale bar of whole sections, 2 mm; scale bar of 10 × images, 100 μm. g, h Representative IF staining and quantification of KRT8+ cells in AT2-KLF2-KO mice (n = 5). Arrows show the KRT8+ cells. Scale bars, 50 μm. i, j The mRNA levels of AT1 cell markers in the lung of AT2-Klf2-KO mice at the differentiation phase in the PNX model. The value of n indicates biologically independent samples (b–d, j). Data of different sections from three biologically independent mice (h). Similar results were repeated in two biologically independent experiments. Throughout, data are mean ± SEM. P-values were obtained by one-way ANOVA with Turkey’s multiple comparison tests.
To examine whether CEBPA is required for TRβ function, we delivered the AAV2/9 viruses expressing Cebpa short hairpin RNA (shCebpa) into the lungs to knock down Cebpa expression (Fig. 8a and Supplementary Fig. 8a, b). The hydroxyproline, lung weight, and WBC/protein in BALF were significantly higher than the control, and GC-1 did not counteract these impacts (Fig. 8b–d). Cebpa knockdown decreased the survival rate (Fig. 8e). The fibrotic markers and AT1 cell markers at protein and mRNA levels, trichrome, micro-CT, and H&E staining corroborated the weakened functions of TRβ activation in the Cebpa deficiency group (Fig. 8f, g and Supplementary Fig. 8d, e). We likewise delivered shCebpa into AT2-lineage mice to confirm the role of CEBPA in AT1 cell differentiation in the PNX model (Fig. 8h). shCebpa dramatically promoted the hyperplasia of AT2 cells and limited their differentiation into AT1 cells (Fig. 8i, j). Corresponding to the results in vitro (Fig. 6i–l), AT2-KLF2-KO substantially reduced Cebpa expression, and the reverse direction of this effect was relatively weakened (Supplementary Fig. 7b and Supplementary Fig. 8c).
a Workflow of AAV-shCebpa delivery, BLM challenge, and GC-1 treatment. b Hydroxyproline level (n = 6, 6, 7, 6, 7, 6). c Total lung wet weight (n = 7, 6, 5, 6, 6, 6). d WBC counts (n = 6, 6, 6, 5, 6, 6) and protein content (n = 7, 7, 7, 6, 7, 6) in BALF. e Line plots of the percent survival from 7 to 21 days (n = 10, 20, 20). f Protein levels of fibrotic markers and AT1 cell markers. g Representative images of trichrome staining. Scale bar of whole sections, 3 mm; Scale bar of 10 × images, 100 μm. h–j Representative images of AT2 cell hyperplasia and quantification of AT1 cell differentiation in AT2-lineage cells after shCebpa delivery in the PNX model (n = 5). The value of n indicates biologically independent samples (b–e). Data of different sections from three biologically independent mice (i). Similar results were repeated in two biologically independent experiments. Scale bar, 50 μm. Throughout bars, mean ± SEM. P-values were obtained by one-way ANOVA with Turkey’s multiple comparison test and log-rank (e), respectively.
In summary, Klf2 deletion or Cebpa deficiency in AT2 cells deteriorated BLM-induced PF, aggravated AT2 cell hyperplasia, blocked AT1 cell differentiation, and blunted the therapeutic benefits of GC-1.
Discussion
Multiple models exhibited a consistently low serum T3 level after lung injury. We found that the reduced synthesis and the conversion from T4 to T3 in peripheral tissue, but not the hypothalamus-pituitary, contributed to the low T3 in serum. Hypoxia and inflammation factors presumably mediated these systemic effects in the acute phase of lung injury19,20,51. Most importantly, the local T3 in the lung is indispensable for lung regeneration in the PNX model. This exquisite and delicate regulation of T3 saved energy expenditure in the whole body while maintaining or enhancing the energy supply for local activation and repair processes, given that alveolar regeneration is an energetically costly process52.
As a progenitor of AT1 cells, AT2 cells are the most pleiotropic and powerful cells in the physiological distal lung, implying that they will be the most intricate and formidable cells in pathological conditions53. AT2 cells concurrently demonstrated the phenotypes of cell proliferation, apoptosis, senescence, and partial epithelial-mesenchymal transition (pEMT) in IPF by reanalyzing the public data2,12,13,46. However, these phenotypes are incompatible with the original conceptions, implying an abnormal cell state and impaired re-epithelialization. These maladaptive AT2 cells may be arrested into a permanent transitional state between AT2 cells and AT1 cells (Krt8+)2,54 or transdifferentiate into a basaloid cell state (KRT17+/KRT5-)43,44,45, exhibiting hyperplasia and metaplasia in pathological remodeling and acting as a driving factor in lung fibrosis by recruiting macrophages and regulating chemokines54. But whether the Krt8+ state and basaloid cell state are the same and their cellular origin except for AT2 cells warrant further validations55. Evolutionarily, lung or other tissue repair requires recapitulation of the developmental pathways (TGF-β, WNT, Notch, etc.) to re-growth56,57, but how to circumvent hyperplasia and metaplasia and return homeostasis is more critical35,58,59. We and others previously substantiated the potential of TH to treat various lung disease models, including ventilator-induced lung injury60, BLM, TGF-β, radiation, and silica-induced fibrosis24,61,62. Moreover, T3 is able to antagonize the TGF-β signaling by reducing phosphorylation of SMADs through TRα or TRβ in vitro and attenuate liver and skin fibrosis in mice model63. In this study, we found that TRα is mainly for lung fibroblast proliferation, while TRβ exerts the main effects on promoting AT1 cell differentiation and constraining AT2 cell hyperplasia. This different role in the regulation of proliferation between TRs is consistent with the recent findings in cardiomyocytes64 and brown adipose tissue39. Therefore, TRβ-specific activation is a more promising way to convert pathological remodeling to regeneration by regulating the epithelial cell differentiation trajectory. As a result, the stimulation effects on fibroblasts by maladaptive AT2 cell paracrine were decreased.
To track the downstream following TRβ, the two TFs, KLF2 and CEBPA, were mined by a series of bio-information, molecular, and literature screen methods. KLF2 is primarily expressed in the lung and has anti-proliferation properties in many cancers65. KLF2 has shown the ability to suppress fibrosis in liver cirrhosis and heart failure and attenuate inflammation and fibroblast activation in lung fibrosis66,67,68. Chimeric KLF2-KO mice showed un-dilated alveoli with abundant AT2 cells and few AT1 cells at embryonic day-18.569,70, indicating that KLF2 is essential for lung maturation. Using AT2-specific knockout mice, we found that KLF2 is essential for AT1 cell differentiation in BLM and PNX models. Consistent with the results in the mouse cell line (MLE12)69, KLF2 directly promotes the expression of AT1 cell markers in the human cell line (A549). In contrast to the enhancer of KLF2 in MLE12, which is a negative TRE69, the human promoter of KLF2 cloned in A549 is a positive TRE in this study. Furthermore, TRβ activation increased KLF2 expression at low doses while suppressing it at high doses, indicating a sophisticated control of TH signaling on KLF2.
