Abstract
Defects at the neuromuscular junction (NMJ) are among the earliest hallmarks of amyotrophic lateral sclerosis (ALS). According to the “dying-back” hypothesis, NMJ disruption not only precedes but also triggers the subsequent degeneration of motoneurons in both sporadic (sALS) and familial (fALS) ALS. Using human induced pluripotent stem cells (iPSCs), we show that the RNA-binding protein HuD (ELAVL4) contributes to NMJ defects and apoptosis in FUS-ALS. HuD overexpression mimics the severe FUSP525L mutation, while its knockdown rescues the FUSP525L phenotypes. In Drosophila, neuronal overexpression of the HuD ortholog, elav, induces motor dysfunction, and its knockdown improves motor function in a FUS-ALS model. Finally, we report increased HuD levels upon oxidative stress in human motoneurons and in sALS patients with an oxidative stress signature. Based on these findings, we propose that HuD plays a role downstream of FUS mutations in fALS and in sALS related to oxidative stress.
Similar content being viewed by others
Introduction
The neurodegenerative disease amyotrophic lateral sclerosis (ALS) is caused by progressive loss of skeletal muscle function due to motoneuron (MN) death1. About 10% of the cases are classified as inherited (familial ALS), while the vast majority are considered sporadic. Pathogenic variants in several genes have been linked to both familial and sporadic ALS cases. Most of these variants map in the C9ORF72 and SOD1 genes, but several ubiquitously expressed RNA-binding proteins (RBPs) have been linked to ALS. In the nervous system, ubiquitous and neural-specific RBPs cooperate in gene expression regulation, and cross-regulation and feedback loops are crucial in establishing proper levels and activity of each RBP2. An extensive crosstalk also exists between ubiquitous ALS-linked RBPs, such as FUS, and other neural RBPs expressed in the MN3,4. Thus, in the MN, ALS-associated genetic variants in one node of the network might impact other RBPs and produce broader, unexpected, effects that could be highly cell-type specific. In this context, we have previously shown that mRNA and protein levels of HuD, encoded by the ELAVL4 gene, are upregulated in MNs derived from FUS mutant human induced pluripotent stem cells (iPSCs) and in a mouse model (Fus-Δ14 knock-in)5. Increased HuD levels in FUSWT MNs produce changes in the transcriptome that are substantially similar to those exerted by the severe FUS p.P525L variant6. A direct consequence of HuD upregulation in FUS mutant iPSC-derived MNs and mouse spinal cord is an increase in the levels of its targets, including Neuritin1 (NRN1; also known as cpg15) and GAP435. HuD plays key roles during nervous system development and recent evidence points to its possible involvement in neurodegenerative processes as well7,8,9.
In this work, we aimed to assess if HuD dysregulation could underlie neuromuscular junctions (NMJs) defects in the context of ALS. Several genes that are commonly altered in FUSP525L MNs and HuD overexpressing MNs are involved in axon projection, cell junction, and synapse formation6,10. ALS is considered a distal axonopathy as early impairment of NMJs has been reported in both animal models and patients. These include SOD1G93A mice, in which endplate denervation, followed by ventral root axon degeneration, occurs way before the death of MNs in the spinal cord11. Notably, such “dying-back” pattern was also evident in a sporadic ALS patient who unexpectedly died few weeks after the diagnosis11, and recently proposed for familial ALS linked to RBP genes, including FUS. Increased apoptosis and lower MNs loss due to toxic gain of function of the mutant protein was observed at birth in a knock-in Fus mouse model (FusΔNLS/ΔNLS)12. Notably, MN loss in FusΔNLS/ΔNLS mice occurred in the absence of FUS protein aggregation and stress granules formation and was rescued by MN-specific reversion of FUS mislocalization12. These findings point to cell-autonomous mechanisms leading to MN death which are due to the gain of toxic cytoplasmatic functions of the FUS mutant protein and are independent from its aggregation. A direct causal link between FUS variants and NMJ defects was demonstrated by Picchiarelli et al., who showed a reduction in the number and area of the endplates in newborn FusΔNLS/ΔNLS mice and progressive denervation and reduction of the endplate size in adult heterozygous mutants (FusΔNLS/+)13. This phenotype, which was observed in human iPSC-derived co-cultures as well, was due to cell-autonomous toxicity of mutant FUS in both skeletal muscle cells (SkMCs) and MNs. In the muscle, innervation induces FUS enrichment in subsynaptic nuclei, where it promotes expression of Chrn genes. However, this cannot be considered the sole underlying mechanism for the observed NMJ defects, as FUS mutation in MNs exerted a similar phenotype upon co-culture with wild-type iPSC-derived SkMCs in the absence of any difference in AChR gene expression in the muscle13. Decreased number of NMJs per myotube and impaired NMJ maturation were also observed in co-cultures of mutant FUS iPSC-derived MNs and human myotubes derived from mesoangioblasts (MAB) from a healthy individual14.
Here we found that HuD gain-of-function can account for most of the cell-autonomous effects of a severe FUS variant in the MN. These include defective maturation of the NMJ and apoptosis, possibly by a “dying-back” pattern, in co-cultures of iPSC-derived MNs and SkMCs. These findings were further supported in vivo using a Drosophila model, in which reduction of the HuD ortholog gene elav is sufficient to rescue the motor phenotype caused by FUS overexpression. Strikingly, in both iPSC-derived co-cultures and Drosophila, HuD (or ELAV) overexpression produces relevant ALS phenotypes per se. Thus, we wondered if HuD-dependent mechanisms might underlie sporadic ALS independently of FUS mutation. Oxidative stress is thought to play a major role as an environmental factor involved in ALS onset and progression1. Notably, increased HuD levels were recently reported in neuroblastoma cells upon oxidative stress and in the motor cortex from few sporadic ALS (sALS) patients8. Here, by reanalyzing the data from a large transcriptome study15, we showed that HuD expression is specifically increased only in sALS cases with an oxidative stress signature (accounting for about 60% of the cohort), while it is decreased or unchanged in the other groups. We propose that HuD-dependent pathological mechanism directly contributes to familial FUS-ALS and in a distinct group of sALS cases associated with oxidative stress.
Results
Establishment of iPSC-derived neuromuscular co-cultures
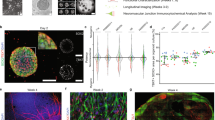
We have previously developed methods to obtain iPSC-derived functional MNs and SkMCs16,17. The protocol for SkMCs generation was improved to reduce the subpopulation of differentiation-resistant cells in more extended cultures (Supplementary Fig. S1A–D). Spinal MN progenitors, generated in 5 days as previously described17, were re-plated onto SkMCs obtained in 13 days with the optimized protocol (Fig. 1A, B). Recombinant agrin and laminin proteins were added to the culture medium to promote the expression of post-synaptic AChR subunit genes in muscle cells through the activation of the agrin–LRP4–MuSK–ERM axis, and pre-synaptic differentiation and neurotransmitter release, respectively14,18. After this step, hereafter considered day 0 of the co-culture, we maintained MNs and SkMCs for long-term culture. In line with proper differentiation, immunostaining of sarcomeric α-actinin showed a striated pattern (Supplementary Fig. S2A, B). Clustering of acetylcholine receptors (AChRs), detected by α-bungarotoxin (α-BTX) staining, suggested the establishment of motor endplates as early as day 14 (Fig. 1C). The co-localization of the presynaptic marker SYNAPTOPHYSIN (SYP) and α-BTX signals further indicated the establishment of neuromuscular junctions (NMJs) (Fig. 1D; Supplementary Fig. S2C). At day 50, SkMCs show few spontaneous contractions, which were significantly increased by addition of glutamate, to stimulate MNs, and subsequently reduced to zero by addition of tubocurarine, a non-depolarizing competitive antagonist of the nicotinic AChR at the NMJ (Fig. 1E). Tubocurarine was also able to block spontaneous contractions observed at a later time point (62 days; Supplementary Video S1 and S2). Notably, SkMCs cultured in the same conditions, but in absence of MNs, never showed contractions.
A Schematic representation of the differentiation protocol to generate co-cultures of spinal MNs and SkMCs. A more detailed SkMC differentiation scheme is shown in Supplementary Fig. S1. B Phase-contrast image of MNs and SkMCs co-culture derived from FUSWT iPSCs at day 14. Examples of MN cell body and SkMC are indicated by arrows. Scale bar: 20 μm. C, D Immunostaining analysis of co-cultures at the indicated time points. α-BTX (magenta) was used as a AChRs clustering marker, TUJ1 (TUBB3, green in (C)) and MyHC (grey) are, respectively, neuronal and muscular markers. SYP (green in (D)) is a presynaptic marker. DAPI (blue) was used for nuclear staining. Scale bar: 15 μm, 10 μm, or 5 μm as indicated. E The graph shows the number of contractions per minute in day 50 co-cultures before treatment, 5′ upon glutamate addition, and 5′ upon further addition of tubocurarine. Contractions were counted in 3 randomly selected fields (each represented by dots connected by a line) of 3 independent co-cultures. One representative monoculture of SkMCs only, showing no contractions in all conditions, is also displayed. Two-way repeated measures ANOVA (performed only among co-cultures): for treatment comparison p = 6.6 × 10−8, for replicate comparison p = 0.0837; post hoc Tukey test p values for multiple comparisons among treatments within each replicate are indicated in the graph. Source data are provided as a Source Data file.
