Abstract
Highly sensitive airborne virus monitoring is critical for preventing and containing epidemics. However, the detection of airborne viruses at ultra-low concentrations remains challenging due to the lack of ultra-sensitive methods and easy-to-deployment equipment. Here, we present an integrated microfluidic cartridge that can accurately detect SARS-COV-2, Influenza A, B, and respiratory syncytial virus with a sensitivity of 10 copies/mL. When integrated with a high-flow aerosol sampler, our microdevice can achieve a sub-single-copy spatial resolution of 0.83 copies/m3 for airborne virus surveillance with an air flow rate of 400 L/min and a sampling time of 30 minutes. We then designed a series of virus-in-aerosols monitoring systems (RIAMs), including versions of a multi-site sampling RIAMs (M-RIAMs), a stationary real-time RIAMs (S-RIAMs), and a roaming real-time RIAMs (R-RIAMs) for different application scenarios. Using M-RIAMs, we performed a comprehensive evaluation of 210 environmental samples from COVID-19 patient wards, including 30 aerosol samples. The highest positive detection rate of aerosol samples (60%) proved the aerosol-based SARS-CoV-2 monitoring represents an effective method for spatial risk assessment. The detection of 78 aerosol samples in real-world settings via S-RIAMs confirmed its reliability for ultra-sensitive and continuous airborne virus monitoring. Therefore, RIAMs shows the potential as an effective solution for mitigating the risk of airborne virus transmission.
Similar content being viewed by others
Introduction
The unprecedented crisis caused by the coronavirus disease 2019 (COVID-19) has cast an immense shadow over global public health1,2. Compelling epidemiological data highlight that the airborne transmission of SARS-CoV-2 contributed significantly to the COVID-19 pandemic3,4,5,6,7. Activities such as exhaling, speaking, or singing by an infected person can all generate virus-laden droplets which rapidly coalesce into aerosol particles with a diameter of less than 5 μm3. These tiny SARS-CoV-2 particles can spread up to 10 m away and have an average half-life of 1–3 h (other common respiratory viruses, such as respiratory syncytial virus and influenza, have similar or potentially longer half-lives)8,9,10,11,12, greatly increasing the risk of infection to nearby people. In the past few years, it has been proved that SARS-CoV-2 can be transmitted by aerosols in various locations, such as hospitals6,13, community settings14, public transportation15,16, schools17,18, bars17, and gymnasiums18, even causing so-called “super-spreading events”19,20. As a result, deploying rapid and sensitive surveillance devices for monitoring contagious bioaerosols in highly crowded places has been gradually regarded as an efficient and non-invasive means to contain the disease spreading without interrupting normal social activities21,22.
Currently, the monitoring of viruses in the air is usually carried out using a two-step analytical protocol: the viruses in aerosols are first collected on-site with a bioaerosol sampler, and then the collected samples are transported to a centralized laboratory and analyzed using conventional reverse transcription-quantitative polymerase chain reaction (RT-qPCR)23,24. While the characteristics of airborne pathogen transmission have been extensively studied using this protocol, it still falls short of embodying the quintessence of an optimal approach for aerosol monitoring. We believe an ideal system for monitoring airborne viruses should possess two key attributes at the same time: ultra-high sensitivity and short turnaround time, preferably enabling ultrasensitive and continuous real-time monitoring of aerosols to allow early risk assessment and prompt interventions with a high spatiotemporal resolution21. Pioneering clinical investigations into SARS-CoV-2 concentrations at different hospital sites have revealed the lowest aerosol concentration could be down to 3 copies/m3 occurred outside the hospital6. The current RT-qPCR has a limit of detection of ~200-500 copies/mL25, which cannot effectively meet the needs for accurate detection of respiratory viruses in aerosols because these aerosol viruses can be considered to be infinitely diluted in the air. Without more sensitive techniques, the current risk assessments for individuals and environments can only be based on theoretical predications. The second consideration of the ideal system revolves around achieving timely evidence-based responses for disrupting the airborne pathogen transmission chain. However, many aerosol detection methods that highly rely on centralized laboratories have inherent limitations6,13,14,17, including the inability to conduct on-site and extensive manual operations. Although continuous aerosol detection devices have been reported26,27,28,29, those systems still face the challenges of coordinating ultra-high sensitivity and specificity simultaneously.
Previously, we have developed a SARS-CoV-2-in-aerosol monitoring system (SIAMs), which can provide an impressive sensitivity of 20 copies/mL30. However, in the subsequent deployment of this system in various scenarios, we found that the SIAMs still fall short in the quantitative and multi-target detection capabilities due to the isothermal amplification biochemistry, and cannot enable continuous and automated monitoring due to the inability to efficiently store reagents on double-sided tape valve-based microfluidic cartridges. In addition, the aerosol collection and the analysis were still performed in two separated devices and the manual sample transfer was inevitable. To overcome these limitations, here we proposed an in-situ PCR chemistry integrated into a brand-new designed, fully-enclosed microfluidic cartridge for ultra-sensitive nucleic acid testing, achieving a remarkable sensitivity of 10 copies/mL (0.01copies/μL)—more than one order of magnitude improvement over commercial assay kits. When integrated with a high-capacity 400 liters per minute (lpm) aerosol sampler, the respiratory virus-in-aerosols monitoring system (RIAMs) can achieve an unparalleled spatial resolution of 0.83 copies/m3 and enable simultaneous detection of SARS-CoV-2 and three other common respiratory viruses. To cater diverse scenarios, we developed three different versions of aerosol monitoring systems, including a multi-site sampling RIAMs (M-RIAMs), a stationary real-time RIAMs (S-RIAMs), and a roaming real-time RIAMs (R-RIAMs). We conducted a comprehensive evaluation of these systems using clinical environmental samples in different settings. The results highlight the superiority of virus-laden aerosol detection as an environment-oriented method over conventional methods. Meanwhile, to verify the feasibility of our system in the real world, we successfully deployed S-RIAMs in several typical locations to analyze aerosol samples. The results demonstrate its excellent clinical applicability, firmly proving it as the preferred method for aerosol monitoring.