CEBPA is also a tumor suppressor gene whose expression is limited to fully differentiated cells. It could coordinate cellular differentiation with growth arrest by repressing the E2F complex71. CEBPA is essential for AT1 cell differentiation in development72,73 and has demonstrated anti-fibrotic effects in liver and lung fibroblasts74,75. Convincingly, GC-1 significantly rescued the decrease of CEBPA in AT2 cells in the fibrotic lung through the direct binding of TRβ on the promoter region of CEBPA. We found that CEBPA knockdown resulted in AT2 cell hyperplasia and blocked AT1 cell differentiation in PNX and BLM models, further exacerbating fibrosis and damping the effects of TRβ activation.
Interestingly, KLF2 promotes the expression of CEBPA, but not vice versa. KLF2 possesses the highest effect on PDPN and HOPX expression, and CEBPA has more potential on AGER expression. Actually, PDPN and HOPX are early AT1 cell markers, whereas AGER is a late AT1 cell marker12, suggesting that KLF2 plays an initial role and CEBPA plays a follower role in driving the program of AT1 cell differentiation, which is consistent with the regulon patterns. Moreover, KLF2 and CEBPA demonstrate synergy by interacting with each other.
Our study raises a series of thinking. For instance, except for TFs, cell fate determination is also regulated by epigenetics and the status of the cell niche. Wnt signaling11, TGF-β signaling76, and inflammatory signals12 play key roles in AT2 cell fate determination, and whether TH regulates these cell milieus needs further detection. Furthermore, whether the noncanonical TH signaling77 affects this process is unclear. The remodeling of chromatin in response to TH was reported78,79, but whether TRβ activation alters the accessibility of AT1 cell genes for KLF2 and CEBPA binding warrants further investigation. In pharmaceuticals, although TRβ agonists demonstrated fewer side effects on tachycardia and bone loss than T341, some concerns remain. The selectivity of TRβ agonists is not absolute, and excessively high doses have positive chronotropic effects80. The tissue distribution may contribute to a part of GC-1 specificity in vivo32,34,42. The risk of unexpected effects on other physiological processes exists when the TRβ agonist is delivered systemically. Therefore, exploring precise dosage, formula (such as aerosol), and cell targeting strategy (such as nanoparticle81) is a direction to promote the therapeutic development of TRβ agonists in lung repair and fibrosis.
In conclusion, our observations support the idea that masking pathologic plasticity and facilitating endogenous differentiation potential of AT2 cells is an inspiring strategy to promote normal regeneration and anti-fibrosis, and TRβ activation is a highly promising strategy to fulfill this intractable task by orchestrating KLF2 and CEBPA (Fig. 9).
Methods
All experimental procedures were approved by the Institutional Animal Care and Use Committee of Henan Normal University (HTU2020-03) and in accordance with the ARRIVE guidelines.
Animals
Wild-type C57BL/6 N mice (8-week-old) purchased from Charles River (Beijing, China) were used throughout this study. Mice were maintained in specific pathogen-free conditions at 22–26 °C, 40–60% humidity, and a 12-hour light cycle. Animals were allowed food (Xietong, Jiangsu, 1010088) and water ad libitum. Sftpc-CreERT2 and Rosa26R-CAG-lsl-tdTomato (R26R-tdT) have been described previously10. Klf2 floxed mice (Klf2f/+) were generated by Cyagen Biosciences Inc. (Suzhou, China) and crossed with Cre-mice in our lab, as described in Fig. 7a. For lineage tracing by Sftpc-CreERT2; R26R-tdT mice and CKO Klf2 by Sftpc-CreERT2; Klf2f/f mice, 3-4 doses of tamoxifen (Sigma-Aldric, dosages: 100 mg/kg, dissolved in Mazola corn oil) were delivered to age-matched mice (8–12 weeks old) via intraperitoneal injection as indicated in the figures. Mice were euthanized when sampling. Sex was not considered as a biological variable. Both male and female mice were included but used separately as repeat experiments due to the differences in body weight and sensitivity to BLM. Male and female mice showed consistent results. All the exhibited results are from male mice.
Cell culture
DMEM/F12 (Hyclone) was used to culture A549 and MLE12, and low-glucose DMEM (Hyclone) was used to culture primary mouse lung fibroblasts and MRC5. All cells were confirmed negative for mycoplasma and cultured with 10% FBS (GIBCO) at 37 degrees in a 5% CO2 incubator, except where indicated.
Primary mouse AT2 cells and fibroblasts isolation
8-12-week-old male mice were used to isolate AT2 cells using a modified method from the previous report2. Briefly, the lung was digested with perfusion of elastase (1 mg/mL; Sigma, E1250) and DNase (1 mg/mL; Sigma, 10104159001) sealed by low-gelling temperature agarose (Sigma, A9045) for 45 min at room temperature. The lung was dissociated in a C tube by the gentleMACS Dissociator (Miltenyi Biotec) with program ‘m-lung-01-02’ before passing through a 100-, 40-, and 20-micron filter (NEST, 258367, and 258369; Millipore, NY2004700), followed by percoll (Solarbio, P8370) gradient centrifugation (300 × g, 20 min) to remove red blood cells and pre-purify. CD45, CD16/32-coated (Biolegend, 103102, and 101302) Petri dishes were used to deplete the immune cells and fibroblasts. Endothelial cells were removed with CD31-specific magnetic beads (Miltenyi Biotec, 130-97-418). AT2 cells were enriched by EpCAM magnetic beads (Miltenyi Biotec, 130-105-958) and were cultured in collagen-coated plates with DMEM/F12 supplemented with 10% FBS for up to 48 h. For primary fibroblasts, the lungs were digested for 30 min at 37 °C with collagenase (1 mg/mL; Sigma, C0130) after being cut into small pieces. Then, the tissues were resuspended in DMEM with 15% FBS following centrifugation at 500 × g for 5 min. The adherent cells were passaged after five days for co-culture or organoid assays.