Collectively, our data suggests a successful development of a functional neuromuscular model system for mechanistic studies.
NMJ defects due to FUS mutation or HuD overexpression in MNs
Impaired NMJ formation upon FUS mutation in SkMCs, MNs, or both had been previously reported13. We aimed to assess whether the detrimental effect of mutant FUS expression in the MN could be due, at least in part, to increased HuD levels. To this aim, we established co-cultures using MNs from iPSCs with 3 different genetic backgrounds: the isogenic FUSWT and FUSP525L, and FUSWT overexpressing HuD under the Synapsin1 promoter (Syn1::HuD; hereafter indicated as “FUSWT + HuD”)5,19 (Fig. 2A). Notably, the Synapsin1 promoter confers overexpression of HuD in the range observed in FUSP525L MNs5. In all subsequent experiments, SkMCs have always been derived from FUSWT iPSCs.
A Schematic representation of co-cultures used for analyses shown in Figs. 2–5. B Representative images of immunofluorescence staining of day 14 co-cultures using the indicated primary antibodies and α-BTX and DAPI. Scale bar for all panels: 50 μm. C The graphs report quantitative analysis of the percentage of α-BTX positive fibers (left) and TUJ1 fluorescence intensity (right), at day 14 and 28 of co-culture. Each dot represents a replicate, consisting of an individual batch of differentiated iPSCs. For each replicate, the following number of randomly selected fields were used for the α-BTX analysis: 6 (day 14 all genotypes, replicates 1 and 2; day 28 FUSWT, replicate 1; day 28 FUSWT + HuD, replicate 3); 5 (day 14 all genotypes, replicate 3; day 28 FUSWT, replicate 3; day 28 FUSP525L, all replicates) or 4 (day 28 FUSWT, replicate 2; day 28 FUSWT + HuD, replicates 1 and 2). 6 randomly selected fields were used for each replicate of the TUJ1 analysis. Error bars indicate standard deviation calculated on the average value of the replicates. Ordinary two-way ANOVA: for α-BTX positive fibers, p = 0.0715 between day 14 and 28, p = 9.7 × 10−10 among genotypes; for TUJ1 fluorescence intensity, p = 1.3 × 10−6 between day 14 and 28, p = 9.7 × 10−7 among genotypes; relevant post hoc Tukey test p values for multiple comparisons among genotypes within each time point are indicated in the graph. Source data are provided as a Source Data file.
After 14 and 28 days, we observed reduction in muscle fibers showing AChR clusters, detected by α-BTX staining, in the co-cultures established with FUSP525L MNs or upon HuD overexpression (Fig. 2B, C). According to our previous observations5, both FUS mutant and HuD overexpressing MNs showed increased neurite network (Fig. 2B, C). Thus, the reduction of skeletal muscle cells forming endplates cannot be explained by a reduction in the neuronal component in FUS mutant, or HuD overexpressing, MN co-cultures.
Detailed analysis of the endplate morphology suggested defective maturation when mutant FUS was expressed in the MN, confirming previous observations13,14. In particular, we observed a reduced fraction of aligned AChR puncta and endplates categorized as “dense” or “clustered” (Fig. 3A, B). Conversely, endplates with diffuse AChR puncta, indicative of an immature formation stage, were significantly increased. Notably, also in this case, HuD upregulation in an otherwise FUS WT genetic background perfectly phenocopied the defects in NMJ maturation produced by the FUS p.P525L variant.
A Representative images of immunofluorescence staining of day 14 co-cultures using the MyHC antibody and α-BTX and DAPI. Images show representative endplates that were categorized based on morphology into diffuse puncta, aligned puncta (indicated by arrows) and dense/cluster. Scale bar: 10 μm. B The graph shows quantification of endplates categories, as a percentage, from co-cultures obtained with FUSWT SkMCs and FUSWT, FUSP525L, or FUSWT + HuD MNs. The results of a blind analysis of 165 (FUS WT), 124 (FUSP525L), and 147 (FUSWT+HuD) total endplates from 3 batches of differentiated cells are shown. Fisher’s exact test with Bonferroni correction for multiple testing. Source data are provided as a Source Data file.
FUS mutation or HuD overexpression in MNs induced cell death by apoptosis upon co-culture with SkMCs
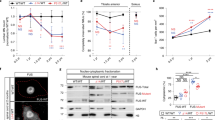
At late time points, beyond day 28, extensive death of both neuronal and muscle cells was observed in co-cultures containing FUSWT SkMCs in presence of FUSP525L or FUSWT + HuD MNs (Fig. 4A). Quantification of dead cells by staining with fluorescent ethidium homodimer-1 in a time course analysis revealed, for both genotypes, significant difference compared to the FUSWT control (Fig. 4B; Supplementary Fig. S3). Notably, we did not find any significant difference between co-cultures made by FUSP525L and FUSWT + HuD MNs. Within each line, a significant increase of dead cells from the initial time point (day 7) was observed for the FUSWT co-cultures only at day 28, while it was evident at earlier time points for FUSP525L (from day 14 onward) and FUSWT + HuD (from day 21 onward) (Fig. 4B, p values table). Increased Cleaved Caspase-3 (CC-3) signal at day 14 and 28 suggests that cell death was a consequence of apoptosis (Fig. 4C–E; Supplementary Fig. S4). From day 14 to day 28, the number of apoptotic cells significantly increased in the co-cultures made by FUSP525L and FUSWT + HuD, but not FUSWT, MNs (Fig. 4E). Importantly, when MNs are cultured in absence of muscle cells, we did not observe differences in CC-3 signal among the 3 genotypes (Fig. 4D).
A Representative brightfield images of co-cultures at day 28. Scale bar: 20 μm. B Quantitative analysis of dead cells using ethidium homodimer-1 at day 7, 14, 21, 28 of co-cultures obtained with FUSWT SkMCs and FUSWT, FUSP525L, or FUSWT + HuD MNs. The graph shows the average (dots) and standard deviation from 3 batches of differentiated cells. For each replicate, 5 fields have been acquired at each time point. Ordinary two-way ANOVA: p = 1.2 × 10−9 for genotypes comparison, p = 3.0 × 10−15 time points comparison, p = 2.0 × 10−7 for their interaction; post hoc Tukey test p values for multiple comparisons among genotypes within each time point, and among day 7 and later time points within each genotype, are indicated in the table. See Supplementary Fig. S3 for representative images of the cells. C Representative merged images of immunofluorescence staining of day 14 co-cultures (left panels) or MN monocultures (right panels) using Cleaved Caspase-3 (CC-3) and MAP2 antibodies, and DAPI. Scale bar: 50 μm. Single panels are shown in Supplementary Fig. S4. D, E The graphs report quantitative analysis of the CC-3 fluorescence intensity at day 14 and 28 of MNs monoculture (D) or co-cultures (E). Each dot represents a replicate, consisting of an individual batch of differentiated iPSCs. For each replicate, 6 fields have been acquired and the dot shows the average value. Error bars indicate standard deviation calculated on the average value of the replicates. Ordinary two-way ANOVA. No significant differences resulted from the comparison of MN monocultures (D). For the co-cultures (E), p = 2.1 × 10−7 for genotypes comparison, p = 0.0003 for time points comparison, p = 0.0324 for their interaction; relevant post hoc Tukey test p values for multiple comparisons are indicated in the graph. Source data are provided as a Source Data file.
To confirm these observations in an independent pair of isogenic FUS lines, considered as a biological replicate, we introduced the p.P525L variant in another iPSC line (EBiSC catalog number WTSIi004-A, male, age of donor 35-39) (Supplementary Fig. S5A). In these lines, hereafter named WTSI-FUSWT and WTSI-FUSP525L, we confirmed upregulation of HuD in FUS mutant MNs (Supplementary Fig. S5B). Moreover, when co-cultured with FUSWT SkMCs, we observed reduction in muscle fibers forming endplates and increased apoptosis in presence of WTSI-FUSP525L MNs, compared with the co-cultures obtained with isogenic WTSI-FUSWT MNs (Supplementary Figs. S5C, D, S6A, B). We then produced a WTSI-FUSWT line containing the Syn1::HuD transgene (hereafter WTSI-FUSWT + HuD). Also in this case, HuD overexpression resulted in a significant reduction of muscle fibers forming NMJs and increase in apoptosis (Supplementary Figs. S5C, D, S6A, B).
Collectively, these results show that, in addition to NMJ formation impairment, HuD overexpression in MNs closely phenocopies the severe FUS p.P525L ALS variant in terms of increased vulnerability by apoptosis, and that such effect emerges only in presence of muscle cells.
