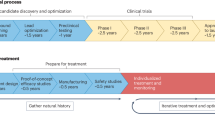
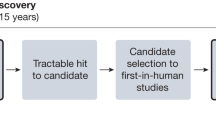
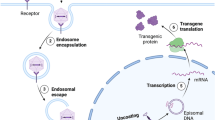
Abstract
Individualized genetic therapies—medicines that precisely target a genetic variant that may only be found in a small number of individuals, as few as only one—offer promise for addressing unmet needs in genetic disease, but present unique challenges for trial design. By nature these new individualized medicines require testing in individualized N-of-1 trials. Here, we provide a framework for maintaining scientific rigor in N-of-1 trials. Building upon best practices from traditional clinical trial design, recent guidance from the United States Food and Drug Administration, and our own clinical research experience, we suggest key considerations including comprehensive baseline natural history, selection of appropriate clinical outcome assessments (COAs) individualized to the patient genotype-phenotype for safety and efficacy assessment over time, and specific statistical considerations. Standardization of N-of-1 trial designs in this fashion will maximize efficient learning from this next generation of targeted individualized therapeutics.
Similar content being viewed by others
Introduction
Maturing platforms for targeted cell and gene-based therapies—including but not limited to antisense oligonucleotides (ASOs), siRNA, mRNA, DNA/RNA editing—are offering great hope for treatment of the over 8000 rare genetic diseases (defined as those affecting less than 1:200,000 individuals). The promise of these therapeutic approaches stems from their ability to directly target the underlying root causes of genetic disease at the level of the RNA or DNA—correcting the impact of genetic mutations, or even the mutations themselves. These technologies have been used under existing regulatory and drug development frameworks to support successful commercial drug development for several of the most prevalent rare genetic diseases1. However, commercial incentives have largely failed to support development of treatments for the “long tail” of the rarest genetic conditions, 85% of which have an estimated prevalence of less than 1:1,000,000.
Recent examples demonstrate that it is possible to use genetically-targeted technologies to develop therapies tailored to an individual’s unique genetic pathology2,3,4,5,6,7. Because of their precise targeting, the resulting drugs are often applicable to vanishingly small “markets” of only a handful of patients (or even just one), turning many facets of traditional drug development on their head2. A series of FDA guidances have been recently drafted to govern the development and investigational clinical deployment of such individualized therapies (“Individualized Antisense Oligonucleotide Drug Products for Severely Debilitating or Life-Threatening Diseases”)8, but specialized approaches to trial design and clinical outcome measures will be required to evaluate them rigorously. Here, we discuss design challenges associated with N-of-1 trials, and propose a clinical framework with which to begin to address them.
N-of-1 trials of individualized, genetically targeted therapies
We refer here to a specific type of N-of-1 trial design involving investigational therapies for which the therapeutic itself is individualized: targeting genetic variants that may be found in only one or a few individuals. We note that these fall in the specific subcategory of single case experimental design (SCED) studies, in which drug and trial design are designed for a single patient. We further note that this is distinct from the traditional concept of N-of-1 trials, a term which has been used to refer to studies involving single patients who are treated with (non-individualized) therapeutics in a repeat cycle of treatment challenge and withdrawal (A-B-A-B)—where non-individualized refers to interventions not specifically designed based on a patient’s unique genetic makeup9. Furthermore, the genetically targeted modalities discussed in this perspective do not naturally lend themselves to crossover clinical trial designs (for instance, due to their long half-life of ASOs, or the single dose nature of AAV or CRISPR therapies).
Challenges of clinical protocol development for N-of-1 clinical trials of individualized, genetically targeted therapies
Protocol development for N-of-1 trials of individualized, genetically targeted therapies involves many careful considerations that are common to the design of any clinical trial (Table 1). These include, but are not limited to, an in-depth understanding of the genotype and phenotype of the patient to be treated, the mechanism of action and tissue distribution of the investigational drug, the development of treatment goals relevant to the patient/caregiver, and the setting of realistic expectations of what may be attainable. However, N-of-1 trials also involve clinical trial design considerations that are more unique to N-of-1 trials, especially as relates to rigorously assessing clinical efficacy, as discussed below.