3D lung organoid culture
The 3D-organoid air-liquid culture was performed following the previous report12. In brief, 1 × 104 freshly isolated tdT+ AT2 cells and 1 × 105 primary fibroblasts were resuspended in DMEM/F12 and mixed in a 1:1 ratio with growth-factor-reduced Matrigel (Corning, 354230). Then, 120 μL mixture was pipetted into a 24-well Transwell insert (0.4 μm; Corning, 3470). 500 μL culture media was added in the lower chamber and changed every other day with or without GC-1 (10 nM). RevitaCell supplement (A2644501, Gibco) was added to the medium for the first two days. The in-well images were captured and stitched by Cytation C10 (Agilent BioTek), and the colony area was calculated using Gen5 software (Agilent BioTek).
Lung injury models and treatments
Mice were anesthetized in the chamber of a small animal anesthesia machine (RWD Life Science, R550, Shenzhen, China) with isoflurane and then perpendicularly suspended by their front teeth on an intratracheal intubation stand. Then, 1 U/kg bleomycin (Hanhui Pharmaceuticals, Zhejiang, China) or 1 mg/kg LPS (sigma, L2630) in 50 μL PBS was delivered to the mouse lung intratracheally by a catheter (22 G). A double dose of bleomycin (2 U/kg) was used to perform the survival analysis in Fig. 3i. For the silicosis model, 100 mg/mL SiO2 (Sigma-Aldric, S5631) in PBS was autoclaved for 30 min and sonicated for 10 min, and 50 μL suspensions were delivered with the same manner as above.
GC-1 (Sigma-Aldric, SML1900) and T3 (sigma, T2877) were dissolved in DMSO at 5 mg/mL for stock and diluted to the desired concentration in PBS. For system administration, 0.2 mL was delivered by intraperitoneal injection to each mouse individually (e.g., the working concentration of 3.75 μg/mL GC-1 was obtained for the dosage of 30 μg/kg in the group of 25 g mean weight, and the DMSO concentration was approximately 1/1333). For lung aerosol, the Buxco Finepointe inhalation system (Data Sciences International) was used. Briefly, 50 μL/mouse GC-1 was pipetted into the nebulizer head (2.5–4 μm), and mice were restricted in the nose-only inhalation tower that allowed simultaneous exposure of 8 mice. The inflow humidity was approximately 50% when the nebulization efficiency was set at 10 μL/min, and the duration was approximately 40 min until the mist stopped forming. Pirfenidone (Continent Pharmaceuticals, Beijing, China) suspensions in 0.2 mL PBS were delivered via oral gavage (100 mg/kg) daily from day 7 after bleomycin in the survival test. Propylthiouracil (Sigma-Aldric, P3755) was dissolved in the drinking water at 0.5 mg/mL with 2% glucose to attenuate the bitter taste. All control groups receive equal-volume vehicles by the same protocol.
Pneumonectomy (PNX) and thyroidectomy (TDX)
Mice were anesthetized on a warm pad with an anesthesia tube placed around their noses. After opening the chest wall at the intercostal ribs, the left lung lobe was pulled out with blunt-tip tweezers and tied with a suture at the left principal bronchus. Then, remove the left lung and close the incision. TDX was performed under the same anesthesia protocol. In brief, the thyroid was clamped and cut off with tweezers and scissors, and the removed tissue was verified by follicle structure with a microscope.
Adeno-associated virus (AAV) delivery
AAV2/9 viruses expressing Cebpa-shRNA (AGCCGAGATAAAGCCAAACAA) or scramble shRNA (CCTAAGGTTAAGTCGCCCTCG) were constructed at Obio Technology (Shanghai, China). 7 days before PNX or bleomycin, each mouse was delivered with 1.2 × 1011 viral genome copies in 50 μL PBS by intubation, same as bleomycin delivery.
Measuring the hydroxyproline or soluble collagen concentration
Lung hydroxyproline concentration was analyzed with a colorimetric assay kit from Sigma-Aldric (MAK008), following the manufacturer’s instructions, and data are expressed as micrograms of hydroxyproline per lung as indicated in figures. Sircol-soluble collagen was assayed using a kit from Biocolor Life Science (S1000).
Lung internal volume and mean linear intercept
The lung internal volume and mean linear intercept measurements follow the previous report82. In brief, after opening the mouse chest and exposing the trachea, a catheter (Introcan, 20 G) was inserted into the lung trachea, tied with a suture, and connected to a pipette with 25 cm of paraformaldehyde (PFA). Record the scale from the beginning and after 1 hour later. The result of subtracting the two records is the lung internal volume. The mean linear intercept was analyzed using 20x H&E staining sections in Photoshop 13.0.
BALF cell count determinations and protein concentration assays
Mouse lungs were lavaged twice with 0.7 mL of saline and pooled. Leukocytes were counted with a hematology analyzer (Beckman Colter, DxH 500). After centrifugation at 1000 × g for 5 min, the protein concentration of BALF supernatant was assayed with the BCA Kit (Solarbio Life Sciences, PC0020).
Immunoblotting
Uncropped images of immunoblots are provided in the source data. RIPA buffers (Beyotime Technology, P0013B) were used to extract the protein from cultured cells and lung tissues. The protein concentrations were tested by BCA kits (Solarbio). After running through the SDS-PAGE gel, proteins were transferred to PVDF membranes. The immunoblotting was captured by a ChemiDoc XRS + System (Bio-Rad) or an ODYSSEY Fc instrument (LI-COR Biosciences). The following primary antibodies were used at 1:1000: COL1A1 (Cell Signaling, 72026), FN1 (Proteintech, 15613), N-Cad (Cell Signaling, 13116), E-Cad (Cell Signaling, 14472), KLF2 (Invitrogen, PA5-120456), KLF2(NOVUS, NBP2-45510), CEBPA (Cell Signaling, 8178), α-SMA (Abcam, ab5694), Vim (Proteintech, 10366), β-Actin (Affinity Biosciences, T0022), CLDN4 (Invitrogen, 32-9400), AGER (Abcam, ab216329), PDPN (Affinity Biosciences, DF12456), CAV1(Proteintech, 16447), HOPX (Proteintech, 11419). The following secondary antibodies were used: Goat anti-Rabbit IgG (H + L) HRP (1:5000, Affinity Biosciences, S0001), Goat anti-Mouse IgG (H + L) HRP (1:5000, Affinity Biosciences, S0002), IRDye 800CW Goat anti-Mouse IgG (H + L) (1:10000, LI-COR).
Histology and immunostaining
The mouse lung was perfused from the right ventricle with 10 mL PBS to remove blood cells, fixed with 4% PFA for 1 h at room temperature under 25 cm pressure as described in lung internal volume testing, and then immersed in PFA overnight at 4 °C, followed by washing in PBS for 3 × 4 h at 4 °C. For paraffin embedding, the lung was dehydrated by an alcohol gradient, as usual. For OCT (Tissue Tek) embedding, the lung was dehydrated through a 15%–30% sucrose gradient at 4 °C. For organoids, the cell colonies were pre-embedded in Histogen (Thermo Scientific), followed by OCT or paraffin embedding. Human lung tissue sections were obtained from the Henan Provincial Chest Hospital (approved No. 2021-03-04). The IPF samples are surgical remnants of lung biopsies or explants. Controls are normal histology tissue obtained from the disease-free margin of lung cancer resection specimens. The written informed consent was obtained from the donors.