Knockdown of HuD or its target genes in FUSP525L co-cultures rescues NMJ defects and apoptosis
Since the effects of HuD overexpression in MNs are strikingly similar to those exerted by the FUSP525L variant, we wondered whether reduction of HuD levels could be beneficial for NMJ formation and survival in co-cultures made by FUSP525L MNs. Small interfering RNAs (siRNAs) transfection effectively reduced HuD mRNA and protein levels in FUSP525L MNs (Supplementary Fig. S7A, B). FUSWT SkMCs and FUSP525L MNs co-cultures were transfected at day 3 and 8 with anti-HuD siRNAs and analyzed at day 14 by α-BTX and CC-3 staining. This analysis revealed significant increase in the fraction of muscle fibers showing AChR clusters and significant reduction of apoptosis upon HuD knockdown (Fig. 5A–D; Supplementary Fig. S7C, D). The same effects were consistently observed upon HuD siRNAs transfection in co-cultures made of WTSI-FUSP525L MNs (Supplementary Fig. S8A, B). We next performed a similar experiment by knocking down, individually, two key HuD targets, NRN1 and GAP43, whose levels are increased in FUSP525L MNs due to overly stabilization by HuD (Supplementary Fig. S7A)5. NRN1 siRNAs significantly increased the fraction of muscle fibers showing AChR clusters and decreased apoptosis to a similar extent as HuD siRNAs (Figs. 5A–D, S7C, D). GAP43 knockdown did not significantly change AChR clustering and produced a milder effect on apoptosis.
A, B Representative merged images of day 14 co-cultures obtained with FUSWT SkMCs and FUSP525L MNs transfected with the indicated siRNAs and stained with the indicated antibodies. Scale bar: 50 μm. Single panels are shown in Supplementary Fig. S7. C, D The graphs report quantitative analysis of the percentage of α-BTX positive fibers (C) and CC-3 fluorescence intensity (D), at day 14 of co-culture as in (A, B). Each dot represents an individual batch of differentiated iPSCs transfected with the indicated siRNAs. The following number of randomly selected fields were used for the α-BTX analysis: 5 (non-targeting siRNA replicate 1 and 2, HuD siRNA replicate 2); 7 (GAP43 siRNA replicate 3); 6 (all other replicates). The following number of randomly selected fields were used for the CC-3 analysis: 8 (non-targeting siRNA replicate 3, HuD siRNA replicate 1, NRN1 siRNA replicate 3); 9 (HuD siRNA replicate 2 and 3; non-targeting siRNA replicate 1); 10 (all other replicates). The dot shows the average value. Error bars indicate standard deviation calculated on the average value of the replicates. Ordinary one-way ANOVA, p = 0.0009 for α-BTX positive fibers, p = 0.0003 for CC-3 immunofluorescence; post hoc Tukey test p-values for multiple comparisons are indicated in the graph. Source data are provided as a Source Data file.
Taken together these results suggest that most, if not all, the cell-autonomous effects of the FUSP525L variant in MNs in terms of NMJ defects and apoptosis could be due to HuD upregulation. In turn, among the tested HuD targets that are also upregulated in FUS mutant MNs5, NRN1 might play a prominent role.
Knockdown of the Drosophila HuD ortholog elav rescues the motor dysfunction caused by FUS overexpression in vivo
Drosophila models represent valuable tools for examining genetic modifiers of neurodegenerative diseases, including FUS-ALS20. In Drosophila melanogaster, the neuronal embryonic lethal abnormal vision (elav) gene encodes for an RBP, ELAV, showing high conservation with the mammalian HuD protein in terms of structure, expression, and functions7. Upon ELAV overexpression in neurons, we observed a strong reduction in the number of flies able to cross a 4 cm threshold in a climbing assay performed at day 6 and 15 (Fig. 6A). We took advantage of fly models selectively overexpressing human FUS WT or p.P525L in neurons (Supplementary Fig. S9A), which show reduced climbing ability21, to assess whether elav modulation might modify such motor dysfunction phenotype. In particular, we attempted a rescue experiment by ELAV reduction by neuronal-specific expression of short hairpin RNAs (shRNAs). We did not observe any difference at day 6, an early age at which neither WT nor mutant FUS overexpression produced motor dysfunction (nearly 100% of flies were able to climb 4 cm in 10 s). Strikingly, elav RNAi significantly rescued the motor dysfunction for both WT and p.P525L FUS at day 15 (Fig. 6B). This effect was further confirmed by analyzing a different motor parameter, climbing velocity, which was significantly improved by ELAV reduction in both WT and p.P525L FUS at day 15 (Fig. 6C). We then analyzed the percentage of mature and collateral boutons in third instar larvae, as an indication of the maturity of NMJs (Fig. 6D; Supplementary Fig. S9B). As shown in Fig. 6E, elav RNAi reduced the percentage of collateral boutons and increased the percentage of mature boutons in FUS WT expressing larvae. However, we did not observe a similar effect in mutant FUS expressing larvae. An opposite trend, reduced mature boutons, and increased collateral boutons, was observed in larvae overexpressing ELAV, however, the differences did not reach statistical significance (Supplementary Fig. S9C).
A The graphs represent the percentage of flies that climb 4 cm in 10 s at day 6 and day 15 in overexpressing elav (elav OE) or Luciferase (luc OE) as a control, respectively. N = 3 experimental replicates (trials) with 8 (day 6, both luc OE and elav OE), 13 (day 15, luc OE) or 7 (day 15, elav OE) flies each. Student’s t-test, unpaired, two tails, p-values are indicated in the graphs. B The graphs show percent flies that climb 4 cm in 10 s at day 6 and day 15 in wild-type or mutant FUS (p.P525L) with or without elav RNAi. N = 3 experimental replicates (trials) with 8 (day 6, FUSWT), 10 (day 6, all other groups), 2 (day 15, FUSWT), 3 (day 15, FUSP525L), or 8 (day 15, all other groups) flies each. Ordinary one-way ANOVA, relevant post hoc Tukey test p-values for multiple comparisons are indicated in the graphs. C The graphs show the climbing velocity (cm/s) of flies as in (B). Each data point represents one fly. The following numbers of flies have been used: 24 (day 6, FUSWT), 28 (day 6, FUSWT - elav RNAi), 30 (day 6, both FUSP525L and FUSP525L - elav RNAi), 6 (at day 15, both FUSWT and FUSP525L), 21 (FUSWT - elav RNAi), 22 (FUSP525L - elav RNAi). Ordinary one-way ANOVA, relevant post hoc Tukey test p-values for multiple comparisons are indicated in the graphs. D Immunohistochemistry analysis of the neuronal marker horseradish peroxidase (HRP) and the synaptic vesicle marker cysteine string protein (dCSP) in third instar larvae expressing FUSWT or FUSP525L, with or without elav RNAi. E The graphs show quantitative analysis of the percentage of satellite and mature boutons in larvae as in (D). N = 8 larvae per genotype. Ordinary one-way ANOVA, relevant post hoc Tukey test p-values for multiple comparisons are indicated in the graphs. In all graphs, bars indicate the mean and standard deviation. Source data are provided as a Source Data file.
Collectively, in line with the results from the in vitro human model, these in vivo experiments in Drosophila show that overexpression of the HuD-related protein ELAV produces per se motor dysfunction, whereas ELAV dampening is sufficient to rescue the motor phenotype produced by FUS completely.