Results
Integrated microfluidic cartridge for RIAMs
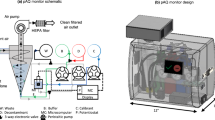
Our RIAMs platform has three different versions of airborne-virus monitoring systems for different application scenarios, including M-RIAMs for multiple-site sampling and on-site batch analysis, S-RIAMs for continuous real-time monitoring of airborne pathogens without any manual intervention in a specific ___location, and R-RIAMs for continuously sampling and analysis of bioaerosols in a large indoor environment (Fig. 1b). Inside of the RIAMs is the central biochemical assay of in-situ PCR which provides the ultra-high sensitivity for virus detection. The in-situ PCR utilizes a piece of chitosan-modified quartz filter (QF) paper for nucleic acid extraction followed by PCR amplification directly on the paper (Fig. 1c)31,32. Unlike the conventional solid-phase nucleic acid extraction methods33, which employ a bind-wash-elute protocol, the QF paper carries positive charges in the acidic condition, leading to the adsorption of negative-charged nucleic acid molecules. After a simple wash with water, PCR reagents are directly loaded to soak the QF paper completely. Then, PCR occurs within the QF paper and all the captured nucleic acids can be used for amplification without any loss (Mechanisms of nucleic acid extraction and in-situ amplification of QF paper can be seen in Supplementary Method 1). This bind-wash-amplification assay not only provides an ultra-high sensitivity for detecting viruses in aerosols, but also simplifies the structures of the integrated microfluidic cartridge for accommodating this process.
a Description of the importance of monitoring airborne virus. (i) Schematic representation of virus-laden aerosols/droplets generated by an infected individual and potential aerosol transmission pathway, (ii) Table of the median half-life of common airborne respiratory viruses. RH: relative humidity. RSV: respiratory syncytial virus. b Schematic diagram of the comprehensive airborne viruses monitoring solution provided by RIAMs, including (i) offline surveillance of airborne respiratory viruses and (ii) continuous surveillance of airborne respiratory viruses. c Schematic diagram of the versatile and ultrasensitive QF paper-centered workflows of in-situ PCR chemistry. QF paper: quartz filter paper. d Exploded schematic of the microfluidic cartridge for fully integrated and automated viral nucleic acid detection. e A series of basic elements of mesoscopic design paradigm, including IN, OUT, MIX for various basic fluidic operations. f Images of the microfluidic cartridge and the compact control and detection instrument.
Once the central biochemistry is determined, we next designed a fully integrated and enclosed microfluidic cartridge to accommodate the assay using the “needle-plug/piston” mesoscopic design paradigm in the “3D extensible” architecture developed previously by our group (Fig. 1d)34,35,36. The mesoscopic design paradigm has a series of basic elements, such as IN, OUT, MIX, etc., for various basic fluidic operations (Fig. 1e). For example, the IN element represents the basic fluidic operation of loading a reagent into the microdevice, while the OUT element is for driving a solution out of the microdevice. In the process of designing this cartridge, a reaction chamber containing an embedded piece of QF paper is first designed for nucleic acid extraction and amplification on a planner plastic chip, which is sealed with a piece of pressure-sensitive membrane. By sequentially linking three IN elements to the reaction chamber, the sample loading, the washing, and the PCR reagent loading can be realized in the cartridge. An OUT element is linked to the reaction chamber as well to function as a waste reservoir. A MIX element can replace the IN element for the PCR mixture loading to realize the reconstitution of the lyophilized reagents in the device. After the initial fluid function verification, the microfluidic cartridge is further modified for injection molding in order to achieve mass production (Supplementary Fig. 1). To operate this microfluidic cartridge, we next built a compact control and detection instrument using a modular design structure. This instrument consists of a fluidic actuation module, a PCR thermal cycle module, and a four-color fluorescence detection module and can process two microfluidic cartridges at the same time (Fig. 1f). Once the cartridge is inserted into the instrument, the entire analysis process can be automatically conducted by the instrument.
Ultrasensitive detection of SARS-COV-2, Influenza A, B, and respiratory syncytial virus
We first tested the performance of the QF paper for RNA extraction and PCR amplification by using a simplified microdevice containing a chamber with a piece of QF paper. Once the RNA capture is finished on the device, the QF paper is taken out from the chamber and put into a tube for PCR amplification (Supplementary Fig. 2b). Here we designed a four-plex PCR system that can simultaneously detect four common human respiratory viruses, including SARS-COV-2, Influenza A, B, and respiratory syncytial virus (RSV) (Table S1, Supplementary Fig. 2a). A concentration gradient of four respiratory viruses ranging from 1000 to 5 copies/mL was prepared to measure the limit of detection (LoD) (Related information of reference materials used in performance evaluation of RIAMs can be seen in Supplementary Method 2). The results showed that our assay achieved the ultra-low LoDs of 10, 5, 5, and 5 copies/mL for SARS-COV-2, influenza A, influenza B, and RSV, respectively (Fig. 2b), with a coefficient of determination of r2 = 0.72 for SARS-COV-2, r2 = 0.78 for Influenza A, r2 = 0.87 for Influenza B, and r2 = 0.71 for RSV (Fig. 2c). We also verified that there are no cross-reactions or non-specific amplifications in the four-plex PCR system using a series of mixture samples (Fig. 2d).
a Screenshots of Supplementary Movie 1 showing the microfluidic cartridge workflow and biochemical principles. b Sensitivity of the in-situ four-plex PCR system. The four target RNAs from 1000 copies/mL to 5 copies/mL were tested, demonstrating an ultra-high sensitivity of 10 copies/mL for SARS-CoV-2, and 5 copies/mL for Influenza A, Influenza B, and RSV. Error bars represent mean ± SD (n = 3 independent experiments on different simplified microdevices). c Linear fitting between the logarithm of virus input concentration of the four target RNAs and the corresponding Ct values. High linearity suggests that the in-situ multiplex PCR system can be used for the quantification of those four target RNAs. Error bars represent mean ± SD (n = 3 independent experiments on different simplified microdevices). d Evaluation of the specificity of the in-situ four-plex RT-qPCR system, showing no cross-reaction or non-specific amplifications of the four-plex PCR assay. Error bars represent mean ± SD (n = 3 independent experiments on different simplified microdevices). e LoDs determination of the microfluidic cartridge assay for ultrasensitive and simultaneous detection of SARS-CoV-2, Influenza A, B, and RSV, measured using pseudovirus quality reference materials of the four target viruses. Error bars represent mean ± SD (n = 3 independent experiments performed on different microfluidic cartridges). NC: negative control. f LoD determination for in-situ RT-qPCR to detect Omicron viral RNA, including the fitted relationship between the logarithm of virus input concentration and Ct value. Error bars represent mean ± SD (n = 3 independent experiments performed on different microfluidic cartridges). g Amplification curves for 10 copies/mL of Omicron at a primer concentration of 0.75 μM. Error bars represent mean ± SD (n = 3 independent experiments performed on different microfluidic cartridges). NC: negative control.