For immunochemistry, after antigen retrieval with tris-EDTA buffer (pH 9.0), the sections were treated with 5% goat serum and 5% donkey serum in 0.3% Triton-X/PBS for 1 h at room temperature and then incubated with the antibodies at 4 °C overnight as follows: SFTPC (1:200, Affinity Biosciences, DF6647), KRT8 (1:200, Abcam, ab53280), KLF2 (1:200, Affinity Biosciences, DF13602), and CEBPA (1:200, Cell Signaling, 8178). Biotin-labeled goat anti-rabbit IgG (H + L) was used to bind the primary antibody before incubation with HRP-labeled streptavidin (Beyotime, P0615), and reacting with DAB. Sections were counterstained with hematoxylin.
For IF, 8–10 μm cryosections were used. After washing and blocking, the primary antibodies were used as follows: KRT8 (1:200, Abcam, ab53280), CEBPA (1:200, Cell Signaling, 8178), Ki67 (1:200, Cell Signaling, 9129), PDPN (1:400, DSHB, 8.1.1) and AGER (1:400, Abcam, ab216329). The secondary antibodies Alexa Fluor-488 (1:1000, Cell Signaling, 4416), Alexa Fluor-647 (1:1000, Invitrogen, A-21451), and Alexa Fluor-594 (1:1000, Cell Signaling, 8889) were incubated at room temperature for 60 min. Nuclei were stained with DAPI, and sections were RI-matched in Antifade Mounting Medium (Beyotime, P0126). Fluorescence images were captured with a confocal microscope (Leica TCS SP8). AT1 cells were counted manually, and other cells were counted by ImageJ based on markers and DAPI.
Quantitative RT-PCR (qPCR)
Total RNA was extracted with TRIzol (Invitrogen) according to the manufacturer’s recommendations. RNA was reverse-transcribed with the 1st strand cDNA synthesis kit (Takara, RR036). cDNA was analyzed using the SYBR Green Master Mix kit (Yesen, 11201) by real-time PCR assay (Roche, LightCycler 480 II). The PCR amplification was performed in triplicate, and the primer sequences are as follows:
Mouse:
Aqp5: F-TCTTGTGGGGATCTACTTCACC, R-TGAGAGcGGGCTGAACCGAT; Pdpn: F-CAAGAAAACAAGTCACCCCAATAG, R-AACAATGAAGATCCCTCCGAC; Cav1: F-TGAGAAGCAAGTGTATGACGC, R-CTTCCAGATGCCGTCGAAAC; Hopx: F-GCAGACGCAGAAATGGTTTAAG, R-GGAGTTCCAAGAGCAAGCT; Ager: F-ACTACCGAGTCCGAGTCTACC, R-CCCACCTTATTAGGGACACTGG; Klf2: F-CTCAGCGAGCCTATCTTGCC, R-CACGTTGTTTAGGTCCTCATCC; Cebpa: F-CAAGAACAGCAACGAGTACCG, R-GTCACTGGTCAACTCCAGCAC; Col1a1: F-CCTCAGGGTATTGCTGGACAAC, R-CAGAAGGACCTTGTTTGCCAGG; Col3a1: F-GACCAAAAGGTGATGCTGGACAG, R-CAAGACCTCGTGCTCCAGTTAG; Fn1: F-CCCTATCTCTGATACCGTTGTCC, R-TGCCGCAACTACTGTGATTCGG; Tnc: F-GAGACCTGACACGGAGTATGAG, R-CTCCAAGGTGATGCTGTTGTCTG; Acta2: F-TGCTGACAGAGGCACCACTGAA, R-CAGTTGTACGTCCAGAGGCATAG; Spp1: F-GCTTGGCTTATGGACTGAGGTC, R-CCTTAGACTCACCGCTCTTCATG; Ctgf: F-TGCGAAGCTGACCTGGAGGAAA, R-CCGCAGAACTTAGCCCTGTATG
Human:
AGER: F-ACTACCGAGTCCGTGTCTACC, R-GGAACACCAGCCGTGAGTT; PDPN: F-GTGCCGAAGATGATGTGGTGAC, R-GGACTGTGCTTTCTGAAGTTGGC; CAV1: F-CCAAGGAGATCGACCTGGTCAA, R-GCCGTCAAAACTGTGTGTCCCT; KLF2: F-GCAAGACCTACACCAAGAGTTCG, R-CATGTGCCGTTTCATGTGC; CEBPA: F-AGGAGGATGAAGCCAAGCAGCT, R-AGTGCGCGATCTGGAACTGCAG
Micro-CT scanning
The mice were examined by Skyscan 1276 (Bruker) under anesthesia with isoflurane. The parameters were set as follows: 0.5 Al filter, 13 μm resolution, 0.4-degree rotation step, and 2 averaging. The image was reconstructed by NRecon software.
Flow cytometry
Mouse lungs from Sftpc-CreERT2; R26R-tdT mice were dissociated as described in AT2 cell isolation, and red blood cells were removed by lysis buffer (Beyotime, C702). Cells were centrifuged at 400 × g for 5 min before being fixed with 4% PFA for 10 min and permeabilized with 0.5% Tween 20 for 10 min at room temperature. KRT8 (1:20, Abcam, ab53280) and PDPN (1:50, DSHB, 8.1.1) were incubated in PBS with 0.1% tween and 3% BSA for 1 h, and Alexa Fluor 647 (Invitrogen, A78962) and Alexa Fluor-488 (Invitrogen, A-11008) were stained for 45 min in the dark. Then, the cells were sorted by flow cytometry (BD FACS Verse). Cells were selected using the FSC/SSC scatter profile to remove debris, and doublets were excluded using the FSC-A/FSC-H to select single cells. Lineage-labeled tdT+ cells were gated to analyze the Krt8+ population.
Proliferation assay
1000 A549 cells were seeded in 6-well plates to assay the colony-forming unit (CFU). GC-1 15 nM changed every other day for 8 days. EdU staining (RIBOBIO, C10310-1) was performed after GC-1 at 15 nM for 36 h, following the manufacturer’s instructions. MLE12 was used to perform the CCK-8 assay (Apexbio, K1018) with GC-1 15 nM changed every day.