HuD might be involved in sporadic ALS with an oxidative stress signature
Our previous work showed increased HuD levels in FUS mutant human iPSC-derived MNs and in the spinal cord of a FUS mutant mouse model5. We and others have also previously detected HuD in pathological aggregates of FUS, TDP-43, and sALS patients3,4. More recently, the Cereda lab reported HuD overexpression in sALS patients’ motor cortex and proposed that this could be due to induction of HuD expression upon oxidative stress, as HuD mRNA levels were increased in neuroblastoma cells treated with H2O28. These findings prompted us to assess whether the same effect could be observed in human MNs. MNs from FUSWT iPSCs were treated with sodium arsenite, an inducer of oxidative stress, for 60 minutes before mRNA analysis by qRT-PCR. We found a small but highly reproducible increase in HuD transcript levels upon oxidative stress (Fig. 7A). We observed a similar effect in FUSWT iPSC-derived co-cultures (Fig. 7B). Upregulation of HuD was likely due to new transcription, as pre-treatment with α-amanitin, a potent and selective inhibitor of RNA polymerase II, abolished the effect of oxidative stress on this transcript (Fig. 7C). All these results were confirmed in WTSI-FUSWT MNs and co-cultures (Fig. 7D, E). Together with the previous findings8, these results suggest that oxidative stress might trigger HuD expression increase in human neurons, including MNs, with possible implications for sALS. To investigate this possibility, we took advantage of a large transcriptome profiling dataset of sALS patients15. In this dataset, ALS patients’ cortex samples could be grouped into 3 distinct clusters based on expression profile. The largest group included patients with hallmarks of oxidative and proteotoxic stress (ALS_Ox). The other two clusters showed signatures of glial activation (ALS_Glia) and high retrotransposon expression (ALS_TE)15. Notably, we found that HuD expression was significantly increased only in the ALS_Ox cluster (Fig. 7G). Conversely, its levels were significantly lower in the ALS_Glia group and no significant change was found in the ALS_TE group. This pattern was also evident for the HuD targets, NRN1 (albeit its increase in ALS_Ox did not reach statistical significance), and GAP43 (Fig. 7G). Moreover, the ELAV family member ELAVL2, encoding for an RBP (HuB) closely related to HuD, also showed significant increased expression in ALS_Ox, decreased expression in ALS_Glia, and no change in ALS_TE (Supplementary Fig. S10A). Finally, SOD1 mRNA, which is a target stabilized by HuD upon oxidative stress8, was also significantly upregulated in the ALS_Ox group (Supplementary Fig. S10A). In order to gain insights into a possible correlation between the levels of HuD (and its targets) and FUS in sporadic ALS patients, we analyzed their expression at individual patient level for all groups, including healthy individuals (CTR) and patients with other neurodegenerative diseases (OND). A matrix reporting the Spearman’s correlation coefficients calculated between the expression levels of FUS and those of HuD, NRN1, and GAP43 is shown in Fig. S10B. This analysis suggests that there is no correlation between FUS and HuD expression levels in OND and ALS_Ox groups, low positive correlation in CTR and ALS_TE groups, and low negative correlation in the ALS_Glia group. For what concerns NRN1 and GAP43, we observed positive correlation with FUS in all groups except for ALS_Glia. Finally, when CTR and ALS groups were merged, we observed no correlation for FUS and HuD, and low positive correlation between FUS and NRN1, and between FUS and GAP43. Collectively, the correlation analysis suggests that altered FUS expression is unlikely the underlying mechanism for HuD upregulation in patients with an oxidative stress signature.
A, B The graphs show quantitative analysis of HuD mRNA levels in control conditions or upon acute stress treatment with 0.5 mM sodium arsenite (ARS) for 1 h in FUSWT iPSCs derived spinal MNs monocultures at day 12 (A) or FUSWT iPSCs derived co-cultures at day 7 (B) (n = 4). C Cells as in (A) were pre-treated or not with α-amanitin before oxidative stress induction with ARS (n = 4). D–F The experiments shown in (A–C) were repeated with the WTSI-FUSWT iPSC line (n = 3 in (D, E); n = 4 in (F)). In (A–F) ATP5O expression was used for normalization and the graphs show the average, standard deviation, and p-values from 3 or 4 replicates, each consisting of a batch of differentiated iPSCs. A, B, D, E: Student’s t-test, unpaired, two tails, p-values are indicated in the graphs. C, F: ordinary one-way ANOVA, p = 1.0 × 10−10 (C), p = 7.5 × 10−6 (F); relevant post hoc Tukey test p-values for multiple comparisons are indicated in the graphs. In all graphs, bars indicate the mean and standard deviation, (G) Violin plots showing the expression levels, reported as log-transformed FPKM values, of HuD, NRN1, and GAP43 in post-mortem sporadic ALS patients’ cortex samples15. The adjusted p-values obtained from differential expression analyses have been corrected for multiple comparisons via the Benjamini-Hochberg procedure. Box boundaries: first and third quartile (Q1 and Q3); central line of box: median; whisker boundaries: the smallest and largest values within 1.5 times the interquartile range from Q1 and Q3. Source data are provided as a Source Data file.
These findings suggest that, in the absence of pathogenic FUS ALS variants, oxidative stress can trigger HuD upregulation in the context of human MNs in vitro. Accordingly, HuD levels are specifically increased in sALS patients with a signature related to oxidative stress.
Discussion
Human in vitro models of the motor unit have shown that MN-restricted expression of ALS-linked FUS variants produces NMJ phenotypes13,14. Here we found that such NMJ defects could be reverted by dampening HuD levels. Moreover, knockdown of HuD decreased mutant FUS-induced apoptosis in vitro, and strikingly, knockdown of the related elav Drosophila gene completely rescued motor phenotypes induced by FUS in vivo. We have recently shown that in MNs FUS mutation or HuD overexpression exert similar effects on the expression of disease-linked genes6. Altogether, these findings point to a crucial role for HuD in the cell-autonomous mechanisms that underlie the death of MNs via a “dying-back” pattern in FUS and, possibly, a subset of sporadic ALS cases with hallmarks of oxidative and proteotoxic stress. To this regard, in both the in vitro and the in vivo model, we found that HuD (or ELAV) overexpression per se phenocopies the severe FUS mutation.
The seminal discovery of early defects at the NMJ in 47-day-old SOD1G93A mice, in which loss of MNs occurs only after 90 days of age, supported the “dying-back” hypothesis which was further reinforced by evidence from a sALS patient and other non-SOD1 ALS animal models11,22. These include the FusΔNLS/+ mice that show reduced endplate area and NMJ denervation as early as 1 month of age, while motor deficits begin around 10 months and MN loss was evident only at 22 months13,23. Neuron-specific expression of mutant human FUS proteins driven by the MAPT regulatory sequences also resulted in early structural and functional defects at the NMJ that preceded MN loss24. These phenotypes were particularly severe in the MAPT-hFUSP525L mouse, which showed significant denervation at day 20 and MN loss at day 30. This evidence suggests that MN-restricted expression of mutant FUS is sufficient in vivo to trigger early retraction of motor axons from the NMJ. Since HuD upregulation was observed in mice carrying a severe FUS mutation in the NLS (Fus-Δ14 knock-in)5, we can speculate that the HuD-dependent circuitry described here could contribute to such NMJ defects due to pre-synaptic mechanisms. For what concerns the Drosophila model used here, the motor phenotype rescue observed upon elav RNAi in neurons of FUSWT overexpressing flies was mirrored by a shift toward more mature NMJs. However, we did not observe a significant change of NMJ types in flies overexpressing mutant FUS, suggesting that other mechanisms might contribute to the beneficial effect of ELAV reduction in vivo.
The evidence of HuD upregulation in the motor cortex of sporadic ALS patients, possibly due to exposure to environmental factors causing oxidative stress8, suggests that this RBP might play a broader role in ALS. To this regard, we and others have previously found HuD in cytoplasmic inclusions of FUS, TDP-43, and sporadic ALS patients3,4. In this work, we further explored the possibility that HuD might be altered in sporadic ALS. We found increased levels of HuD and some of its targets in a subset of sporadic ALS patients characterized by a signature related to oxidative stress, who account for the largest subgroup (about 60% of the cohort)15. Intriguingly, a significant decrease was instead observed in the cluster with a signature related to glial activation and inflammation (about 20% of the cohort). Thus, taken together, altered HuD levels in either direction would be associated with most sporadic ALS cases. So far, to the best of our knowledge, no known genetic variants in the HuD gene have been conclusively linked to ALS. However, from a survey of the Project MinE (http://databrowser.projectmine.com) we noticed 4 variants of uncertain significance (VUS) found in ALS patients but not in controls (Supplementary Fig. S11A, B). Future work will be needed to clarify if such VUS have any effect on HuD protein levels and/or activity. The molecular mechanisms underlying HuD upregulation upon oxidative stress are unknown. The fact that HuD mRNA levels increase upon either arsenite (present work) or H2O2 treatment8 points to eIF2α-dependent and -independent pathways25. While regulation of HuD by FUS occurs at the post-transcriptional level4,5,26, here we show that HuD mRNA increase upon oxidative stress is likely due to transcriptional regulation. A mechanistic link with FUS is therefore unlikely in this latter case. Moreover, we did not find any correlation between FUS and HuD expression at individual patients’ levels in the oxidative stress cluster. Thus, altogether, our previous findings4,5,26 and present work point to FUS-dependent (in FUS-ALS) and FUS-independent (in sALS) mechanisms leading to HuD dysregulation in ALS.
Among known HuD targets, in this work, we focused on NRN1 and GAP43. Notably, in postmortem ALS patients both GAP43 and NRN1 have been found upregulated in regions of the nervous system with implications in ALS and frontotemporal lobar degeneration (FTLD), including the anterior horn of the spinal cord and the frontal cortex area 827,28,29. This is in line with the known role of HuD in stabilizing (and localizing) GAP43 and NRN1 mRNAs through AU-rich elements (ARE)30,31. We found that knockdown of NRN1 or GAP43 produced a partial rescue of NMJ and/or apoptosis phenotypes in co-cultures containing FUS mutant MNs. This effect might be due to their known roles in developing neurons. To this regard, aberrant axon guidance is among the proposed mechanisms underlying the “dying-back” phenomenon32,33. For instance, it has been shown that increased expression of the axon guidance protein Semaphorin 3A (Sema3A) by terminal Schwann cells (tSCs) might result in axonal denervation at the NMJ due to repulsion of the motor axon in SOD1G93A mice34. Notably, GAP43 can modulate axon guidance signals triggered by Sema3A, and exposure to Sema3A induced neuronal death by apoptosis in a GAP43-dependent manner35. Accordingly, loss of GAP43 conferred resistance to apoptotic stimuli in cultured mouse neurons. On the other hand, overexpression of GAP43 in adult mice resulted in prolonged sprouting and death of MNs36,37. In line with these studies in animal models, we show here that knockdown of GAP43 by RNAi partially reverts apoptosis in iPSC-derived co-cultures made by mutant FUS MNs. In this case, however, we speculate that increased HuD levels would mediate GAP43 gain of function in a cell-autonomous manner, rather than by Sema3A signaling, as our in vitro model does not contain tSCs.