Clinical outcome assessments
For many of the rare diseases that are the subject of N-of-1 trials, there are no pre-established disease-relevant clinical outcome assessments (COAs), and formal development of such a measure may not be possible in advance due to the small number of patients, the heterogeneity in symptoms, limited characterization of the natural history, and the global distribution of the patients. N-of-1 clinical trials can follow many of the principles outlined in the FDAs Patient-Focused Drug Development (PFDD) Guidance documents for selection of Clinical Outcome Assessments (COAs)10. COAs should be selected based on the patient’s lived experience and reflect the current state of their condition. While traditional clinical trial design often utilizes focus groups and exit interviews in early phase trials to determine the appropriateness of COAs, N-of-1 trials only have to account for one or a small number of patient symptoms. Rigorous qualitative interview structures may be deployed at the drug development stage to ensure there is a comprehensive disease concept model that identifies the main areas of impairment and represents each patient’s experience. COAs may be identified to capture the identified symptoms and structured data collection can begin ideally well before the intervention. It is essential that clinical investigators treating patients with ultra-rare disease have a deep understanding of disease pathophysiology and the patients’ unmet treatment needs to therefore identify the most appropriate outcome measures to track safety and efficacy of investigational individualized therapies.
Defining relevant changes
Once COAs are selected, clinical trials for individualized drugs still face the challenge of defining meaningful change. Traditionally this is captured via the concept of the trial’s minimal clinically important difference (MCID)—the smallest response that would be considered clinical impactful—which can be then used in an objective and quantifiable way to classify patients into responders or non-responders11. With sufficient patient population data, statistical methods can be applied to establish a standardized MCID12—e.g., for Duchenne Muscular Dystrophy, a threshold of 30 meters on the 6-min walk test, as determined by assessing more than 150 patients in a Phase 2B clinical trial13. However, it is often challenging or impossible to implement this approach to current individualized N-of-1 trials. This is due to the fact that current individualized N-of-1 trials involve ultra-rare diseases for which this data typically does not exist. Furthermore, even if a standardized MCID has been established for the disease population in question, its applicability may be limited by clinical heterogeneity, since an individualized drug may only target a small subset of patients (or a single patient) with a specific mutation, whose clinical trajectories may differ from the group.
While a standard statistical approach to the determination of MCID is not possible in an N-of-1 trial as discussed here, the diligent collection of individualized ‘natural history data’ prior to treatment may allow for determination of test-retest and longitudinal stability of measures with longer baseline data collection providing more data points. One of the methods of determination of MCID outlined by the PFDD Guidance document four is to use hypothetical scenarios measured within a particular set of COAs to understand a group of patients assessment of meaningfulness through qualitative interviews. This could be a useful method for determining an “a priori” categorization of what a meaningful outcome would be for an individual patient prior to initiating an N-of-1 trial14.
This general approach of collecting “individual natural history” data also usefully addresses several inherent challenges to N-of-1 trials, namely genotype-phenotype heterogeneity15 and the scarcity of characterized and validated biomarkers16. For instance, among individuals diagnosed with the same disease, there may be great variability in the manifestation of symptoms, amplifying the challenge of selecting relevant COAs. As an example, patients with pathogenic SCN2A variant related disorders and developmental epileptic encephalopathies (DEEs) can exhibit variable symptoms ranging across the phenotypic spectrum, with differing symptoms and ages of onset depending on the genetic variant (i.e., gain of function, loss of function, etc.), including infantile spasms, neonatal onset medically refractory seizures, neurodevelopmental delay, choreoathetosis, intellectual disability, autism, ataxia, and significant gastrointestinal and autonomic dysfunction17. Genetic diseases often impact multiple domains, and the heterogeneity within and between these domains adds another layer of complexity18,19 which further requires N-of-1 trial design tailored to the individual with appropriate outcome and endpoint measures.
A suggested approach to N-of-1 trial design
Implementing this strategy, we suggest that designing N-of-1 trials is usefully subdivided into consideration of markers of target engagement, clinical biomarkers, and outcome assessments that can reliably measure aspects of function, development and behavior relevant and individualized to the patient and underlying disease. These assessments should be informed by what is known about the toxicity profiles for the investigational agent or class of agent deployed. Objective and quantitative clinical assessments are of special value, but qualitative tools remain imperative for capturing clinical impacts and tracking meaningfulness to the patient in terms of disease morbidity and quality of life. We highlight several subdomains of special relevance to current N-of-1 trials in the discussion below.