Next, we test the sensitivity of the assay conducted in the cartridge using the compact instrument. The analytical process is shown as follows (Fig. 2a and Supplementary Movie 1): 1-mL sample in lysis buffer is first driven through the modified QF paper, where RNA is captured, by the actuation plunger in the instrument. After that, 1 mL of DEPC water is washed through the paper followed by the loading of 50-μL PCR mixture into the chamber. Finally, the OUT-waste container is closed and the thermal cycling is conducted in the chamber. The amplification curves can be read out through the fluorescence intensity emitted by the QF paper during the amplification reaction. We prepared gradient samples containing varying amounts of pseudoviruses of these four respiratory viruses ranging from 1000 copies down to zero. The on-cartridge results showed that our system achieved a remarkable sensitivity with a LoD of 10 copies/mL for all four respiratory viruses (Fig. 2e, and Supplementary Fig. 2c). We also tested the detection of the Omicron variant of SARS-CoV-2 using the RIAMs, showing that 20 copies/mL of Omicron could be detected with a good linear correlation between Ct values and RNA concentration (Fig. 2f and Supplementary Fig. 2d). With further optimization of the primers, this sensitivity can be improved to 10 copies/mL (Fig. 2g).
M-RIAMs for ultrasensitive virus-laden aerosol monitoring with sub-single-copy spatial resolution
To analyze airborne viruses in aerosols, we employ a cyclone aerosol sampler with a high air flow rate of 400 lpm to collect aerosol particles30,37. As the air swirls in the cyclone pipe, the particles hit the pipe wall due to the centrifugal force generated by the swirling motion and fall into the bottom collection tube containing virus lysis buffer (The working principal of the aerosol sampler can be seen in Supplementary Method 3 and Supplementary Fig. 4a). By using polystyrene latex (PSL) microspheres with various diameters, we confirmed that the sampler is capable of collecting particles as small as 200 nm (Supplementary Fig. 4b). We further measured the total collection efficiency of the sampler using a cyclone pipe with a smooth inner surface manufactured by injection molding (Supplementary Fig. 4c) (Refers specifically to the number of particles collected in the aerosol sampler)38. We found that when the diameter exceeds 0.5 μm, the total collection efficiency surpasses 80%, representing a significant improvement over our previously reported total collection efficiencies, which were all noticeably below 25%30,37. In addition, when the particle size exceeds 2 μm, the total collection efficiency approaches 100% (Fig. 3b, Supplementary Fig. 4d, and Supplementary Method 4). By using a slender silencer at the air outlet of the sampler, we further reduced its working noise to alleviate the disturbance to relevant personnel. We found that when the distance away from the sampler is 2 m, the working noise can be significantly reduced to less than an acceptable level of 65 dB (Fig. 3c and Supplementary Fig. 4e).
a Workflow of M-RIAMs from aerosol sample collection to on-site or centralized nucleic acid testing. b Physical collection efficiency curves of the aerosol sampler when collecting polystyrene latex microspheres with different sizes ranging from 0.5, 0.8, 1.0, 1.5, and 2.0 μm. Error bars represent mean ± SD (n = 3 collection efficiency values from independent aerosol particle generation and collection experiments). c Working noise of the aerosol sampler with and without silencer at different distances. Error bars represent mean ± SD (n = 3 independently measured noise values). d Performance assessment of M-RIAMs to detect mock samples to test potential impurities in real aerosol samples. Error bars represent mean ± SD (n = 8 independently collected aerosol samples). ND: not detected. e Performance re-assessment of M-RIAMs that adds a piece of filter cotton to the air inlet of the cyclone sampler to detect mock samples collected in the parking lots. Error bars represent mean ± SD (n = 8 independently collected aerosol samples). ND: not detected. f Schematic diagram of sampling sites in wards for SARS-CoV-2 patients. g Comparison of the virus concentration of aerosol samples collected before and after indoor ventilation conditions in the same patient ward. Solid lines represent the mean values. (n = 4 independently collected aerosol samples). h Violin plot of Ct values of positive test results at seven environmental sampling sites, revealing that aerosol samples show lower mean Ct values, i.e. better environmental virus risk assessment. i Virus concentration in wards where aerosol samples were collected. j Positive detection rates of the seven environmental samples, highlighting the highest positive detection rate of aerosol samples. k Comparison of positive detection rates of environmental samples from the same patient ward collected and tested on day 1 and day 3. l Positive detection rates of environmental samples in two wards for severe COVID-19 patients and two wards for mild COVID-19 patients indicating that aerosol samples can better reflect the patients’ disease status (in terms of individual viral shedding).
Here we proposed a multi-site sampling RIAMs (M-RIAMs) containing multiple aerosol samplers for the application where the aerosols can be collected in several locations and then analyzed in a batch. Since the samples must be transported from the sampling to the analysis site, we designed that the sample container of the cartridge can be directly used as the collection tube in the sampler. As a result, once the aerosol collection is completed and the sample container is capped at the sampling site, there is no need to open the cap again in the rest of the analysis to guarantee the safety of the operator and to eliminate the risk of sample cross-contamination (Fig. 3a). The spatial virus resolution of RIAMs can be calculated by Eq. (1):
The spatial resolution of virus detection is dependent on sampling conditions. When the sampling buffer is 1 ml (all used for detection) and the sampling time is 30 min, and assuming that there is no wall particle adsorption under ideal conditions (or we can compensate for the efficiency by extending the sampling time), the spatial virus resolution reaches the maximum value with a sub-single-copy spatial resolution of 0.83 copies/m3, which is the highest spatial resolution reported to date.