We found that NRN1 knockdown in FUS mutant MNs increased AChR clusters. Previous literature on a possible role for NRN1 in NMJ formation is scarce. A relevant study in Xenopus laevis tadpoles showed that overexpression of the ortholog gene cpg15 promoted spinal MN axon growth by increasing branch addition and maintenance and decreasing branch retraction38. This dysregulation could eventually impact some crucial processes such as extension, refinement, and pruning. Indeed, in such developmental context, new branches formed upon NRN1/cpg15 overexpression were apposed to AChR clusters. However, the authors did not provide information on further maturation of the new endplates, and whether increased branching during development would be followed by productive formation of mature NMJs remains unclear. It should be noted that the adult human spinal cord is among the nervous system districts with the lowest expression of NRN15. We might speculate that aberrantly increased levels of this growth-promoting factor might be detrimental, rather than beneficial, in the adult motor system, generating excess of axonal extensions, which would be less able to establish functional connections.
In the co-cultures, we observed a significant increase on cell death in both neural and muscle populations upon MN-restricted FUS mutation or HuD overexpression. Both in vivo and in vitro, MNs are recognized for their ability to release trophic factors that support the survival and growth of nearby cells. The presence of MNs in the co-culture can thus enhance the survival of skeletal muscle cells and contribute to their overall health. We hypothesize that upon increased FUSP525L and FUSWT + HuD MNs death and/or failure to form mature NMJs, their support to muscle survival in vitro fails, thus explaining the increased apoptosis of muscle fibers.
The case of the sporadic ALS patient who died unexpectedly11, despite anecdotal, suggests that at the time of diagnosis (first signs of motor symptoms) MN cell bodies might still be intact. Thus, early intervention aimed to restore NMJs might result in an effective therapeutic approach to prevent irreversible subsequent MN degeneration. To this regard, our work shows that individual knockdown of HuD and one of its key targets, NRN1, can rescue endplate formation and apoptosis in vitro. Moreover, in vivo, evidence from the Drosophila model shows that reduction of the HuD-related gene elav expression is sufficient to rescue the motor phenotype caused by FUS completely. Here we took advantage of RNA interference. However, recent advances in the development of antisense oligonucleotides (ASOs) for therapy, also in the context of ALS39, provide the opportunity to translate these findings to patients in the near future.
Methods
Our research complies with all relevant ethical regulations. All procedures for the care and treatment of animals were in accordance with the Animals (Scientific Procedures) Act 1986 Amendment Regulations 2012. Materials are available upon request.
iPSC culture, transfection, and differentiation
Human iPSC lines were maintained in Nutristem-XF medium (Biological Industries), with 0.1X Penicillin/Streptomycin (Merck Life Sciences) (Nutristem P/S), in Matrigel-coated dishes (hESC-qualified Matrigel; Corning) and passaged every 4–5 days with 1 mg/ml Dispase (Gibco). Lines were tested for mycoplasma contamination every three months resulting negative. The human FUSWT line, its isogenic FUSP525L line, and the FUSWT + HuD line had been previously derived from the WT-I line5,19. As indicated in the original study, informed consent had been obtained from all patients involved prior to cell donation. Sapienza Ethics Committee for Transdisciplinary Research (CERT) approval number 5/2022. The FUSWT iPSCs line WTSIi004-A (WTSI-FUSWT) was obtained by the European Bank for induced pluripotent Stem Cells (EBiSC; hPSCreg name: WTSIi004-A, RRID:CVCL_AI02; as stated by EBiSC, informed consent was obtained from the donor, NRES Committee Yorkshire & The Humber—Leeds West approval number 15/YH/0391). This line was used to generate an independent set of iPSC lines, considered here as a biological replicate of the WT-I-based set of iPSC lines. From these cells we generated the isogenic WTSI-FUSP525L iPSC line by TALEN-directed mutagenesis as previously described19. Both mutant and control lines went through the gene editing process. To generate the iPSC line WTSI-FUSWT overexpressing HuD (WTSI-FUSWT + HuD), cells were co-transfected with 4.5 μg of transposable vector epB-Puro-TT-SYN1-HuD (SYN1::HuD) and 0.5 μg of the piggyBac transposase using the Neon Transfection System (Life Technologies) as described5. Stable and inducible iPSCs for spinal MN differentiation (NIL iPSCs) were obtained as previously described17,40. Briefly, iPSCs were transfected with 4.5 μg of NIL piggyBac construct and 0.5 μg of transposase vector using the Neon Transfection System (Life Technologies) and then selected with 5 μg/ml blasticidin in Nutristem-XF medium containing 0.1X P/S for 10 days. KOLF-1 (FUSWT) iPSCs (as indicated in the original study, informed consent was obtained from the donor, NRES Committee Yorkshire & The Humber—Leeds West approval number 15/YH/0391), stably transfected with inducible transposable vectors encoding for BAF60c and MyoD141 (MB iPSCs), were used to generate SkMCs. Briefly, this line was generated by co-transfecting iPSCs with 2.25 μg each of the ePB-Bsd-TT-BAF60c and ePB-Puro-TT-m-MyoD vectors, and 0.5 μg of piggyBac transposase, and then selected with 1 μg/ml puromycin and 5 μg/ml blasticidin in Nutristem P/S for 10 days, as previously described17,40.
Differentiation of spinal MNs and SkMCs and co-cultures
Spinal MNs were obtained as previously described17,40. Briefly, NIL iPSCs were dissociated with Accutase (Thermo Fisher Scientific) to single cells and replated in Nutristem P/S supplemented with 10 μM Y-27632 ROCK inhibitor (Enzo Life Sciences). The next day differentiation was induced in DMEM/F12 (Dulbecco’s Modified Eagle’s Medium/ Nutrient Mixture F-12 Ham; Merck Life Sciences), 1X Glutamax (Thermo Fisher Scientific), 1X NEAA (Thermo Fisher Scientific), 0.5X P/S and doxycycline 1 μg/ml (Thermo Fisher Scientific) for 2 days. On the third day, medium was replaced by Neurobasal/B27 (Neurobasal medium, Thermo Fisher Scientific; 1X B27, Thermo Fisher Scientific; 1X Glutamax; 1X NEAA; 0.5X P/S) supplemented with 5 μM DAPT, 4 μM SU5402 (both from Merck Life Sciences) and 1 μg/ml doxycycline for 3 days. At day 5, MN progenitors were dissociated with Accutase and counted. 1 × 106 cells were plated onto Matrigel-coated 35 mm dishes for RNA analyses, and 5 × 104 cells per well were plated onto Matrigel-coated μ-Slide 8 Well (ibidi) for immunofluorescence analyses. 10 μM of Y-27632 ROCK inhibitor was added for 24 h after dissociation. Neurons were maintained in Neurobasal/B27 supplemented with 20 ng/ml L-ascorbic acid (Merck Life Sciences), 20 ng/ml BDNF (PreproTech) and 10 ng/ml GDNF (PreproTech).
To obtain SkMCs, we adapted a method to convert iPSCs into SkMCs upon ectopic expression of MyoD and BAF60c16,42. This protocol has been previously developed in our lab for short-term cultures (up to 9 days), however, we noticed that a subpopulation of differentiation-resistant cells persisted in longer cultures. Over time, these mitotically active cells outnumbered terminally differentiated SkMCs, thus hindering the establishment of long-term co-cultures. To overcome this problem, we included a step consisting of the transient addition of cytosine arabinoside (AraC), which successfully reduced the number of proliferating undifferentiated cells, as follows. MB iPSCs were dissociated with Accutase (Thermo Fisher Scientific) to single cells and replated in Nutristem P/S supplemented with 10 μM Y-27632 in 35 mm dishes (2 × 105 cells). In the following two days, cells were maintained with Growth Medium consisting of high glucose DMEM (Merck Life Sciences), 1X Glutamax, 1X P/S, 20% FBS (Merck Life Sciences), 50 μg/ml Insulin (Roche), 25 ng/ml bFGF (Corning) and 10 ng/ml hEGF (Corning) and 200 ng/ml doxycycline. At day 2, medium was replaced by Skeletal Muscle Differentiation Medium (Promega), 1X P/S, 200 ng/ml doxycycline, and 2 μM AraC (Merck Life Sciences). At day 4, this medium was replaced by Skeletal Muscle Differentiation Medium, 1X P/S, and 200 ng/ml doxycycline. The medium was then changed every other day.
