Abstract
Functional cellular heterogeneity in tumours often underlies incomplete response to therapy and relapse. Previously, we demonstrated that the growth of the paediatric brain malignancy, sonic hedgehog subgroup medulloblastoma, is rooted in a dysregulated developmental hierarchy, the apex of which is defined by characteristically quiescent SOX2+ stem-like cells. Integrating gene expression and chromatin accessibility patterns in distinct cellular compartments, we identify the transcription factor Olig2 as regulating the stem cell fate transition from quiescence to activation, driving the generation of downstream neoplastic progenitors. Inactivation of Olig2 blocks stem cell activation and tumour output. Targeting this rare OLIG2-driven proliferative programme with a small molecule inhibitor, CT-179, dramatically attenuates early tumour formation and tumour regrowth post-therapy, and significantly increases median survival in vivo. We demonstrate that targeting transition from quiescence to proliferation at the level of the tumorigenic cell could be a pivotal medulloblastoma treatment strategy.
Similar content being viewed by others
Introduction
Medulloblastoma (MB) is a paediatric malignant tumour of the cerebellum and consists of four molecular subgroups with varying clinical features and prognosis1,2. A large subset of MB is characterised by constitutive activation of the sonic hedgehog (SHH) signalling pathway3 and has been associated with the worst outcome of the four subgroups4,5. In particular, high risk SHH-TP53 mutant tumours6,7 are associated with highly aggressive disease, treatment resistance, and relapse8,9,10. Despite the tremendous advances made in genomic characterisation of SHH-subgroup MB (SHH-MB)11,12,13,14,15,16,17,18, targeted treatment against the SHH pathway has not been effective in preventing local relapse19,20, and concerns regarding the impact of prolonged SHH inhibition on the central nervous system (CNS) as well as extra-CNS development exist21.
Within individual tumours, the neural cell compartment of MB consists of heterogeneous populations of undifferentiated and more differentiated cells within specific cerebellar lineages, that have distinct tumorigenic potentials22,23,24,25. This view of these tumours as disrupted developmental lineages is reinforced by single-cell RNA sequencing (scRNAseq) studies of patient samples16,17,26. We previously demonstrated in a mouse model of SHH-MB that the different tumour cell states are functionally distinct, and represent a related developmental lineage, with a rare compartment of largely quiescent tumour cells expressing the stem cell marker SRY-box transcription factor 2 (Sox2), capable of driving long-term tumour growth in vivo by giving rise to tumour bulk consisting of progeny with progenitor and differentiated cell states24. This tumour hierarchy recapitulates the normal developmental process of the granule neuron lineage27. Despite this increasing recognition of the variety of MB cell transcriptional identities, the regulation of cell state transitions within specific neoplastic developmental lineages has been relatively underexplored. For this study, we hypothesised that defining mechanisms of cell fate decisions and transitions in the medulloblastoma hierarchy could hold a key to unlocking novel therapeutic approaches in this disease.
In this work, we address a major and specific unanswered question in the medulloblastoma hierarchy: how the quiescent, self-renewing, and tumorigenically potent neoplastic stem cells transition into rapidly cycling progenitor cells to generate tumour bulk. Targeting the large fraction of rapidly proliferating but downstream progenitors alone is insufficient to prevent tumour regrowth24, as a residual persister SOX2+ population is capable of regenerating the more differentiated and proliferative bulk. By defining the molecular transitions from quiescence to proliferation at the level of the SOX2+ population, we identify a targetable transcription factor, whose action is rare in the MB neural lineage, but where inhibition blocks growth and generation of tumour bulk from the SOX2+ population both in early tumour formation and in the persister cells following treatment of full-blown tumours.
Results
Quantitative modelling of proliferation dynamics of SHH-MB subpopulations
As existing human cellular models of MB do not well reflect patient tumours28,29, we studied the clinically relevant Ptch1 heterozygous irradiated transgenic mouse model (Ptc) of SHH-MB30, where we had previously defined at least three distinct cellular compartments: 1) a rare but slowly cycling population of stem cells expressing Sox2 2) rapidly cycling and common doublecortin (Dcx) positive progenitors, and 3) abundant postmitotic cells expressing neuronal nuclei (NeuN). The hierarchical organisation of these cell states was definitively shown by the Sox2 lineage trace24. To determine the proliferation kinetics of these three populations, the loss of 5-ethynyl-2’-deoxyuridine (EdU), following its short and longer-term incorporation in a pulse-chase assay was previously assessed24 (Supplementary Fig. 1a). Performing a quantitative analysis of this data, we demonstrate the presence of a quiescent (slow-cycling) stem cell population, dividing largely asymmetrically to self-renew while giving rise to progenitor cells at an estimated cell division rate of approximately once every 20 days (for details of analysis see Supplementary Note). Notably, progenitor cells also self-renew, dividing asymmetrically at a rate of approximately once every 15 hours, giving rise to NeuN+ cells. These postmitotic cells, which comprise a large fraction of the tumour bulk, constitute a transient population becoming lost due to apoptosis at a comparably high rate. These results are consistent with a proliferative hierarchy in which slowly cycling SOX2+ cells (S) divide to give rise to intermediate progenitor-like cells (P) expressing Dcx, which in turn generate short-lived non-cycling postmitotic NeuN+ cells (N) (Fig. 1a). It follows that the self-renewing cells at the apex of the hierarchy (S) must exit their dormant state to generate more differentiated progenitor cells (P), and therefore exist in quiescent as well as activated cycling states.