We investigated whether the detection results could be affected by unknown airborne impurities in different environments. We conducted the aerosol collections for 30 minutes in several locations, including public restroom, offices, corridors, and parking lots. Then, we added 1 μL of 50 copies/μL SARS-CoV-2 pseudovirus into a collection tube with 1 mL of lysis buffer to generate a positive sample with a final concentration of 50 copies/mL (Supplementary Fig. 5). We found that only the samples collected from parking lots showed some false negative results (Fig. 3d), which can be resolved by adding a piece of filter cotton to the air inlet of the cyclone sampler (Fig. 3e).
Large-scale analysis of COVID-19 environmental samples using M-RIAMs
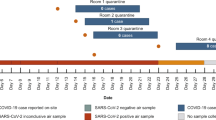
We conducted a comprehensive evaluation of the M-RIAMs with aerosol and surface swabbing samples collected from COVID-19 patient wards in Peking University First Hospital (Fig. 3f and Supplementary Method 6). We first collected aerosol samples using the cyclone aerosol sampler before and after the enhanced ventilation in the same patient ward. We then analyzed these samples and found that all four samples were positive before the enhanced ventilation, while only one out of four tests remained positive after ventilation, and the mean virus concentration of collected aerosol samples dropped significantly, suggesting that the aerosol sample analysis can effectively reflect the risk of aerosol transmission (Fig. 3g). Next, we collected various environmental samples in wards to assess the effectiveness of the aerosol analysis compared with the conventional surface swabbing method for the assessment of infection risks. In each room, the aerosol samples and six surface swabbing samples from the bedside table, the patient collar, the bedding, the patient skin, the medical monitor, and the pager were repeatedly collected 5 times at 1-hour intervals. Ultimately, we tested a total of 210 environmental samples from five wards on different days. From the violin plots of Ct values of those collected samples, we noticed that the median Ct values of the positive aerosol samples was smaller than those of surface swabbing samples (typically 1-2 units lower than those of the swabbing samples) (Fig. 3h). Through calculation, we found that the virus concentrations of most positive aerosol samples ranged from 1.98–71.6 copies/m3 (only one sample reached a virus concentration of 127.05 copies/m3) (Fig. 3i), which are similar to those reported in literatures (Supplementary Table 4)6,13,18,39,40,41,42,43,44. In addition, we found that indoor aerosol samples showed the highest positivity rate of 60% (Fig. 3j), suggesting that aerosol monitoring could be a more sensitive tool for detecting environmental viruses than surface swabbing method18,45. The aerosol samples can achieve the full coverage of the space and are not affected by sampling site bias, allowing a more objective assessment of the infection risks due to aerosol exposure.
In addition, we looked into the positive detection rates of the samples collected on day 1 and 3 of the same ward and found that the positive detection rate of the aerosols matched the patient’s condition very well. As the patient’s condition improved from severe to mild (as assessed by medical staff based on the patient’s clinical symptoms)46,47, the positive detection rate dropped from 100% to 20%, while other environmental samples did not show a similar discernible pattern (Fig. 3k). Furthermore, we found that the aerosol samples collected from the wards with severe COVID-19 patients have a higher positive detection rate than those from the wards with mild COVID-19 patients, while this correlation was not observed in other environmental samples (Fig. 3l). These results highlight the potential of the aerosol monitoring for accurately reflecting the patients’ disease status (in terms of individual viral shedding).
S-RIAMs: design and performance evaluation for real-time virus-laden aerosol monitoring
We next developed a stationary real-time RIAMs (S-RIAMs) for semi-continuously monitoring of airborne pathogens in a specific ___location, with manual intervention required for system cleanup for a positive test result. The S-RIAMs consists of four modules: a cyclone bioaerosol sampler, an automated sampling unit, a loading tray which can store up to 8 cartridges, and a control and detection system (Fig. 4a, b and Supplementary Fig. 6). The S-RIAMs can be modified and assembled on a robotic chassis to form the roaming real-time RIAMs (R-RIAMs) for continuously sampling and analysis of bioaerosols in a large indoor environment (Fig. 4c and Supplementary Fig. 7). Unlike the M-RIAMs which collects aerosols into a sample tube, the automated sampling unit in the S-RIAMs and R-RIAMs can quantitatively inject the sample liquid from the collection tube of the sampler into the sample container of the cartridge via a needle using a peristaltic pump (Fig. 4d). Once the sample is loaded, the loading tray can move the cartridge to the multiplex PCR detection system for the virus detection (see Supplementary Movie 2 and Supplementary Method 5). We verified the adsorption of nucleic acids on the surface of the silicone tubing and other components used for liquid transfer within S-RIAMs and R-RIAMs. Results showed that the Ct values of these samples did not change significantly, indicating no nucleic acid adsorption within the sampling system (Fig. 4e). The total costs of the S-RIAMs and R-RIAMs instrumentations as well as the disposable microfluidic cartridge are listed in Supplementary Table 5. The cost of a single test includes a microfluidic cartridge with PCR reagents, which is approximately $ 2.75. We believe that the relatively low cost of each test can definitely support the actual deployment of RIAMs in large-scale applications.
a Structural overview of S-RIAMs and exploded schematic of its functional parts, including a cyclone bioaerosol sampler unit (i), a microfluidic cartridge sampling unit (ii), microfluidic cartridges loading tray (iii), and a control and detection system for performing fluid actuation, PCR temperature control, and fluorescence signal read-out (iv). b Optical image of S-RIAMs showing its 3D dimensions. c Optical image of R-RIAMs showing its 3D dimensions. d Schematic illustration of the microfluidic cartridge sampling process. e Ct values of viral RNA samples before and after transfer through the sampling system, verifying that there is no adsorption of viral RNA within the sampling systems inside the S-RIAMs and R-SIAMs. Error bars represent mean ± SD (n = 3 independently transferred or not-transferred samples).