To generate co-cultures of spinal MNs and SkMCs we adapted previously published protocols14,43. MB iPSCs were dissociated and replated in 35 mm dishes (2 × 105 cells) or μ-Slide 8 Well (ibidi; 2 × 104 cells per well) and then SkMC differentiation was induced as described above. Spinal MNs differentiation started independently and in parallel when SkMCs were at day 7 of the muscle differentiation protocol. At day 5, MNs progenitors were replated (at approximately 1:1 ratio) in the 35 mm dishes (5 × 105 cells) or μ-Slide 8 Well (ibidi; 5 × 104 cells per well) containing SkMCs, which at this stage had reached day 13 of differentiation, in Neurobasal/B27 medium containing 20 ng/ml L-ascorbic acid, 20 ng/ml BDNF, 10 ng/ml GDNF, 200 ng/ml doxycycline and 10 μM Y-27632. The next day, medium was replaced with Neurobasal/B27, 20 ng/ml L- ascorbic acid, 20 ng/ml BDNF, 10 ng/ml GDNF, 200 ng/ml doxycycline, 20 μg/ml laminin (Merck Life Sciences) and 0.01 μg/ml agrin (R&D system). Co-cultures were maintained for 14, 28, or 50 days, changing medium every 4 days.
Functional assays in iPSC-derived cells
Induced functional activity in co-cultures and muscle monocultures at day 50 was assessed by adding 50 μM glutamate (Gibco) to cells previously plated in μ-Slide 8 Well (ibidi) supports and maintained in Neurobasal/B27 medium containing 20 ng/ml L-ascorbic acid, 20 ng/ml BDNF and 10 ng/ml GDNF. Five minutes after the addition of glutamate, the number of muscle contractions per minute in 3 randomly selected fields was counted under Carl Zeiss Axio Vert.A1 Microscope. To verify muscle contraction inhibition, 10 μM tubocurarine (Merck Life Sciences) was added to the medium containing glutamate. Five minutes after the addition of tubocurarine, the same co-culture fields were identified for measuring muscle contractions per minute. Spontaneous functional activity, in the absence of glutamate, was assessed in co-cultures at day 62 before and after addition of 10 μM tubocurarine. Video recording of spontaneous contractions was assessed in cells previously plated in 12-well multi-dishes. Light brightfield videos were recorded with Carl Zeiss Axio Vert.A1 Microscope equipped with 20X air objectives. These experiments were performed in 3 independent batches of differentiated iPSCs.
Real-Time qRT-PCR
Total RNA was extracted using E.Z.N.A Total RNA Kit I (Omega BIO-TEK) and retrotranscribed with iScript Supermix (Bio-Rad Laboratories) according to manufacturers’ instructions. qRT-PCR was performed with the iTaq Universal SYBR Green Supermix kit (Bio-Rad Laboratories) on a ViiA 7 system (Thermo Fisher Scientific). ATP5O and TUBB3 were employed as calibrator genes. Full list of primers is reported in Supplementary Data S1.
Cell immunofluorescence and confocal imaging
Cells were fixed in 4 % PFA (Electron microscopy sciences) and 30 mM sucrose (Sigma-Aldrich) for 10 min at room temperature (RT), then washed with PBS with calcium chloride and magnesium chloride (Merck Life Sciences) and permeabilized with a solution of 0.5% BSA, 10% Horse Serum (HRS), 0.2% Triton X-100 and PBS (Merck Life Sciences) for 15 min. Cells were then washed and treated with a blocking solution composed of 0.5% BSA, 10% HRS, and PBS for 30 min. For NMJ staining, anti-MyHC (1:50; MAB4470, R&D system) and anti-SYP (1:200; ab32127, Abcam) were diluted in blocking solution and incubated at 4 °C overnight. Anti-TUJ1 (1:1000; T2200, Merck Life Sciences) was diluted in the same solution and incubated for 1 h at RT. Cells were then washed with PBS and incubated with α-bungarotoxin-488 (1:350, B13422, Thermo Fisher Scientific) for 1 h at RT. Secondary anti-mouse Alexa Fluor 647 (1:250, Immunological Science) and anti-rabbit Alexa Fluor 594 (1:250, Immunological Sciences) antibodies were then incubated for 1 h. DAPI (1:2000; Sigma-Aldrich) diluted in 0.5% BSA, 10% HRS in PBS was added for 5 min. Cells were washed in PBS and ibidi Mounting Medium (50001, ibidi) was added. Confocal image stacks were acquired using an inverted Olympus iX73 equipped with an X-light V3 spinning disc head (Crest Optics), a LDI laser illuminator (89 North), Prime BSI sCMOS camera (Photometrics), and MetaMorph software (Molecular Devices) with 40X air or 100X oil objectives (Olympus). For skeletal muscle with or without 48 h Ara-C treatment, fixed cells were incubated with anti-MyHC (1:50; MAB4470, R&D system) and anti-NANOG (1:200; PA1-097X; Thermo Fisher Scientific) primary antibodies overnight. Then, anti-mouse Alexa Fluor 647 (1:250, Immunological Science), anti-rabbit Alexa Fluor 488 (1:250, Immunological Science), and DAPI were used and mounted with ibidi Mounting Medium (50001, ibidi). Confocal image stacks were acquired using an Olympus FV1200 confocal laser scanning microscope with 20X air objective. For α-BTX positive fibers analysis, randomly selected fields for each batch of differentiated iPSCs, considered as a replicate, were used to count the total number of muscle fibers (based on the MyHC staining), and the total number of muscle fibers showing α-BTX signal to calculate the percentage. For TUJ1 analysis, we calculated the average value of randomly selected fields for each replicate.
Cell-based assays and confocal imaging
For apoptosis analysis, fixed cells were incubated with anti-CC-3 (1:400; 9661, Cell signaling) overnight, anti-MAP2 (1:2000; ab5392, Abcam) for 1 h at RT. Anti-chicken Alexa Fluor 488 (1:250; Immunological Sciences) and anti-rabbit Alexa Fluor 594 (1:250; Immunological Sciences) secondary antibodies were then used. DAPI was added for 5 minutes. Confocal image stacks were acquired using an Olympus FV10i laser scanning confocal microscope with 10X air objective at 2x zoom or the X-light V3 spinning disc confocal microscope described before with 20X air objective (Olympus). Phase-contrast images were acquired using Carl Zeiss Axio Vert.A1 Microscope with 20X or 40X air objectives. For dead cell quantification, live co-cultures were incubated (5% CO2 at 37 °C) with Neurobasal/B27, 20 ng/ml L-ascorbic acid, 20 ng/ml BDNF and 10 ng/ml GDNF medium supplemented with 0.6 μl ethidium homodimer-1 594 (Thermo Fisher Scientific) according to manufacturer’s protocol. After 30 minutes cells were washed with PBS and maintained in Neurobasal/B27, 20 ng/ml L-ascorbic acid, 20 ng/ml BDNF, and 10 ng/ml GDNF medium during acquisition. Confocal image stacks were acquired using Olympus FV10i laser scanning confocal microscope with 10x air objective at 1.5x zoom.
RNA interference (RNAi) in iPSC-derived in co-cultures
Lyophilized siRNAs were resuspended in nuclease-free water at 20 μM stock concentration. HuD, NRN1, GAP-43, and non-targeting siRNAs (siRNA-SMARTpool and ON-TARGET plus Non-targetingPool, Dharmacon) were transfected at 10 nM concentration using siLentFect Lipid Reagent (Bio-Rad Laboratories) according to manufacturer’s instructions. Medium was replaced 5 h after transfection.
Western blot
Whole-cell protein extracts were prepared from iPSC-derived cells lysed in RIPA buffer. Protein extracts were separated by electrophoresis with NuPAGE 4-12% Bis-Tris gels (Thermo Fisher Scientific) in MOPS-SDS buffer and electroblotted onto PVDF membrane (Protran). Filters were incubated with the following antibodies: anti-HuD (1:1000; sc-48421, Santa Cruz Biotechnology), anti-GAPDH (1:10000; sc-32233, Santa Cruz Biotechnology) primary antibodies, and goat anti-mouse IgG (H + L) secondary antibody (1:10000; 32430, Thermo Fisher Scientific). For Drosophila melanogaster, fly heads were collected from each genetic cross and snap-frozen on dry ice. Western blots were run in triplicate using biological replicates. Heads were crushed on dry ice and lysed with RIPA. Lysates were sonicated and centrifuged to remove debris. Supernatants were boiled in Laemmli Buffer for 5 min and loaded onto 4-12% NuPage Bis-Tris gels. The gels were transferred onto the nitrocellulose membrane using the iBlot2. The blots were blocked with milk solution and further incubated in primary antibodies overnight. Primary antibodies used were β-Actin (D6A8) Rabbit mAb (1:1000; 4967 L, Cell Signaling Technology) and mouse anti-HA (1:1000; H3663-100UL, Sigma-Aldrich). Blots were washed and incubated at RT for one hour with goat-anti-mouse IRDye 680RD (1:10000; 925-68070, Licor) and goat-anti-rabbit DYLight 800 (1:10000; SA5-35571, Thermo Fisher Scientific), and imaged on a Licor imager. Protein levels were quantified using NIH Image J software. Statistical analysis was performed with GraphPad Prism 8 software. Uncropped images of the western blots are shown in Supplementary Fig. S12.