Wound healing assays
6-well plates were drawn serval lines 5 mm apart at the outside bottom to mark the ___location for image capture. 1 × 106 MRC5 were seeded in the well with 1% FBS and scratched with a 10 μL pipette tip after adherence, followed by washing three times. Pictures were obtained 24 and 48 hours after drug treatment and analyzed by ImageJ.
Luciferase assay
The cross-species conserved promoter regions (KLF2: + 99 ~ − 980bp; CEBPA: TSS − 373 ~ + 456 bp; PDPN: + 127 ~ − 1001bp; CAV1: + 1500 ~ − 76bp; HOPX: + 1000 ~ − 136bp; AGER: + 991 ~ − 100bp) were cloned into the pGL3.0 luciferase reporter vector. The overexpression plasmids pcDNA 3.1 (600 ng), reporter plasmid (600 ng), and TK (100 ng) were transfected into A549 cells with Lipofectamine 3000 transfection reagent (Invitrogen, L3000015) in 24-well plates. 10 nM GC-1 was added to the TRβ group after 24 h, and the cells were harvested at 36 h. The lysate luciferase activity was detected with a substrate kit (YEASEN, 11402) in a microplate reader (Synergy LX, BioTek).
The cell-based agonist specificity was tested using a modified method from the previous report83. Briefly, human TRα1 and TRβ1 mammalian expression plasmids were constructed with pcDNA3.1. A luciferase reporter plasmid was constructed by inserting a TRE sequence (DR4: TAAGGTCATTTAAGGTCATTTAAGGTCATT TAAGGTCA) in pGL4.23. A549 Cells were maintained in phenol-red free DMEM/F12 medium (Thermo, 11039021) and supplemented with 10% charcoal-stripped FBS (Sigma, C6241). Cells were transfected with expression and reporter plasmid in white 96-well plates (Thermo, 165306) for 24 h and treated with the indicated range of concentrations of either T3 or GC-1 for another 16 h. Subsequently, luciferase activity was determined by the Dual Glo Luciferase Assay Kit (YEASEN, 11405E). EC50 values were determined by nonlinear regression analysis (GraphPad Prism). The fold of selectivity was calculated by (GC-1 EC50 on TRα/T3 EC50 on TRα)/ (GC-1 EC50 on TRβ/T3 EC50 on TRβ).
CoIP and ChIP qPCR
A549 cells were transfected with THRB, KLF2, and CEBPA in a 10 cm culture dish individually. The cells were crosslinked with 1% freshly prepared formaldehyde (Sigma, 252549) for 10 min and quenched with 0.125 M glycine for 5 min. Nuclei were extracted and lysed with buffer containing 1% SDS, 10 mM EDTA, 50 mM Tris-Cl, pH8.1, and a protease inhibitor cocktail, and sheared to an average DNA fragment size of 500 bp with a sonic dismembrator (Fisherbrand, Model 120). Aliquots of the chromatin were diluted and incubated with 5 μg IgG (Beyotime, A7028, A7016), HA (Sigma, H3663), and Flag (Cell Signaling, 14793) for immunoprecipitation overnight at 4 °C. Protein A/G magnetic beads (Thermo Scientific, 88802) were used to precipitate the antibody-chromatin complexes, followed by washing, elution, and reversal of DNA crosslinks at 60 °C for 6 h. DNA fragments were purified with a QIAquick PCR purification kit (Qiagen, 28104), assayed by qPCR, and normalized to input.
For CoIP, KLF2 and CEBPA overexpression plasmids were co-transfected into A549 cells with a 10 cm culture dish for 48 h. The lysates were passed through a needle attached to a 1 mL syringe six times, followed by an aliquot of 500 μL with or without 2.5 μ DNAse I (Promega, M6101) at 37 °C for 30 min. The immunoprecipitation process was similar to ChIP, and the elution protein was detected by immunoblotting.
The ChIP-qPCR primers used are as follows:
KLF2: F-TCCCATCCATCCAGGGTTCT, R-TCAGAGACTCTCAGGGGAGC; CEBPA: F-GACTTCTACGAGGCGGAGC, R-ATGTCGATGGACGTCTCGTG; AGER: F-TCAGAGCCCCCGATCCTATT, R-ACAGTCTGGCCTGAACCCTA; HOPX: F-TCAGGACTTGACTGTCGCTG, R-GGTACATTCTGCCGGGACAT; PDPN: F-GGGACGTTCAGGAGGGCTTG, R-GAGGCCTCTTCCAGTCCAGTG; CAV1: F-CACGGAAAAGGGGATTGGGGT, R-GGACTCGGGTAAATAACACCTCC; KRT5- TTCATCGACAAGGTGAGCTACG, R-CCAGGGCACAGAAACAAACGG
Collagen gel contraction assay and AT2/fibroblast co-culture
Mouse lung fibroblasts were suspended in serum-free medium and mixed with neutralized rat tail collagen I (Corning, 354236) to obtain a mixture with a final cell density of 1 × 105 and a final collagen concentration of 1 mg/mL. Then, pipet the mixture into 24-well plates and put the plates in an incubator at 37 °C for 1 h to coagulate the gel. The edge of the gel was detached from the well walls with a pipet tip before 2 × 105 primary AT2 cells with 1 mL medium were added. After culture for 48 h, pictures of the gels were captured, and the gel area was analyzed using ImageJ software (US National Institutes of Health).
For immunofluorescence and immunoblotting (Fig. 4k, l), fibroblasts were seeded in the low chamber of the 24-well plate with a coverglass or 6-well plate, and 2 × 105 or 1 × 106 AT2 cells were seeded in an insert for the 24-well or 6-well individually (0.4μm; Corning, 3470, 3450). After 48 h of culture, the fibroblast samples were obtained for tests.
Measurement of serum T3, TSH, ALT, AST, and CREA levels
Peripheral blood was centrifuged at 1000 × g for 10 min, and the hormone levels of mice were measured using an ELISA kit (TSH, Solarbio Life Sciences, Beijing, China, SEKM-0272; T3, Jianglai, Shanghai, China, JL11497), following the manufacturer’s instructions. The ALT, AST, and CREA levels of mice were analyzed by a clinical chemistry analyzer (Beckman Colter, AU5800).
RNA-Seq preparation and analysis
RNA integrity was assessed using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, CA, USA). Then, mRNA was purified and fragmented, followed by cDNA synthesis. Adapters with a hairpin-loop structure were ligated to prepare for hybridization. Then, PCR products were purified (AMPure XP system), and library quality was also assessed. After cluster generation, the library preparations were sequenced on an Illumina Novaseq platform, and 150-bp paired-end reads were generated. All the downstream analyses were based on clean and high-quality data. Reads were mapped using Hisat2 v2.0.5 to the reference genome. FeatureCounts v1.5.0-p3 was used to count the read numbers mapped to each gene.