We then evaluated the capability of the S-RIAMs for monitoring the airborne transmission of SARS-CoV-2 using a Whole-Body Inhalation Exposure System (WIES), which can simulate real-world aerosol environment by generating consistent aerosol particles through the compression of a medical nebulizer (Fig. 5a). Using the S-RIAMs, we successfully detected pseudovirus samples with a gradient of concentrations of SARS-CoV-2 generated by WIES with a coefficient of determination r² = 0.81, validating the reliability of the S-RIAMs in aerosol monitoring (Fig. 5b, c).
a Photograph of the experimental setup of the generation and simultaneous detection of SARS-CoV-2 aerosols in a biosafety cabinet. b A histogram of Ct values of mock aerosol samples with varying viral concentrations in the aerosolized liquid stock. Error bars represent mean ± SD (n = 3 independently aerosolized and collected samples). c The fitted relationship between the logarithm of virus input concentration and Ct values. Error bars represent mean ± SD (n = 3 independently aerosolized and collected samples). d Results of 6-day continuous aerosol COVID-19 monitoring using S-RIAMs in an office workplace affected by the COVID-19 epidemic, showing the positive detection rates and the absolute amount of airborne virus in the space. Solid lines represent mean values. (n = 8 independently collected and tested aerosol samples). e Comparison of the positive detection rates and the virus concentrations on day 1 and day 4 of airborne viruses monitoring results around infected individuals within the student dormitory affected by the COVID-19 pandemic, indicating a gradual increase in the number of infected people. Solid lines represent mean values. (n = 10 independently collected and tested aerosol samples). f A scatter plot of the virus concentrations of aerosol RSV samples collected from neonatal RSV-positive areas, including 10 all-positive aerosol samples from 5 neonatal RSV-positive wards.
Real-world deployment of S-RIAMs for ultrasensitive aerosol surveillance of SARS-CoV-2 and RSV
To validate the applicability of the S-RIAMs for monitoring SARS-CoV-2 and other respiratory aerosol viruses in real-world settings, we deployed the S-RIAMs in several real-world locations, including the office workplace, the centralized dormitories, and the pediatric wards with RSV-positive infants in the hospital (Details of those samples can be seen in Supplementary Method 6). During the COVID-19 epidemic in China at the end of 2022, we deployed the S-RIAMs in an office space and configured the system to run continuously for 6 consecutive days from 8 a.m. to 8 p.m with a monitoring interval of 1.5 hour. In total, 48 aerosol samples were collected and analyzed. From the plot of summarized virus concentrations, we observed that the mean concentration of spatial virus in each day initially increased and then decreased, reaching a maximum on the fourth day, which is 228.6 copies/m3. The change of the positive detection rate followed a similar pattern, highlighting the capability of the S-RIAMs to continuously and ultra-sensitively detect SARS-CoV-2 in aerosols (Fig. 5d). Next, we conducted the aerosol monitoring around infected individuals within the student dormitory affected by the COVID-19 pandemic. The results showed the increasing trend of the spatial virus concentration and positive detection rate on day 1 and 4, indicating a gradual increase of the number of infected people (Fig. 5e). The results demonstrated that the monitoring of airborne SARS-CoV-2 can reflect the spread and the infection rate in a specific area. We also deployed S-RIAMs at different locations that were confirmed virus-free, testing a total of 16 samples. No false positives were detected for any of the four viruses in these samples (Supplementary Fig. 8).
Other than SARS-CoV-2, RSV primarily causes respiratory infections in infants and young children, especially those under two years of age48. Due to the issue of the uncomfortable sampling via nasopharyngeal swab for infants, we believe the aerosol testing could be used as a non-invasive monitoring method of RSV infections. We totally tested 10 aerosol samples from 5 neonatal RSV-positive wards using the S-RIAMs at the Second Affiliated Hospital of Wenzhou Medical University and the all-positive results demonstrated it is possible to monitor the infection spread of RSV. Overall, the monitoring results of S-RIAMs in these real-world settings convincingly demonstrate its capability for highly sensitive and continuous airborne virus detection, enabling the accurate assessment of the viral risk in a specific ___location.
Discussion
Airborne human infectious viruses pose a significant threat to public health. Surveillance of SARS-CoV-2 and other respiratory viruses in aerosols represents an alternative strategy that liberates the detection from individuals. Previously, the aerosol surveillance remains challenging due to the lack of monitoring equipment with a high spatial resolution and a rapid turnaround time, significantly reducing the effectiveness of early virus risk assessment. In response, we have developed the RIAMs, a system capable of achieving ultra-sensitive and continuous monitoring of SARS-CoV-2 and other respiratory viruses in aerosols. Compared to the SIAMs developed previously (Supplementary Table 3)30, the RIAMs has three key updates: the high-flow aerosol sampler with an improved collection efficiency designed using a dry-wall centrifugal mechanism, an ultra-sensitive detection chemistry based on in-situ four-plex PCR, and a newly developed microfluidic cartridge using the “needle-plug/piston” mesoscopic design paradigm36. As a result, RIAMs demonstrates a superb sensitivity of 10 copies/mL, which is an order of magnitude higher than that of commercial kits. When the aerosol sampling module operates at 400 lpm, the RIAMs can achieve an unparalleled sub-single-copy spatial virus resolution of 0.83 copies/m³, making it the most sensitive airborne virus detection device known to date.
To better meet the needs of different aerosol application scenarios, we have developed three different forms of aerosol monitoring systems: M-RIAMs, S-RIAMs, and R-RIAMs. Together, these three sub-systems enable the RIAMs to support both offline and online multi-mode aerosol monitoring. Using the M-RIAMs, we conducted the comprehensive evaluation of the system using 210 environmental samples (including 30 aerosol samples) collected from COVID-19 patient wards and demonstrated the superiority of aerosol virus surveillance over other environmental surface swabbing samples. Additionally, we tested 78 aerosol samples using the S-RIAMs, showing the excellent capability for ultrasensitive monitoring of SARS-CoV-2 and other respiratory viruses in aerosols. We envision the widespread use of the RIAMs in the following application scenarios so that relevant personnel can take timely preventive measures to cut off the transmission line of severe respiratory infectious diseases: (i) aerosol monitoring in hospitals, such as fever clinics, wards, and public spaces, to reduce the risk of nosocomial infections; (ii) aerosol monitoring in locations with vulnerable people, such as kindergartens, primary schools, and nursing homes, to safeguard their lives and health; (iii) aerosol monitoring at airports, railway stations, border ports, and other relevant sites to prevent cross-border spread of infectious diseases.