Drosophila melanogaster stocks
All Drosophila melanogaster stocks were maintained on standard cornmeal medium at 25 °C in light/dark-controlled incubators. The UAS-FUS WT and UAS-FUS p.P525L lines were generated through site-specific insertion of the transgene at BestGene Inc. using the (attp2) integration vector and were previously described44,45. The TRIP control lines, the UAS-Luciferase (luc OE, # 35789) line, and the UAS-elav (elav OE, stock # 38400) line were obtained from Bloomington Drosophila Resource Center (BDSC). The UAS-elav RNAi (#37915) was obtained from the Vienna Drosophila Resource Center (VDRC). The Elav-GeneSwitch-Gal4 is a gift from Dr. Haig Keshishian at Yale University.
Drosophila melanogaster motor function assays
The motor function phenotypes of all control and diseased conditions were collected at 25 °C. Elav-GS-Gal4 (neuron-specific driver) was used to express FUS-WT and FUS-P525L with and without knockdown of ELAV. For performing the climbing assay, flies were crossed in the absence of RU486 (mifepristone, Cayman Chemical, #10006317) on standard food. Day 1 adult females were transferred to RU486 (5 mM) drug food and placed at 25 °C for 6–15 days before performing the motor function assay at the time points indicated. 6–10 flies were transferred to empty rounded vials and allowed to acclimate for 20 min in the new vials. Flies were knocked to the bottom of the vials by tapping each vial against the laboratory bench three times, and a video camera was used to record flies climbing up the walls. The percentage of flies that climbed 4 cm in 10 s and the velocity of each fly were quantified and analyzed using GraphPad PRISM 8.0 software. Three experimental replicates (trials) were performed for each group.
Drosophila melanogaster NMJ analysis
All the genetic crosses were crossed with the inducible pan-neuronal driver, Elav-GS-Gal4, at 25 °C on standard media fed with 5 μM final concentration of RU-486 drug (#10006317, Cayman Chemicals). Third-instar larvae were collected for NMJ analysis. For immunofluorescence, the larvae were rinsed with PBS and dissected along the dorsal midline to expose the NMJs. Dissected Drosophila tissues were washed with PBS (Lonza) and fixed with 4% paraformaldehyde (Sigma-Aldrich, P6148) for 20 min at room temperature. The tissues were washed three times with PBS following fixation and blocked with blocking buffer: 5% normal goat serum (NGS, Abcam, AB7681) in PBS with 0.1% Triton-X (PBST). The samples were incubated with primary antibodies overnight at 4 °C and washed four times with 0.1% PBST, incubated with secondary antibodies for 2 h at room temperature followed by four washes with PBST. Samples were mounted with Fluoroshield (Sigma-Aldrich, F6057) onto glass slides. Primary and secondary antibodies were prepared in a blocking buffer. Primary antibodies: Cy3 conjugated Horse Radish Peroxidase (1:200; Jackson ImmunoResearch, 123-165-021), mouse Cysteine String Protein (dCSP) (1:100; Developmental Studies Hybridoma Bank). Secondary antibodies: Goat anti-mouse Alexa Fluor 488 (1:1000; Invitrogen, A11008); Goat anti-rabbit Alexa Fluor-568 (1:100; Invitrogen, #651727). Confocal images were acquired in Z-stacks using a Nikon A1-T216.3 confocal microscope at 60X (oil) magnification. NMJs innervating Muscle 4 at hemi-segments A3-A4 were used for analyzing synaptic bouton quantities. For each NMJ, the number of synaptic boutons per NMJ was normalized to the surface area of the innervating muscle. The bouton number and the NMJ area were quantified using ImageJ (NIH).
Oxidative stress induction and transcription block
Cells were left untreated or treated with 0.5 mM sodium arsenite (Merck Life Sciences) to induce acute oxidative stress for 1 h. For transcription inhibition, 25 μl/ml α-amanitin solution (Merck Life Sciences) was added to the medium for 8 h; in the last hour, 0.5 mM sodium arsenite was added to induce oxidative stress.
Bioinformatics analysis of sporadic ALS RNA-seq datasets
The raw count matrix relative to the RNA-seq analysis of ALS patient samples and individuals with and without other neurological disorders produced by the NYGC consortium15 was retrieved from the Gene Expression Omnibus (GEO) database46 (accession number: GSE124439), while the metadata information was taken from the original publication. Starting from this matrix, sample-specific gene-level FPKM (Fragments Per Kilobase of exon per Million mapped reads, a normalized expression measure) values were calculated using the “fpkm” function from the DESeq2 v1.32.0 R package47 and the exonic gene size information retrieved from the GENCODE v42 annotation48. Differential expression analysis between different conditions was performed using the “DESeq” function, controlling for sex and tissue type.
Statistics and reproducibility
Statistical analysis, graphs, and plots were generated using GraphPad PRISM 8.0 (GraphPad Software). The statistical test used is indicated in each figure legend. Sample size and the definition of replicates for each experiment are also indicated in the figure legends. We deemed statistically significant p-values at a threshold of less than 0.05. Images in Figs. 1B–D and 3A are representative micrographs from an experiment repeated 3 times with similar results. The adjusted p-values obtained from differential expression analyses shown in Fig. 7G and Supplementary Fig. S10A have been corrected for multiple comparisons via the Benjamini-Hochberg procedure.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data generated or analyzed during this study are included in the manuscript and supporting files. Source data are provided with this paper. Public sequencing data relative to the RNA-seq analysis of ALS patient samples and individuals with and without other neurological disorders produced by the NYGC consortium were acquired from the Gene Expression Omnibus (GEO) with accession number GSE124439 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE124439). Source data are provided with this paper.
References
Brown, R. H. & Al-Chalabi, A. Amyotrophic lateral sclerosis. N. Engl. J. Med. 377, 162–172 (2017).
Dassi, E. Handshakes and fights: the regulatory interplay of RNA-binding proteins. Front. Mol. Biosci. 4, 67 (2017).
Blokhuis, A. M. et al. Comparative interactomics analysis of different ALS-associated proteins identifies converging molecular pathways. Acta Neuropathol. 132, 175–196 (2016).
De Santis, R. et al. Mutant FUS and ELAVL4 (HuD) aberrant crosstalk in amyotrophic lateral sclerosis. Cell Rep. 27, 3818–3831.e5 (2019).
Garone, M. G. et al. ALS-related FUS mutations alter axon growth in motoneurons and affect HuD/ELAVL4 and FMRP activity. Commun. Biol. 4, 1025 (2021).
Garone, M. G., Salerno, D. & Rosa, A. Digital color-coded molecular barcoding reveals dysregulation of common FUS and FMRP targets in soma and neurites of ALS mutant motoneurons. Cell Death Discov. 9, 33 (2023).
Bronicki, L. M. & Jasmin, B. J. Emerging complexity of the HuD/ELAVl4 gene; implications for neuronal development, function, and dysfunction. RNA 19, 1019–1037 (2013).
Dell’Orco, M. et al. HuD regulates SOD1 expression during oxidative stress in differentiated neuroblastoma cells and sporadic ALS motor cortex. Neurobiol. Dis. 148, 105211 (2021).
Silvestri, B., Mochi, M., Garone, M. G. & Rosa, A. Emerging roles for the RNA-binding protein HuD (ELAVL4) in nervous system diseases. Int J. Mol. Sci. 23, 14606 (2022).
Garone, M. G. et al. Proteomics analysis of FUS mutant human motoneurons reveals altered regulation of cytoskeleton and other ALS-linked proteins via 3’UTR binding. Sci. Rep. 10, 11827 (2020).
Fischer, L. R. et al. Amyotrophic lateral sclerosis is a distal axonopathy: evidence in mice and man. Exp. Neurol. 185, 232–240 (2004).
Scekic-Zahirovic, J. et al. Toxic gain of function from mutant FUS protein is crucial to trigger cell autonomous motor neuron loss. EMBO J. 35, 1077–1097 (2016).
Picchiarelli, G. et al. FUS-mediated regulation of acetylcholine receptor transcription at neuromuscular junctions is compromised in amyotrophic lateral sclerosis. Nat. Neurosci. 22, 1793–1805 (2019).
Stoklund Dittlau, K. et al. Human motor units in microfluidic devices are impaired by FUS mutations and improved by HDAC6 inhibition. Stem Cell Rep. 16, 2213–2227 (2021).