Differential expression analysis of two groups was performed using the DESeq2 v1.20.0 R package. The resulting P-values were adjusted using Benjamini and Hochberg’s approach for controlling the false discovery rate. GO enrichment analysis of differentially expressed genes was implemented by the ClusterProfiler v4.4.4 R package, in which gene length bias was corrected. GO terms with a corrected P-value less than 0.05 were considered significantly enriched by differentially expressed genes. The pheatmap v1.0.12 R package was used to visualize AT1 cell markers.
FactoMineR v2.4.0 was used for PCA to obtain the top 30 TFs of variance and visualized by factoextra v1.0.7 R packages. The Eigenvalue ≥ 1 was considered a significant PC. The importance of variance was assessed by Cos2 and contributions.
Processing of scRNA-seq data
The 10X Cell Ranger computational pipeline was used as previously described for the single-cell data from BLM-induced fibrosis model84. Unqualified cells, with a high proportion (> 15%) of transcript counts derived from mitochondria-encoded genes and a high proportion (> 5%) of transcript counts derived from hemoglobin-encoded genes, were removed from downstream analysis. Cells with a high number of UMI counts (> 5000) may represent doublets and be removed. Genes were only considered if they were expressed in at least three cells in the dataset. Feature selection and data dimensionality reduction were performed using Scanpy or Seurat. Trajectory analysis was conducted using Monocle2, while RNA velocity analysis was completed using Velocyto. Cytotrace was employed for predicting cell lineage and calculating cellular differentiation chronology. The activity of transcription factor regulons was computed using pySCENIC85. The signatures of four cell subtypes were established using the claimed marker sets of previously determined IS, ADI, DATPs, and PATS. The score of the four cell signatures was calculated by “AddModuleScore” and presented in UMAP plots.
Statistical analysis
All data are presented as mean ± SEM, and differences were considered statistically significant at p < 0.05. Interventions were not blinded, but data collection and analysis of mouse samples were. There was no blinding in cell culture experiments. No data were excluded from the analyses. The efficacy experiments in vivo were assigned to at least 10 mice per group and were performed twice independently. All the in vitro experiments were repeated at least three times, independently. Two-tailed unpaired student’s t test was used to compare two groups with a normal distribution, and the U test for comparisons of two groups with a non-normal distribution. One-way ANOVA with Turkey’s multiple comparison tests was used for the comparison of three or more groups. Two-way ANOVA with sidak multiple comparisons test was used for two categorical variables. Kaplan-Meier survival curves were analyzed with the log-rank (Mantel-Cox) test. Statistical analyses were performed in GraphPad Prism 9.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data associated with this study are present in the paper or the Supplementary Materials. Bulk RNA-seq and scRNA-seq data are deposited on GEO under accession code GSE246654, and raw data is linked to SRA number PRJNA1033428. Source data are provided in this paper.
References
Schiller, H. B. et al. The human lung cell atlas: A high-resolution reference map of the human lung in health and disease. Am. J. Respir. Cell Mol. Biol. 61, 31–41 (2019).
Strunz, M. et al. Alveolar regeneration through a Krt8+ transitional stem cell state that persists in human lung fibrosis. Nat. Commun. 11, 3559 (2020).
Picard, M. Why do we care more about disease than health? Phenomics 2, 145–155 (2022).
Xu, Y. et al. Single-cell RNA sequencing identifies diverse roles of epithelial cells in idiopathic pulmonary fibrosis. JCI Insight 1, e90558 (2016).
Zacharias, W. J. et al. Regeneration of the lung alveolus by an evolutionarily conserved epithelial progenitor. Nature 555, 251–255 (2018).
Kumar, P. A. et al. Distal airway stem cells yield alveoli in vitro and during lung regeneration following H1N1 influenza infection. Cell 147, 525–538 (2011).
Martinez, F. J. et al. Idiopathic pulmonary fibrosis. Nat. Rev. Dis. Prim. 3, 17074 (2017).
Basil, M. C. et al. The cellular and physiological basis for lung repair and regeneration: Past, present, and future. Cell Stem Cell 26, 482–502 (2020).
Fehrenbach, H. Alveolar epithelial type II cell: defender of the alveolus revisited. Resp. Res. 2, 1–20 (2001).
Barkauskas, C. E. et al. Type 2 alveolar cells are stem cells in adult lung. J. Clin. Investig. 123, 3025–3036 (2013).
Nabhan, A. N., Brownfield, D. G., Harbury, P. B., Krasnow, M. A. & Desai, T. J. Single-cell Wnt signaling niches maintain stemness of alveolar type 2 cells. Science 359, 1118–1123 (2018).
Choi, J. et al. Inflammatory signals induce AT2 cell-derived damage-associated transient progenitors that mediate alveolar regeneration. Cell Stem Cell 27, 366–382 (2020).
Kobayashi, Y. et al. Persistence of a regeneration-associated, transitional alveolar epithelial cell state in pulmonary fibrosis. Nat. Cell Biol. 22, 934–946 (2020).
Kasper, M. & Haroske, G. Alterations in the alveolar epithelium after injury leading to pulmonary fibrosis. Histol. Histopathol. 11, 463–483 (1996).
Melms, J. C. et al. A molecular single-cell lung atlas of lethal COVID-19. Nature 595, 114–119 (2021).
Delorey, T. M. et al. COVID-19 tissue atlases reveal SARS-CoV-2 pathology and cellular targets. Nature 595, 107–113 (2021).
Lv, T. et al. Defense of COVID-19 by human organoids. Phenomics 1, 113–128 (2021).
Boelen, A., Kwakkel, J. & Fliers, E. Beyond low plasma T3: local thyroid hormone metabolism during inflammation and infection. Endocr. Rev. 32, 670–693 (2011).
Fliers, E., Bianco, A. C., Langouche, L. & Boelen, A. Thyroid function in critically ill patients. Lancet Diabetes Endocrinol. 3, 816–825 (2015).
de Vries, E. M., Fliers, E. & Boelen, A. The molecular basis of the non-thyroidal illness syndrome. J. Endocrinol. 225, R67–81, (2015).
Gereben, B., McAninch, E. A., Ribeiro, M. O. & Bianco, A. C. Scope and limitations of iodothyronine deiodinases in hypothyroidism. Nat. Rev. Endocrinol. 11, 642–652 (2015).
Ortiga-Carvalho, T. M., Sidhaye, A. R. & Wondisford, F. E. Thyroid hormone receptors and resistance to thyroid hormone disorders. Nat. Rev. Endocrinol. 10, 582–591 (2014).