One limitation of the current form of the RIAMs is that it can only detect four airborne viruses simultaneously in each run. The diverse range of pathogenic microorganisms surviving in the air, including bacteria, fungi, and viruses, necessitates the future efforts to increase the number of detection targets to provide a more objective and comprehensive assessment of the infection risk for airborne pathogens. In addition, necessary real-world detection studies of influenza and its subtypes, as well as more RSV samples, are needed to further demonstrate the ability of RIAMs. Other improvements of the system that need to be considered include the reduce of the cost, the shortening of the analytical time of each test and the completion of automated system cleanup of RIAMs. Furthermore, the effects of adjustable flow mode on sampling efficiency, cutoff size, spatial virus resolution, etc., as well as the changes in spatial virus depiction caused by different sampling efficiencies can be studied49. Besides, regarding aerosol particle collection, this study focused on the direct collection of particles within the cyclone pipe. Further studies need to be conducted to measure the “total” and “actual” collection efficiency, which can be achieved using tracer aerosols, isokinetic sampling, or comparison with another reference sampler38, to more accurately evaluate the actual collection efficiency of the sampler.
Methods
Reagents and materials
Detailed information about reagents and materials, including their functions and commercial vendors, are provided in Supplementary Table 2. Detailed item numbers of important reagents and materials are mentioned in the description of the supplementary methods, including the item numbers of polystyrene latex (PSL) microspheres used to measure collection efficiency of the aerosol sampler and the item numbers of RNA standards and pseudovirus reference materials used for assay performance evaluation.
Fabrication and operation of the microfluidic cartridge
All the components of the microfluidic cartridge were designed using AutoCAD 2019 and CATIA P3 V5R21 software for 2D and 3D drawings. The microfluidic cartridge used in the validation experiments was fabricated from polymethylmethacrylate (PMMA) using a CNC milling machine. For mass production, the final version of the cartridge was produced using an injection molding process by Shenzhen Hechuan Medical Technology Co., Ltd., China. During the biological experiment, the cartridge was pre-loaded with all necessary reagents, including 1 mL of lysis buffer in the sample chamber, 1 mL of DEPC water in the washing chamber (purchased from Shanghai Beyotime Biotechnology Co., Ltd., China.), and 50 μL of PCR reagent in the upper part of the PCR reagent tube. The PCR mix was provided as lyophilized powder and stored in the lower part of the PCR reagent tube (purchased from Zhuhai Bao Rui Biotechnology Co., Ltd., China.).
Preparation of chitosan-modified quartz filter paper
Chitosan with low molecular weight was purchased from Sigma-Aldrich (St. Louis, MO). MES (2-(N-morpholino)-ethanesulfonic acid) and GPTMS (3-glycidyloxypropyltrimethoxysilane) were purchased from Shanghai Macklin Biochemical, China. The quartz filter (QF) paper was produced from Ø 47 mm diameter glass fiber filter paper (QMA, Whatman, GE Healthcare, Pittsburgh, PA). All solutions were prepared in water purified to 18.2 MΩ·cm by Milli-Q Advantage A10 (Millipore, Massachusetts, MA). The modified procedure, which has been previously published31,32, involves the mixing of a 0.1% MES solution with a 1% chitosan solution, the adding of the filter paper to a Petri dish containing the mixed reagents, and then the adding of the GPTMS solution to the Petri dish. After the incubation at 75 °C for 24 hours, the filter paper was washed 3-4 times with ultrapure water and dried at 50 °C to obtain the final chitosan-modified filter paper. Before use, the modified filter paper can be stored in a dry container to avoid absorbing water.
Four-plex PCR assay
The primers and the probes of SARS-CoV-2, Influenza A, B, and RSV in the four-plex PCR were designed using the online Primer-BLAST program (https://www.ncbi.nlm.nih.gov/tools/primer-blast). And the sequences of the primers and the probes can be found in Supplementary Table 1. All the primers and the probes were synthesized and purified using HPLC by Shanghai Sangon Biotech, China. All primers, including forward and reverse primers for each respiratory virus, were at a concentration of 0.5 μM, and the concentration of the probes were 0.3 μM.
LoDs determination using quality control reference materials
To determine the LoDs of the four-plex PCR amplification, we added 2 μL of viral RNA at various concentrations into 1-mL lysis buffer, with final concentrations of 1,000, 200, 100, 50, 20, 10, and 5 copies/mL, respectively. RNA extraction was conducted using the simplified microfluidics chips embedded with Ø 3.5 mm QF papers. After the nucleic acid capture, these filter papers were then washed with 1 mL of DEPC water to eliminate impurities from the lysis buffer. Subsequently, the filter paper was transferred to a 25 μL RT-qPCR amplification system and RT-qPCR was performed on an Applied Biosystems 7500 real-time PCR system. The amplification protocol consisted of a 5-min RT step at 52 °C, followed by 45 cycles of denaturation at 95 °C for 5 s, and annealing and extension at 60 °C for 30 s. To determine the LoDs provided by the microfluidic cartridge, 2 μL of pseudoviruses at different concentrations was added to the sample tube of the microfluidic cartridge. Once the cartridge is transferred to the instrument, the bind-wash-in-situ amplification step can be performed automatically in 45 minutes. All information of the reference materials used in RIAMs performance evaluation experiments can be found in Supplementary Method 2.
PCR specificity testing
Pseudoviruses of the four viruses were added to 1 mL of lysis buffer without one of the four viruses. Those carrying all four viruses and those without the virus were used as the positive and the negative controls. The concentration of each virus is 200 copies/mL. The viral RNA extractions were performed in the simplified microfluidic chips described above and the RNA amplifications were performed on the 7500 real-time PCR system.
Performance verification of aerosol sampler
A customized air filter screen with a particle size of 100 μm (Item No. F25F-102-102, Shenzhen Biocomma Biotechnology Co., Ltd. China) was installed in the air inlet of the sampler to remove large particles (≥100 µm) in specific occasions, such as parking lots, where relatively large amounts of dust exist.
The Whole-Body Inhalation Exposure System (WIES) was manufactured by PARI, Germany (purchased from Shanghai Yuyan Scientific Instrument Co., Ltd., China.). Monodisperse green fluorescent polystyrene microspheres with diameters of 0.2, 0.5, 1, and 2 μm were purchased from Wuxi Ruiger Biotechnology Co., Ltd., China. To verify the capability of the aerosol sampler to collect particles of different sizes, 3.5 mL of microspheres of four different diameters were added to the reagent tubes of the WIES aerosol generation system. Particle size analysis and statistical evaluation were performed using a BD Aria III flow cytometer after half an hour of aerosol generation and simultaneous parallel aerosol collection. It is important to note that due to the chamber’s volume of 0.8 m³ and the fact that it is not sealed, additional air could be drawn into the chamber to dilute the samples during the aerosol collection process.