Safety
Safety outcome measures for N-of-1 trials need to be matched for potential modes of toxicity that may be possibly associated with the therapeutic in question. This includes both the toxicity profiles of the specific investigational agent in question (as established in the course of either preclinical testing or prior clinical experience), as well as known or suspected class effects associated with the therapeutic modality. For example, typical safety-related clinical biomarkers for intrathecal ASO trials may include CSF sampling for cell count, protein, glucose, microbial culture, surveillance neuroimaging and symptom screening for ventricular changes, blood testing for basic chemistries platelets/liver function tests, and urinalysis for proteinuria. What safety assessments are most appropriate for a particular class of therapeutic intervention would be expected to evolve as we learn more about the safety profile of that class from traditional clinical trials as well as N-of-1 trials.
Clinical outcome measures
For central nervous system disorders, relevant clinical outcome measures may include seizure logs, quantitative or qualitative electroencephalograms (EEGs), electromyography/nerve conduction studies (EMG/NCVs), qualitative and/or quantitative volumetric neuroimaging, MR spectroscopy, quantitative sensory testing, neurodevelopmental assessments, and other structured neurologic rating scales. Of particular interest are wearable biometric sensors capable of capturing longitudinal data relevant to symptoms such as motor strength, coordination, sleep, or seizures. Validated devices may prove preferable when assessing an individual with significant cognitive impairment or developmental delays as they acquire data passively and do not depend upon patient understanding or cooperation with testing, such as inertial sensors for ataxic gait measurement.
Many current N-of-1 trials target central nervous system diseases for which neurodevelopmental delays are an especially prominent disease feature impacting quality of life. Neurodevelopmental COAs can include specific assessments of cognitive, academic, adaptive behavior, executive function, sensorimotor, and other behavioral, mood and social concerns. These assessments need to be tailored to individual patients’ specific diagnoses and needs and be developmentally age-appropriate. Curation of the patient’s neurodevelopmental test battery should incorporate consideration of test validity and reliability (demonstrated in neurotypical and if available, similar disease cohorts). Although these assessments are typically ordinal, many have still been shown to reflect an MCID or MDD during serial assessment in larger trials20. Since these instruments will at best be validated only for disease groups with shared clinical features to the N-of-1 trial subject, multiple scales and perspectives (e.g., patient, caregiver, clinician) may be required to test the same ___domain in order to reduce bias and optimize objectivity.
Fluid/tissue biomarkers
In a perfect world, an interventional trial would ideally incorporate measurement of fluid and tissue biomarkers that demonstrate target engagement of the investigational drug (example: Huntington protein lowering in response to treatment with a HTT knockdown ASO), but these do not exist and are not feasible for the majority of N-of-1 trials. This is unsurprising given the rarity of the conditions studied in these trials, combined with the fact that currently, most N-of-1 trials target the CNS where tissue sampling is not possible. Furthermore, some N-of-1 trials employ drugs that may act in a mutation- or allele-specific manner, requiring correspondingly mutation- or allele-specific measures of target engagement. Nonetheless, other fluid and tissue biomarkers may provide helpful guidance to investigators managing patients enrolled in N-of-1 trials when available. For neurodegenerative diseases, plasma or CSF neurofilament levels may correlate with rates of neuronal injury, potentially informing clinical decision making. In addition, electrophysiologic biomarkers on EEG or EMG/NCV studies, if identified, may provide an objective metric of disease course and progression as it relates to the biological underpinnings of the genetic disease. Ideally, a reliable and informative biomarker can serve in a predictive and/or prognostic capacity.
Future statistical study design considerations
Statistical analyses are a seeming challenge given the nature of an individualized intervention, as traditional methods may not readily apply. These challenges are not entirely new, however, as structured approaches towards these issues have been developed in other fields such as oncology21,22. There are various statistical methods available for analyzing individual data from N-of-1 trials and other single-case designs. While full discussion is beyond the scope of this paper, we suggest that the clinical design considerations outlined here may serve as a first step towards meeting this challenge for rare genetic disease, paving the way towards the development of master clinical protocols to allow data sharing and aggregation of signals for meta-analyses across multiple N-of-1 interventions23,24,25,26.