Luongo, C., Dentice, M. & Salvatore, D. Deiodinases and their intricate role in thyroid hormone homeostasis. Nat. Rev. Endocrinol. 15, 479–488 (2019).
Yu, G. et al. Thyroid hormone inhibits lung fibrosis in mice by improving epithelial mitochondrial function. Nat. Med. 24, 39–49 (2018).
Oldham, J. M. et al. Thyroid disease is prevalent and predicts survival in patients with idiopathic pulmonary fibrosis. Chest 148, 692–700 (2015).
Bano, A. et al. Thyroid function and the risk of fibrosis of the liver, heart, and lung in humans: A systematic review and meta-analysis. Thyroid 30, 806–820 (2020).
Zhang, Y. et al. Mendelian randomisation highlights hypothyroidism as a causal determinant of idiopathic pulmonary fibrosis. EBioMedicine 73, 103669 (2021).
Chaker, L. et al. Hypothyroidism. Nat. Rev. Dis. Prim. 8, 30 (2022).
Forhead, A. J. & Fowden, A. L. Thyroid hormones in fetal growth and prepartum maturation. J. Endocrinol. 221, R87–R103 (2014).
Pascual, A. & Aranda, A. Thyroid hormone receptors, cell growth and differentiation. Biochim. Biophys. Acta 1830, 3908–3916 (2013).
Chiellini, G. et al. A high-affinity subtype-selective agonist ligand for the thyroid hormone receptor. Chem. Biol. 5, 299–306 (1998).
Grover, G. J. et al. Effects of the thyroid hormone receptor agonist GC-1 on metabolic rate and cholesterol in rats and primates: selective actions relative to 3,5,3’-triiodo-L-thyronine. Endocrinology 145, 1656–1661 (2004).
Lin, J. Z. et al. Pharmacological activation of thyroid hormone receptors elicits a functional conversion of white to brown fat. Cell Rep. 13, 1528–1537 (2015).
Trost, S. U. et al. The thyroid hormone receptor-beta-selective agonist GC-1 differentially affects plasma lipids and cardiac activity. Endocrinology 141, 3057–3064 (2000).
Peng, T. et al. Hedgehog actively maintains adult lung quiescence and regulates repair and regeneration. Nature 526, 578–582 (2015).
Liu, Z. et al. MAPK-Mediated YAP activation controls mechanical-tension-induced pulmonary alveolar regeneration. Cell Rep. 16, 1810–1819 (2016).
Sikkema, L. et al. An integrated cell atlas of the lung in health and disease. Nat. Med. 29, 1563–1577 (2023).
Han, X. et al. Mapping the mouse cell atlas by microwell-seq. Cell 172, 1091–1107 (2018).
Liu, S. et al. Triiodothyronine (T3) promotes brown fat hyperplasia via thyroid hormone receptor alpha mediated adipocyte progenitor cell proliferation. Nat. Commun. 13, 3394 (2022).
Kelly, M. J. et al. Discovery of 2-[3,5-dichloro-4-(5-isopropyl-6-oxo-1,6-dihydropyridazin-3-yloxy)phenyl]-3,5-dioxo-2,3,4,5-tetrahydro[1,2,4]triazine-6-carbonitrile (MGL-3196), a Highly Selective Thyroid Hormone Receptor beta agonist in clinical trials for the treatment of dyslipidemia.J. Med. Chem. 57, 3912–3923 (2014).
Baxter, J. D. & Webb, P. Thyroid hormone mimetics: potential applications in atherosclerosis, obesity and type 2 diabetes. Nat. Rev. Drug Discov. 8, 308–320 (2009).
Ferrara, S. J. et al. Ester-to-amide rearrangement of ethanolamine-derived prodrugs of sobetirome with increased blood-brain barrier penetration. Bioorg. Med. Chem. 25, 2743–2753 (2017).
Kathiriya, J. J. et al. Human alveolar type 2 epithelium transdifferentiates into metaplastic KRT5(+) basal cells. Nat. Cell Biol. 24, 10–23 (2022).
Adams, T. S. et al. Single-cell RNA-seq reveals ectopic and aberrant lung-resident cell populations in idiopathic pulmonary fibrosis. Sci. Adv. 6, eaba1983 (2020).
Habermann, A. C. et al. Single-cell RNA sequencing reveals profibrotic roles of distinct epithelial and mesenchymal lineages in pulmonary fibrosis. Sci. Adv. 6, https://doi.org/10.1126/sciadv.aba1972 (2020).
Wu, H. et al. Progressive pulmonary fibrosis is caused by elevated mechanical tension on alveolar stem cells. Cell 180, 107–121 (2020).
Plataki, M. et al. Expression of apoptotic and antiapoptotic markers in epithelial cells in idiopathic pulmonary fibrosis. Chest 127, 266–274 (2005).
Lomas, N. J., Watts, K. L., Akram, K. M., Forsyth, N. R. & Spiteri, M. A. Idiopathic pulmonary fibrosis: immunohistochemical analysis provides fresh insights into lung tissue remodelling with implications for novel prognostic markers. Int. J. Clin. Exp. Pathol. 5, 58 (2012).
Raghu, G. et al. Diagnosis of idiopathic pulmonary fibrosis. An official ATS/ERS/JRS/ALAT clinical practice guideline. Am. J. Respir. Crit. Care Med. 198, e44–e68 (2018).
Kuo, C. T. et al. The LKLF transcription factor is required for normal tunica media formation and blood vessel stabilization during murine embryogenesis. Genes Dev. 11, 2996–3006 (1997).
Warner, M. H. & Beckett, G. J. Mechanisms behind the non-thyroidal illness syndrome: an update. J. Endocrinol. 205, 1–13 (2010).
Wang, Z. et al. Enhanced glycolysis-mediated energy production in alveolar stem cells is required for alveolar regeneration. Cell Stem Cell 30, 1028–1042 (2023).
Desai, T. J., Brownfield, D. G. & Krasnow, M. A. Alveolar progenitor and stem cells in lung development, renewal and cancer. Nature 507, 190–194 (2014).
Wang, F. et al. Regulation of epithelial transitional states in murine and human pulmonary fibrosis. J. Clin. Invest. 133, https://doi.org/10.1172/JCI165612 (2023).
Lang, N. J. et al. Ex vivo tissue perturbations coupled to single-cell RNA-seq reveal multilineage cell circuit dynamics in human lung fibrogenesis. Sci. Transl. Med. 15, eadh0908 (2023).
Chanda, D. et al. Developmental pathways in the pathogenesis of lung fibrosis. Mol. Asp. Med. 65, 56–69 (2019).
Selman, M., Lopez-Otin, C. & Pardo, A. Age-driven developmental drift in the pathogenesis of idiopathic pulmonary fibrosis. Eur. Respir. J. 48, 538–552 (2016).