a Visualisation of the approximate lineage hierarchy in SHH-MB based on quantitative analysis of proliferation dynamics (for details, see Supplementary Note); based on data from Vanner et al.24; created in BioRender.com. b Unsupervised clustering using Uniform Manifold Approximation and Projection (UMAP) performed on 6739 single cells from four mouse medulloblastoma primary Ptc tumours; OPC oligodendrocyte progenitor cells. c Scatter plot of the cell cycle phases in SOX2+ cells computationally extracted from the scRNAseq data of 4 Ptc mouse tumours. d Genomic annotation distribution calculated using cis-regulatory element annotation system (CEAS)89 of the genome (left), of the catalogue of chromatin accessible regions identified through ATACseq of SOX2+ and SOX2− subpopulations (centre) and of the top 10% most variable chromatin accessible regions across SOX2+ and SOX2− subpopulations (right). e Relative normalised chromatin accessibility between SOX2+ and SOX2− subpopulations for each of the regions significantly correlated to the Sox2 promoter calculated using C3D (‘Methods’); n = 4 biologically independent samples; the box of the box and whiskers plots extends from the 25th to 75th percentiles; the centre line is plotted at the median; whiskers indicate min. and max. values; two-tailed unpaired t-test; *p < 0.05, **p < 0.01, ***p < 0.001; L–R p-values: 0.0025, 0.0009, 0.0085, 0.0353, 0.0432, 0.0245, and 0.0280. f Gene Ontology Biological Process (GO BP) enrichment analysis of the genes associated with significantly differentially accessible regions across SOX2+ and SOX2– subpopulations. The nodes represent gene sets, and the edges represent mutual overlap. Clusters, representing overlapping gene sets, were enriched in either SOX2+ population-associated gene sets (green nodes), SOX2– population-associated gene sets (grey nodes), or a mixture of both; n = 4 biologically independent samples. g HOMER motif analysis of uniquely accessible regions in the SOX2+ and SOX2– subpopulations; AT2=AT2G33550; n = 4 biologically independent samples; motif enrichment is calculated using cumulative binomial distributions. Source data are provided as a Source Data file.
Integration of whole-genome datasets reveals an epigenetically distinct cycling stem cell population
In order to interrogate this SHH-MB stem cell state transition, we first performed single-cell transcriptomic analysis on four mouse tumours from Ptc mice of SHH-MB. Unsupervised clustering using Uniform Manifold Approximation and Projection (UMAP) was performed on the individual cell transcriptomes of 6739 cells, and 16 unique clusters were identified (Fig. 1b; Supplementary Fig. 1b). Cluster identities were assigned using expression of established markers combined with the scCATCH method of automatic annotation on cell types of clusters from scRNAseq data31, and included progenitor and mature neural cell populations, as well as immune cells (Fig. 1b; Supplementary Fig. 1b; Supplementary Data 1). The cellular markers are expressed in frequencies and patterns observed previously24 (Fig. 1a; Supplementary Fig. 1c, e), with rare Sox2-expressing cells observed distributed across all phases of the cell cycle (Fig. 1c; Supplementary Fig. 1d), highlighting that a fraction of the SOX2+ stem cells are actively proliferating or in an activated state before transitioning into progenitors.