Regulatory considerations
The current regulatory landscape allows for the development of individualized investigational drugs for ultra-rare diseases via the submission of N-of-1 research INDs10,14. While pathways to approval or reimbursement of individualized drugs have not yet been established, incorporating the clinical trial design principles discussed here will support the rigorous clinical science necessary to meet these standards27.
Discussion
Individualized interventions, made possible through recent advances in genetically targeted therapeutic technologies, are now a reality. They represent opportunities to meet unmet medical needs while also posing interesting challenges to clinical trial designs as traditionally implemented. These challenges specifically require collaborative engagement across entities, including manufacturers, investigators, regulatory bodies, institutional review boards, patient advocacy organizations, funding grant agencies and other relevant stakeholders. Tackling the N-of-1 trial design within the framework discussed herein is critical to unlocking the path forward, for the potential benefit of those with rare genetic disease.
References
Baylot, V., Le, T. K., Taïeb, D., Rocchi, P. & Colleaux, L. Between hope and reality: treatment of genetic diseases through nucleic acid-based drugs. Commun. Biol. 7, 489 (2024).
Woodcock, J. & Marks, P. Drug regulation in the era of individualized therapies. N. Engl. J. Med. 381, 1678–1680 (2019).
Marks, P. & Witten, C. Toward a new framework for the development of individualized therapies. Gene Ther. 28, 615–617 (2021).
Kim, J. et al. Patient-customized oligonucleotide therapy for a rare genetic disease. N. Engl. J. Med. 381, 1644–1652 (2019).
Tran, H. et al. Suppression of mutant C9orf72 expression by a potent mixed backbone antisense oligonucleotide. Nat. Med. 28, 117–124 (2022).
Korobeynikov, V. A., Lyashchenko, A. K., Blanco-Redondo, B., Jafar-Nejad, P. & Shneider, N. A. Antisense oligonucleotide silencing of FUS expression as a therapeutic approach in amyotrophic lateral sclerosis. Nat. Med. 28, 104–116 (2022).
Kim, J. et al. A framework for individualized splice-switching oligonucleotide therapy. Nature 619, 828–836 (2023).
Fda, U. Nonclinical testing of individualized antisense oligonucleotide drug products for severely debilitating or life-threatening diseases. Draft Guidance. pdf. (2021).
Guyatt, G. et al. Determining optimal therapy–randomized trials in individual patients. N. Engl. J. Med. 314, 889–892 (1986).
Center for Drug Evaluation & Research. FDA Patient-Focused Drug Development Guidance Series for Enhancing the Incorporation of the Patient’s Voice in Medical Product Development and Regulatory Decision Making. U.S. Food and Drug Administration https://www.fda.gov/drugs/development-approval-process-drugs/fda-patient-focused-drug-development-guidance-series-enhancing-incorporation-patients-voice-medical (2024).
Jaeschke, R., Singer, J. & Guyatt, G. H. Measurement of health status. Ascertaining the minimal clinically important difference. Control. Clin. Trials 10, 407–415 (1989).
Copay, A. G., Subach, B. R., Glassman, S. D., Polly, D. W. Jr & Schuler, T. C. Understanding the minimum clinically important difference: a review of concepts and methods. Spine J. 7, 541–546 (2007).
McDonald, C. M. et al. The 6-minute walk test and other clinical endpoints in Duchenne muscular dystrophy: reliability, concurrent validity, and minimal clinically important differences from a multicenter study. Muscle Nerve 48, 357–368 (2013).
Center for Drug Evaluation & Research. Patient-focused drug development: Methods to identify what is important to patients. U.S. Food and Drug Administration https://www.fda.gov/regulatory-information/search-fda-guidance-documents/patient-focused-drug-development-methods-identify-what-important-patients (2022).
Vinkšel, M., Writzl, K., Maver, A. & Peterlin, B. Improving diagnostics of rare genetic diseases with NGS approaches. J. Community Genet. 12, 247–256 (2021).
Bax, B. E. Biomarkers in rare diseases 2.0. Int. J. Mol. Sci. 23, 4582 (2022).