Hogan, B. L. et al. Repair and regeneration of the respiratory system: complexity, plasticity, and mechanisms of lung stem cell function. Cell Stem Cell 15, 123–138 (2014).
Beers, M. F. & Morrisey, E. E. The three R’s of lung health and disease: repair, remodeling, and regeneration. J. Clin. Invest. 121, 2065–2073 (2011).
Barca-Mayo, O. et al. Role of type 2 deiodinase in response to acute lung injury (ALI) in mice. Proc. Natl. Acad. Sci. USA 108, E1321–E1329 (2011).
Li, L. et al. Aerosolized thyroid hormone prevents radiation induced lung fibrosis. Front. Oncol. 10, 528686 (2020).
Yang, M. et al. Triiodothyronine ameliorates silica-induced pulmonary inflammation and fibrosis in mice. Sci. Total Environ. 790, 148041 (2021).
Alonso-Merino, E. et al. Thyroid hormones inhibit TGF-beta signaling and attenuate fibrotic responses. Proc. Natl. Acad. Sci. USA 113, E3451–E3460 (2016).
Ferdous, A. et al. FoxO1-Dio2 signaling axis governs cardiomyocyte thyroid hormone metabolism and hypertrophic growth. Nat. Commun. 11, 2551 (2020).
Taniguchi, H. et al. Silencing of Kruppel-like factor 2 by the histone methyltransferase EZH2 in human cancer. Oncogene 31, 1988–1994 (2012).
Marrone, G. et al. KLF2 exerts antifibrotic and vasoprotective effects in cirrhotic rat livers: behind the molecular mechanisms of statins. Gut. 64, 1434–1443 (2015).
Li, H. et al. Endothelial Klf2-Foxp1-TGFbeta signal mediates the inhibitory effects of simvastatin on maladaptive cardiac remodeling. Theranostics 11, 1609–1625 (2021).
Shi, J. et al. KLF2 attenuates bleomycin-induced pulmonary fibrosis and inflammation with regulation of AP-1. Biochem. Biophys. Res. Commun. 495, 20–26 (2018).
Pei, L. et al. Thyroid hormone receptor repression is linked to type I pneumocyte-associated respiratory distress syndrome. Nat. Med. 17, 1466–1472 (2011).
Wani, M. A., Wert, S. E. & Lingrel, J. B. Lung Kruppel-like factor, a zinc finger transcription factor, is essential for normal lung development. J. Biol. Chem. 274, 21180–21185 (1999).
Nerlov, C. C/EBPalpha mutations in acute myeloid leukaemias. Nat. Rev. Cancer 4, 394–400 (2004).
Didon, L., Roos, A. B., Elmberger, G. P., Gonzalez, F. J. & Nord, M. Lung-specific inactivation of CCAAT/enhancer binding protein α causes a pathological pattern characteristic of COPD. Eur. Respir. J. 35, 186–197 (2010).
Martis, P. C. et al. C/EBPalpha is required for lung maturation at birth. Development 133, 1155–1164 (2006).
Reebye, V. et al. Gene activation of CEBPA using saRNA: preclinical studies of the first in human saRNA drug candidate for liver cancer. Oncogene 37, 3216–3228 (2018).
Liu, W. et al. Targeted regulation of fibroblast state by CRISPR-mediated CEBPA expression. Respir. Res. 20, 281 (2019).
Riemondy, K. A. et al. Single cell RNA sequencing identifies TGFbeta as a key regenerative cue following LPS-induced lung injury. JCI Insight 5, https://doi.org/10.1172/jci.insight.123637 (2019).
Hones, G. S. et al. Noncanonical thyroid hormone signaling mediates cardiometabolic effects in vivo. Proc. Natl. Acad. Sci. USA 114, E11323–E11332 (2017).
Praestholm, S. M. et al. Multiple mechanisms regulate H3 acetylation of enhancers in response to thyroid hormone. PLoS Genet. 16, e1008770 (2020).
Grontved, L. et al. Transcriptional activation by the thyroid hormone receptor through ligand-dependent receptor recruitment and chromatin remodelling. Nat. Commun. 6, 7048 (2015).
Sjouke, B. et al. Eprotirome in patients with familial hypercholesterolaemia (the AKKA trial): a randomised, double-blind, placebo-controlled phase 3 study. lancet Diabetes Endocrinol. 2, 455–463 (2014).
Mitchell, M. J. et al. Engineering precision nanoparticles for drug delivery. Nat. Rev. Drug Discov. 20, 101–124 (2021).
Liu, Z., Fu, S. & Tang, N. A standardized method for measuring internal lung surface area via mouse pneumonectomy and prosthesis implantation. J. Vis. Exp. 125, https://doi.org/10.3791/56114 (2017).
Hu, L. et al. Discovery of highly potent and selective thyroid hormone receptor beta agonists for the treatment of nonalcoholic steatohepatitis. J. Med. Chem. 66, 3284–3300 (2023).
Zheng, G. X. Y. et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017).
Van de Sande, B. et al. A scalable SCENIC workflow for single-cell gene regulatory network analysis. Nat. Protoc. 15, 2247–2276 (2020).
Acknowledgements
This study was supported by National Key R&D Program of China (2019YFE0119500 to G.Y.), Key R&D Program of Henan province, China (231111310400 to G.Y.), Zhongyuan scholar, Henan, China (244000510009 to G.Y.), The 111 Project, China (D20027 to G.Y.), and Henan Project of Science and Technology, China (232102521025 and GZS2023008 to L.W.). We thank the Instrumental Analysis Center, College of Life Science, Henan Normal University, and The High Performance Computing Center of Henan Normal University for technical assistance.
Author information
Authors and Affiliations
Contributions
Conceptualization: X.P. and G.Y.; Investigation: X.P., C.L., L.L., Y.L., M.X., J.P., M.W., M.L., S.G, and P.Y.; Methodology: X.P., J.Y., and C.X.; Software: X.P., L.W., C.D., and Z.L.; Formal Analysis and Visualization: X.P. and Z.L.; Resources: X.P., Y.L., J.P., and M.W. Writing - Original Draft: X.P. Writing - Review & Editing: X.P., J.Y., L.W., C.D., I.O.R., and G.Y. Supervision, Project Administration, and Funding Acquisition: G.Y.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Subburaman Mohan, Gregory Brent, and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Pan, X., Wang, L., Yang, J. et al. TRβ activation confers AT2-to-AT1 cell differentiation and anti-fibrosis during lung repair via KLF2 and CEBPA. Nat Commun 15, 8672 (2024). https://doi.org/10.1038/s41467-024-52827-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-52827-z