We also hypothesised that the gene expression, functional, and tumorigenic differences within the MB lineage24 are orchestrated by distinct states of chromatin organisation. Using the Ptc model containing a Sox2-eGFP reporter32 (Supplementary Fig. 1f), we profiled the chromatin accessibility landscapes within enriched tumorigenic SOX2+ (GFP+) and non-tumorigenic SOX2− (GFP−) tumour fractions24 (Supplementary Fig. 1g) following fresh sorting of MB tumour cells for GFP using the assay for transposase-accessible chromatin using sequencing (ATACseq)33. Of the top 0.5% most variable chromatin accessible sites across the SOX2+ and SOX2− tumour fractions, a large majority (85.8%) were more accessible in the stem cells (Supplementary Fig. 1i), reminiscent of the normal developmental pattern of stem cells being more epigenetically pliable than their more differentiated progeny34,35. The majority of the differentially accessible regions between the two subpopulations were associated with distal regulatory elements, in large part enhancers36,37,38,39,40 (Fig. 1d; Supplementary Fig. 1h), suggesting that distal regulatory elements govern the highly dynamic transitions across cellular hierarchies. This observation was further highlighted when we identified the set of regulatory elements associated with the Sox2 promoter using cross-cell-type correlation in analysis of DNase I hypersensitivity (C3D)41,42,43 (Supplementary Fig. 1j, k; ‘Methods’). We observed up to 3-fold greater chromatin accessibility at the Sox2 enhancers compared to the promoter across SOX2+ and SOX2− tumour populations (Fig. 1e), emphasising the role of enhancers in regulating lineage identities44.
Analysis of the most significantly differentially accessible regions revealed an enrichment of the gene ontology (GO) terms 'regulation of cell differentiation', 'regulation of metabolic process' and 'MAPK cascade' (Fig. 1f), in SOX2+ cells, in keeping with the MB stem cells recapitulating normal developmental programmes24,27 (Supplementary Fig. 1l). Chromatin accessibility patterns at transcription factor gene promoters mirrored differences in their expression across the tumour subpopulations (Supplementary Fig. 1m). To identify preferentially enriched regulators in SOX2+ stem cells compared to SOX2– tumour bulk, we performed a motif enrichment analysis in the set of uniquely accessible regions in each group (Fig. 1g) and compared it to their expression patterns across the subpopulations (Supplementary Fig. 1n, o). Sox9, Sox10, Sox3 and Sox4 were among the most significantly enriched transcription factor (TF) motifs in the SOX2+ stem cells. In the SOX2– fraction, motifs of basic helix-loop-helix (bHLH) lineage specification TFs (Neurod1, Atoh1, Neurog2, Olig2 and Tal1) were significantly enriched.
Olig2 a regulator of stem cell proliferation in the MB lineage
To further dissect the molecular mechanisms regulating this SOX2+ cell state transition from quiescence to activation, we crossed Sox2-eGFP with Mki67-RFP mice45 on the Ptc background and sorted for GFP/RFP, identifying cycling (MKI67+) and non-cycling (MKI67–) SOX2+ stem cells in SHH-MB tumour primary cells (Supplementary Fig. 2a, b). To determine regulators of the transition between these enriched populations of non-cycling and cycling SOX2+ stem cells, we performed ATACseq followed by a motif enrichment analysis in the set of uniquely accessible regions in each group (Fig. 2a). Sox9 was the only TF motif that was significantly enriched in the non-cycling stem cell fraction, and has been implicated in promoting self-renewal and relapse in SHH-MB46,47. In the cycling stem cells, motifs of bHLH lineage specification TFs (Neurod1, Atoh1, Neurog2, and Olig2) were significantly enriched. Notably, most of these motifs were also enriched in the SOX2− bulk (predominantly cycling) tumour cells (Fig. 1g), providing a transcriptional regulation link between the activated stem cell and rapidly cycling progenitor states.