Prototype design of M-RIAMs, S-RIAMs, and R-RIAMs
Prototype processing instruments for M-RIAMs, S-RIAMs, and R-RIAMs were developed to demonstrate the automation feasibility of the system by integrating off-the-shelf components, custom-designed components using SolidWorks, custom electronic boards, and custom software written in C. Custom-designed components were fabricated using standard manufacturing methods, including commercial CNC machining of aluminum components and stereolithography (SLA) 3D printing. Commercially available components used in buffer transfer of S-RIAMs and R-RIAMs included two Ditron-tech peristaltic pumps (S100-2B, Ditron-tech Co., Ltd., Baoding, China), five BEION solenoid pinch valves (P20NC12-02, Beionfluid Co., Ltd., Shanghai, China), an Elveflow bubble remover, and two non-touch liquid-level sensors (XKC-Y28-NC, XKC Technology Co., Ltd., Shenzhen, China). The amplification protocol for the in-situ four-plex PCR within the RIAMs device is the same as that of the Applied Biosystems 7500 RT-PCR system.
Ethics declaration
All experiments conducted in real-world scenarios were approved by the Science and Technology Ethics Committee of Tsinghua University (Project Number: THU01-20230093). In the comparative experiment using M-RIAMs on clinical aerosol samples in the ward and other environmental samples, 210 clinical samples were sampled by colleagues from Peking University First Hospital. All samples were then analyzed using our in-situ ultra-sensitive detection methods. Similarly, for clinical aerosol RSV samples monitoring in wards using S-RIAMs, experiments were conducted on-site at the Second Affiliated Hospital of Wenzhou Medical University by colleagues from Wenzhou Medical University. Of note, all patients from whom samples were collected signed informed consent for the collection and analysis of various environmental samples, including aerosol samples, from their respective wards. The remaining experiments involving the use of the S-RIAMs to analyze and detect infected office and school aerosol samples were conducted with informed consent from all individuals involved.
Data availability
The source data underlying Fig. 2b, d, e, f, Fig. 3b, c, d, e, g, h, i, Fig. 4e, Fig. 5b, d, e, f, Supplementary Fig. 8 are provided as a Source data file. Source data are provided with this paper.
References
Zhu, N. et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 382, 727–733 (2020).
Hsiang, S. et al. The effect of large-scale anti-contagion policies on the COVID-19 pandemic. Nature 584, 262–267 (2020).
Wang, C. C. et al. Airborne transmission of respiratory viruses. Science 373, eabd9149 (2021).
Greenhalgh, T. et al. Ten scientific reasons in support of airborne transmission of SARS-CoV-2. Lancet 397, 1603–1605 (2021).
Stadnytskyi, V., Bax, C. E., Bax, A. & Anfinrud, P. The airborne lifetime of small speech droplets and their potential importance in SARS-CoV-2 transmission. Proc. Natl. Acad. Sci. 117, 11875–11877 (2020).
Liu, Y. et al. Aerodynamic analysis of SARS-CoV-2 in two Wuhan hospitals. Nature 582, 557–560 (2020).
Jaumdally, S. et al. Frequency, kinetics and determinants of viable SARS-CoV-2 in bioaerosols from ambulatory COVID-19 patients infected with the Beta, Delta or Omicron variants. Nat. Commun. 15, 2003 (2024).
Setti, L. et al. Airborne transmission route of COVID-19: why 2 meters/6 feet of inter-personal distance could not be enough. Int. J. Environ. Res. Public. Health 17, 2932 (2020).
Van Doremalen, N. et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 382, 1564–1567 (2020).
Rechsteiner, J. & Winkler, K. C. Inactivation of respiratory syncytial virus in aerosol. J. Gen. Virol. 5, 405–410 (1969).
Weber, T. P. & Stilianakis, N. I. Inactivation of influenza A viruses in the environment and modes of transmission: a critical review. J. Infect. 57, 361–373 (2008).
Loosli, C. G., Lemon, H. M., Robertson, O. H. & Appel, E. Experimental air-borne influenza infection. I. Influence of humidity on survival of virus in air. Exp. Biol. Med. 53, 205–206 (1943).
Chia, P. Y. et al. Detection of air and surface contamination by SARS-CoV-2 in hospital rooms of infected patients. Nat. Commun. 11, 2800 (2020).
Raymenants, J. et al. Indoor air surveillance and factors associated with respiratory pathogen detection in community settings in Belgium. Nat. Commun. 14, 1332 (2023).
Moriarty, L. F. et al. Public health responses to COVID-19 outbreaks on cruise ships—worldwide, February–March 2020. MMWR. Morb. Mortal. Wkly. Rep. 69, 347–352 (2020).
Hadei, M. et al. Presence of SARS-CoV-2 in the air of public places and transportation. Atmos. Pollut. Res. 12, 302–306 (2021).
Ramuta, M. D. et al. SARS-CoV-2 and other respiratory pathogens are detected in continuous air samples from congregate settings. Nat. Commun. 13, 4717 (2022).
Zhang, X. et al. Monitoring SARS-CoV-2 in air and on surfaces and estimating infection risk in buildings and buses on a university campus. J. Exposure Sci. Environ. Epidemiol. 32, 751–758 (2022).
Miller, S. L. et al. Transmission of SARS‐CoV‐2 by inhalation of respiratory aerosol in the Skagit Valley Chorale superspreading event. Indoor Air 31, 314–323 (2021).
Shen, Y. et al. Community outbreak investigation of SARS-CoV-2 transmission among bus riders in eastern China. JAMA Intern. Med. 180, 1665 (2020).
Qiu, G. et al. On-site airborne pathogen detection for infection risk mitigation. Chem. Soc. Rev. 52, 8531–8579 (2023).
Morawska, L. & Milton, D. K. It is time to address airborne transmission of coronavirus disease 2019 (COVID-19). Clin. Infect. Dis. 71, 2311–2313 (2020).