Wolff, M., Brunklaus, A. & Zuberi, S. M. Phenotypic spectrum and genetics of SCN2A-related disorders, treatment options, and outcomes in epilepsy and beyond. Epilepsia 60, S59–S67 (2019).
Tandon, P. K. & Kakkis, E. D. The multi-___domain responder index: a novel analysis tool to capture a broader assessment of clinical benefit in heterogeneous complex rare diseases. Orphanet J. Rare Dis. 16, 183 (2021).
Lenderking, W. R. et al. Measuring health-related quality of life in patients with rare disease. J. Patient Rep. Outcomes 5, 61 (2021).
Mishra, B. et al. Minimal clinically important difference (MCID) in patient-reported outcome measures for neurological conditions: review of concept and methods. Ann. Indian Acad. Neurol. 26, 334–343 (2023).
Gouda, M. A., Buschhorn, L., Schneeweiss, A., Wahida, A. & Subbiah, V. N-of-1 trials in cancer drug development. Cancer Discov. 13, 1301–1309 (2023).
Biankin, A. V., Piantadosi, S. & Hollingsworth, S. J. Patient-centric trials for therapeutic development in precision oncology. Nature 526, 361–370 (2015).
Punja, S. et al. N-of-1 trials can be aggregated to generate group mean treatment effects: a systematic review and meta-analysis. J. Clin. Epidemiol. 76, 65–75 (2016).
Lekstrom-Himes, J. et al. Moving away from one disease at a time: Screening, trial design, and regulatory implications of novel platform technologies. Am. J. Med. Genet. C Semin. Med. Genet. 193, 30–43 (2023).
Lekstrom-Himes, J. et al. Data sharing to advance gene-targeted therapies in rare diseases. Am. J. Med. Genet. C Semin. Med. Genet. 193, 87–98 (2023).
Augustine, E. F., Adams, H. R. & Mink, J. W. Clinical trials in rare disease: challenges and opportunities. J. Child Neurol. 28, 1142–1150 (2013).
Pian, J. M. Y. et al. How to pay for individualized genetic medicines. Nat. Med. https://doi.org/10.1038/s41591-024-03071-x (2024).
Acknowledgements
This work was supported in part by grant funding from California Institute of Regenerative Medicine (CIRM): CLIN2-15085 (O.K.M.) and CIRM-DISC2-13469 (J.G.), as well as the Chan Zuckerberg Initiative (W.Y., T.Y.), SFARI (T.Y.), and the Termeer Foundation (W.Y.).
Author information
Authors and Affiliations
Contributions
All authors supported content generation and contributed to the development and revision of the manuscript. Olivia Kim-McManus, Timothy Yu, Richard Finkel, and Erika Augustine formulated the initial concept for the manuscript. Olivia Kim-McManus was the primary content generator and organized the scope of the manuscript. Joseph G. Gleeson, Laurence Mignon, Amena Smith Fine, Winston Yan, Nicole Nolen, Scott Demarest, Elizabeth Berry-Kravis, Stefanie Leonard, Samuel Finlayson, Gholson J. Lyon, Rebecca Schule, and Timothy Yu supported content generation and contributed to the development of the manuscript. Olivia Kim-McManus and Timothy Yu supervised quality review and manuscript revisions.
Corresponding author
Ethics declarations
Competing interests
Joseph Gleeson serves as a consultant for Ionis Pharmaceuticals, Neurocrine Pharmaceuticals, and the n-Lorem Foundation. Winston Yan is a founder, employee, and shareholder of Arbor Biotechnologies, and serves as President and Board Member of the N=1 Collaborative. Timothy Yu has received research support from EveryONE Medicines, and volunteer Board Member for the N=1 Collaborative, the Oligonucleotide Therapeutics Society, and the Society for RNA Therapeutics, and a volunteer advisor to several rare disease foundations. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Suzanne Mcdonald, Liam Smeeth and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Kim-McManus, O., Gleeson, J.G., Mignon, L. et al. A framework for N-of-1 trials of individualized gene-targeted therapies for genetic diseases. Nat Commun 15, 9802 (2024). https://doi.org/10.1038/s41467-024-54077-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-54077-5
This article is cited by
-
Toward an Extensible Regulatory Framework for N-of-1 to N-of-Few Personalized RNA Therapy Design
Therapeutic Innovation & Regulatory Science (2025)