Tam, O. H. et al. Postmortem cortex samples identify distinct molecular subtypes of ALS: retrotransposon activation, oxidative stress, and activated glia. Cell Rep. 29, 1164–1177.e5 (2019).
Lenzi, J. et al. Differentiation of control and ALS mutant human iPSCs into functional skeletal muscle cells, a tool for the study of neuromuscolar diseases. Stem Cell Res 17, 140–147 (2016).
Garone, M. G. et al. Conversion of human induced pluripotent stem cells (iPSCs) into functional spinal and cranial motor neurons using PiggyBac vectors. J. Vis. Exp. 147, e59321 (2019).
Ichikawa, N. et al. Identification of neurite outgrowth active sites on the laminin alpha4 chain G ___domain. Biochemistry 44, 5755–5762 (2005).
Lenzi, J. et al. ALS mutant FUS proteins are recruited into stress granules in induced pluripotent stem cells (iPSCs) derived motoneurons. Dis. Model Mech. 8, 755–766 (2015).
Casci, I. & Pandey, U. B. A fruitful endeavor: modeling ALS in the fruit fly. Brain Res. 1607, 47–74 (2015).
Marrone, L. et al. FUS pathology in ALS is linked to alterations in multiple ALS-associated proteins and rescued by drugs stimulating autophagy. Acta Neuropathol. 138, 67–84 (2019).
Alhindi, A., Boehm, I. & Chaytow, H. Small junction, big problems: neuromuscular junction pathology in mouse models of amyotrophic lateral sclerosis (ALS). J. Anat. 241, 1089–1107 (2021).
Scekic-Zahirovic, J. et al. Motor neuron intrinsic and extrinsic mechanisms contribute to the pathogenesis of FUS-associated amyotrophic lateral sclerosis. Acta Neuropathol. 133, 887–906 (2017).
Sharma, A. et al. ALS-associated mutant FUS induces selective motor neuron degeneration through toxic gain of function. Nat. Commun. 7, 10465 (2019).
Emara, M. M. et al. Hydrogen peroxide induces stress granule formation independent of eIF2α phosphorylation. Biochem. Biophys. Res. Commun. 423, 763–769 (2012).
De Santis, R. et al. FUS mutant human motoneurons display altered transcriptome and microRNA pathways with implications for ALS pathogenesis. Stem Cell Rep. 9, 1450–1462 (2017).
Parhad, I. M., Oishi, R. & Clark, A. W. GAP-43 gene expression is increased in anterior horn cells of amyotrophic lateral sclerosis. Ann. Neurol. 31, 593–597 (1992).
Ikemoto, A., Hirano, A. & Akiguchi, I. Increased expression of growth-associated protein 43 on the surface of the anterior horn cells in amyotrophic lateral sclerosis. Acta Neuropathol. 98, 367–373 (1999).
Andrés-Benito, P., Moreno, J., Aso, E., Povedano, M. & Ferrer, I. Amyotrophic lateral sclerosis, gene deregulation in the anterior horn of the spinal cord and frontal cortex area 8: implications in frontotemporal lobar degeneration. Aging 9, 823–851 (2017).
Wang, Z.-H. et al. HuD regulates the cpg15 expression via the 3’-UTR and AU-rich element. Neurochem. Res. 36, 1027–1036 (2011).
Yoo, S. et al. A HuD-ZBP1 ribonucleoprotein complex localizes GAP-43 mRNA into axons through its 3’ untranslated region AU-rich regulatory element. J. Neurochem. 126, 792–804 (2013).
Pasterkamp, R. J. & Giger, R. J. Semaphorin function in neural plasticity and disease. Curr. Opin. Neurobiol. 19, 263–274 (2009).
Dadon-Nachum, M., Melamed, E. & Offen, D. The ‘dying-back’ phenomenon of motor neurons in ALS. J. Mol. Neurosci. 43, 470–477 (2011).
de Winter, F. et al. The expression of the chemorepellent Semaphorin 3A is selectively induced in terminal Schwann cells of a subset of neuromuscular synapses that display limited anatomical plasticity and enhanced vulnerability in motor neuron disease. Mol. Cell Neurosci. 32, 102–117 (2006).
Gagliardini, V., Dusart, I. & Fankhauser, C. Absence of GAP-43 can protect neurons from death. Mol. Cell Neurosci. 16, 27–33 (2000).
Aigner, L. et al. Overexpression of the neural growth-associated protein GAP-43 induces nerve sprouting in the adult nervous system of transgenic mice. Cell 83, 269–278 (1995).
Harding, D. I., Greensmith, L., Mason, M., Anderson, P. N. & Vrbová, G. Overexpression of GAP-43 induces prolonged sprouting and causes death of adult motoneurons. Eur. J. Neurosci. 11, 2237–2242 (1999).
Javaherian, A. & Cline, H. T. Coordinated motor neuron axon growth and neuromuscular synaptogenesis are promoted by CPG15 in vivo. Neuron 45, 505–512 (2005).
McCampbell, A. et al. Antisense oligonucleotides extend survival and reverse decrement in muscle response in ALS models. J. Clin. Investig. 128, 3558–3567 (2018).
De Santis, R. et al. Direct conversion of human pluripotent stem cells into cranial motor neurons using a piggyBac vector. Stem Cell Res. 29, 189–196 (2018).
Tiago, T. et al. Small heat-shock protein HSPB3 promotes myogenesis by regulating the lamin B receptor. CDDis 12, 452 (2021).
Caputo, L. et al. Acute conversion of patient-derived Duchenne muscular dystrophy iPSC into myotubes reveals constitutive and inducible over-activation of TGFβ-dependent pro-fibrotic signaling. Skelet. Muscle 10, 642 (2020).
Guo, X., Gonzalez, M., Stancescu, M., Vandenburgh, H. H. & Hickman, J. J. Neuromuscular junction formation between human stem cell-derived motoneurons and human skeletal muscle in a defined system. Biomater 32, 9602–9611 (2011).
Casci, I. et al. Muscleblind acts as a modifier of FUS toxicity by modulating stress granule dynamics and SMN localization. Nat. Commun. 10, 5583 (2019).
Anderson, E. N. et al. Traumatic injury induces stress granule formation and enhances motor dysfunctions in ALS/FTD models. Hum. Mol. Genet. 27, 1366–1381 (2018).
Edgar, R., Domrachev, M. & Lash, A. E. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 (2002).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Harrow, J. et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 22, 1760–1774 (2012).
Acknowledgements
The authors wish to thank the Imaging Facility at Center for Life Nano and Neuro-Science, Fondazione Istituto Italiano di Tecnologia, for support and technical advice. We thank Dr. Tiziana Santini and Dr. Ersilia Fornetti for advice on NMJ staining and analysis. We thank Dr. R. De Santis, Prof. I. Bozzoni, Prof. S. Di Angelantonio, and the members of the Rosa lab for helpful discussion. The EBiSC Bank acknowledges Wellcome Trust Sanger Institute / HipSci as the source of the human induced pluripotent cell line WTSIi004-A which was generated with support from the EBiSC project. The EBiSC has received support from the Innovative Medicines Initiative (IMI) Joint Undertaking (JU) under grant agreement n°115582 and from the IMI-2 JU under grant agreement No 821362, resources of which are composed of financial contribution from the European Union’s Seventh Framework Programme (FP7/2007-2013), European Union’s Horizon 2020 research and innovation programme and EFPIA. Project funded in the framework of the National Center for Gene Therapy and Drugs Based on RNA Technology, National Recovery and Resilience Plan (NRRP), Mission 4 “Education and Research”, Component 2 “From Research to Business”, Investment 1.4 “Strengthening research structures for supporting the creation of National Centres, national R&D leaders on some Key Enabling Technologies”, funded by the European Union - Next Generation EU, Project CN00000041, CUP B93D21010860004, Spoke 3, to M.B. and A.R. This work has been supported by National Institute of Health (NIH) grant R01NS081303 to U.B.P., by Human Frontier Science Program (HFSP) Postdoctoral Fellowship (LT0024/2022-L) to M.G.G., by Sapienza University (RM12117A5DE7A45B) and PRIN 2022 – Progetti di Rilevante Interesse Nazionale (2022BYB33L, CUP: B53D23016100006) to M.B., and by Sapienza University (RP123188EC9F2349) to A.R.
Author information
Authors and Affiliations
Contributions
Conceptualization, B.S., A.R.; Formal analysis, B.S., M.M., D.M., A.C., M.B.; Investigation, B.S., D.M.; Methodology, B.S., M.M., A.C., B.B., V.d.T., M.Medici, E.N.A., C.P.Z., M.S., M.G.G.; Project administration, A.R.; Supervision, M.B., U.B.P., A.R.; Writing - original draft, A.R.; Writing - Review & Editing, B.S., U.B.P., A.R. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Silvestri, B., Mochi, M., Mawrie, D. et al. HuD impairs neuromuscular junctions and induces apoptosis in human iPSC and Drosophila ALS models. Nat Commun 15, 9618 (2024). https://doi.org/10.1038/s41467-024-54004-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-54004-8