Rahmani, A. R., Leili, M., Azarian, G. & Poormohammadi, A. Sampling and detection of corona viruses in air: A mini review. Sci. Total Environ. 740, 140207 (2020).
Rastmanesh, A., Boruah, J. S., Lee, M.-S. & Park, S. On-Site Bioaerosol Sampling and Airborne Microorganism Detection Technologies. Biosensors 14, 122 (2024).
Hu, X. et al. Evaluation of the clinical performance of single-, dual-, and triple-target SARS-CoV-2 RT-qPCR methods. Clin. Chim. Acta 511, 143–148 (2020).
Puthussery, J. V. et al. Real-time environmental surveillance of SARS-CoV-2 aerosols. Nat. Commun. 14, 3692 (2023).
Cho, Y. S. et al. Continuous surveillance of bioaerosols on-site using an automated bioaerosol-monitoring system. ACS Sens. 5, 395–403 (2020).
Heo, K. J., Ko, H. S., Jeong, S. B., Kim, S. B. & Jung, J. H. Enriched aerosol-to-hydrosol transfer for rapid and continuous monitoring of bioaerosols. Nano Lett 21, 1017–1024 (2021).
Lee, I. et al. Integrated bioaerosol sampling/monitoring platform: field-deployable and rapid detection of airborne viruses. ACS Sens. 5, 3915–3922 (2020).
Li, S. et al. An ultrasensitive and rapid “sample-to-answer” microsystem for on-site monitoring of SARS-CoV-2 in aerosols using “in situ” tetra-primer recombinase polymerase amplification. Biosens. Bioelectron. 219, 114816 (2023).
Gu, Y. et al. Modular-based integrated microsystem with multiple sample preparation modules for automated forensic DNA typing from reference to challenging samples. Anal. Chem. 91, 7435–7443 (2019).
Gan, W. et al. Chitosan-modified filter paper for nucleic acid extraction and “ in situ PCR” on a thermoplastic microchip. Anal. Chem. 89, 3568–3575 (2017).
Emaus, M. N. et al. Nucleic acid extraction: fundamentals of sample preparation methodologies, current advancements, and future endeavors. Trac Trends Anal. Chem. 130, 115985 (2020).
Geng, Z., Gu, Y., Li, S., Lin, B. & Liu, P. A fully integrated in vitro diagnostic microsystem for pathogen detection developed using a “3d extensible” microfluidic design paradigm. Micromachines 10, 873 (2019).
Geng, Z. et al. “Sample-to-answer” detection of rare ctDNA mutation from 2 mL plasma with a fully integrated DNA extraction and digital droplet PCR microdevice for liquid biopsy. Anal. Chem. 92, 7240–7248 (2020).
Lin, B. et al. Needle‐Plug/Piston‐Based Modular Mesoscopic Design Paradigm Coupled With Microfluidic Device for Point‐of‐Care Pooled Testing. Adv. Sci. 2406076. https://doi.org/10.1002/advs.202406076 (2024).
Li, X. et al. A robot assisted high-flow portable cyclone sampler for bacterial and SARS-CoV-2 aerosols. Aerosol Air Qual. Res. 21, 210130 (2021).
Mainelis, G. Bioaerosol sampling: classical approaches, advances, and perspectives. Aerosol Sci. Tech. 54, 496–519 (2020).
Santarpia, J. L. et al. The size and culturability of patient-generated SARS-CoV-2 aerosol. J. Expo. Sci. Environ. Epidemiol. 32, 706–711 (2022).
Stern, R. A. et al. Characterization of hospital airborne SARS-CoV-2. Respir. Res. 22, 73 (2021).
Birgand, G. et al. Assessment of air contamination by SARS-CoV-2 in hospital settings. JAMA Netw. Open. 3, e2033232 (2020).
Buonanno, G. et al. Link between SARS-CoV-2 emissions and airborne concentrations: closing the gap in understanding. J. Hazard. Mater. 428, 128279 (2022).
Stern, R. A., Al-Hemoud, A., Alahmad, B. & Koutrakis, P. Levels and particle size distribution of airborne SARS-CoV-2 at a healthcare facility in Kuwait. Sci. Total Environ. 782, 146799 (2021).
Zhou, L. et al. Breath-, air- and surface-borne SARS-CoV-2 in hospitals. J. Aerosol Sci. 152, 105693 (2021).
Ang, A. X. et al. Airborne SARS‐CoV‐2 surveillance in hospital environment using high‐flowrate air samplers and its comparison to surface sampling. Indoor Air 32 (2022).
He, X. et al. Clinical symptom differences between mild and severe COVID-19 patients in China: a meta-analysis. Front. Public Health 8, 561264 (2021).
Song, J.-W. et al. Immunological and inflammatory profiles in mild and severe cases of COVID-19. Nat. Commun. 11, 3410 (2020).
Karron, R. A. Preventing respiratory syncytial virus (RSV) disease in children. Science 372, 686–687 (2021).
Luhung, I. et al. Experimental parameters defining ultra-low biomass bioaerosol analysis. npj Biofilms Microbiomes 7, 37 (2021).
Acknowledgements
This work was financially supported by the National Key Research and Development Program of China (Grant No. 2022YFC2704902 [Liu]) and the National Natural Science Foundation of China (Grant No. 32001021 [Gu]).
Author information
Authors and Affiliations
Contributions
B. Li and B. Lin designed and performed experiments, analyzed the data, and prepared the manuscript. Y.W. and Y.S. helped design and perform experiments. P.L. conceived the project, designed experiments, provided overall supervision for this work and wrote the manuscript. G.W. and G.X. provided clinical samples and co-supervised this work. W.Z. helped design the microfluidic cartridge and the instrument of M-RIAMs, S-RIAMs, and R-RIAMs. H.G., H.C. and X.Z. helped provide clinical samples. Y.G. provided the QF paper for in-situ amplification. Y.Z. helped perform experiments of clinical samples. C.L. helped revise the manuscript. All authors commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Irvan Luhung, Monika Redlberger-Fritz, Alistair Walsham and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, B., Lin, B., Wang, Y. et al. Multi-scenario surveillance of respiratory viruses in aerosols with sub-single-copy spatial resolution. Nat Commun 15, 8770 (2024). https://doi.org/10.1038/s41467-024-53059-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-53059-x