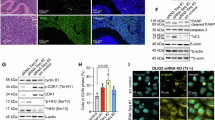
a HOMER motif analysis of uniquely accessible regions in the cycling (MKI67+) and non-cycling (MKI67−) fractions of the stem (SOX2+) cells; n = 2 biologically independent samples; motif enrichment is calculated using cumulative binomial distributions. b Overlap of OLIG2 binding peaks in mNSCs (data obtained from Mateo et al.53) with ATACseq peaks of SOX2+/− (top pie chart) as well as SOX2+/MKI67+ and SOX2+/MKI67– (bottom pie chart) tumour fractions. c Gene Ontology Biological Process (GO BP) analysis of genes associated with the overlaps in (b). d, e Immunocytochemistry of Ptch1+/−;Trp53−/− mouse tumour cells (d), quantification of EdU+ cells after a 1-hour pulse averaged across three biological replicates (e); n = 3, error bars denote mean ± SEM; two-tailed unpaired t-test; ns p ≥ 0.05, ****p < 0.0001; p > 0.9999 (EdU+ vs. OLIG2+), p < 0.0001 (EdU+ vs. EdU+/OLIG2+), p < 0.0001 (OLIG2+ vs. EdU+/OLIG2+). f Phase object confluence of adherently grown Ptch1+/−;Trp53−/− CRISPR-Cas9 control or Olig2-KO mouse tumour cells measured across 14 days using live cell imaging; data shown are representative of three independent experiments; error bars denote mean ± SEM; two-tailed unpaired t-test performed at mid-exponential phase time point (144 hours); p = 0.0021 (control vs. Olig2-KO). g Limiting dilution analysis (LDA) performed on Ptch1+/−;Trp53−/− CRISPR-Cas9 control or Olig2-KO cells; n = 3 biological replicates; individual replicates were assessed by goodness of fit testing using extreme limiting dilution analysis (ELDA) software (see Methods); error bars denote mean ± SEM; unpaired t-test; two-tailed p-value; p = 0.0182. h Tumour volumes in NOD-scid-gamma (NSG) mice subcutaneously engrafted in both flanks with either Ptch1+/−;Trp53−/− CRISPR-Cas9 control or Olig2-KO cells. n = 4 mice (8 flanks) for control and n = 3 (6 flanks) for Olig2-KO condition; error bars denote mean ± SEM; chi-square test; p = 0.0157; f–h: *p < 0.05, **p < 0.01. Source data are provided as a Source Data file.
Pioneer factors can be identified by their DNA sequence motifs at sites that become more chromatin accessible during lineage progression48,49,50. Consistently, Olig2 expression is significantly higher in the stem cell population (Supplementary Fig. 1m, o; Supplementary Data 1) and the Olig2 DNA motif is among the most significantly enriched in uniquely accessible chromatin regions in the cycling stem cells and in the bulk tumour (Supplementary Fig. 1m). Known to have pioneer factor activity in the developmental context51,52, Olig2 potentially acts by reorganising the chromatin landscape in stem cells to govern the transition of a neoplastic stem cell from a quiescent to downstream activated states. To test this further, we computationally overlapped publicly available chromatin immunoprecipitation followed by sequencing (ChIPseq) data of OLIG2 in mouse neural stem cells (NSCs)53 with the ATACseq data of the SHH-MB tumour compartments (Fig. 2b). We found OLIG2 binding sites in both the accessible and inaccessible chromatin regions, and determined the associated gene promoters using C3D. OLIG2 binding sites in chromatin regions closed in SOX2+ cells but open in the SOX2– bulk tumour fraction were associated with 'repressing transcription factor binding', consistent with reports of its role as a transcriptional repressor54,55, and suggesting compartmentally restricted regulation by OLIG2. By contrast, OLIG2 binding sites in regions accessible across the tumour compartments were associated with 'genes involved in transcription' and 'notch signalling pathway', indicating the multifunctional roles played by OLIG2 in NSCs to regulate proliferation (Fig. 2c). To confirm this, we found nearly 100% of OLIG2+ cells were actively dividing as indicated by incorporation of EdU (Fig. 2d, e), as determined by immunocytochemistry of MB cells derived from tumours with a heterozygous mutation in Ptch1 and a null mutation in Trp53 (PTCP53 304), modelling an aggressive form of SHH-MB with robust in vitro tractability23,56. We also found Olig2 expression to be significantly enriched in activated as compared to quiescent stem cells based on analyses of publicly available transcriptomic datasets of mouse NSCs57,58,59 (Supplementary Fig. 2c–e).
Genes associated with uniquely accessible regions in cycling stem cells were enriched in signalling pathways such as glutamate receptor and hedgehog signalling in a KEGG pathway analysis, whereas the non-cycling stem cell-associated genes were enriched for negative regulation demonstrating that hedgehog signalling contributes to cellular proliferation in SHH-MB (Supplementary Fig. 2k, l). To probe the effects of inhibiting the SHH pathway, we mined a publicly available dataset20 to assess the impact of Vismodegib (a SHH pathway inhibitor) on key markers (Sox2, Olig2, Mki67 and Dcx) (Supplementary Fig. 2m, n). Interestingly, Olig2 expression was most abrogated after treatment, even compared with proliferative markers (Mki67 and Dcx), suggesting that Olig2 is a key regulator of cellular proliferation mediated by SHH signalling. To identify its targets in the context of medulloblastoma tumours, we inferred the functional regulatory network of Olig2 in primary mouse MB primary tumour samples using C3D. Anchoring our analysis to the Olig2 promoter and using accessible chromatin regions defined in the primary tumour samples, we identified 358 putative gene targets associated with the Olig2 promoter with high stringency (r > 0.9) (Supplementary Fig. 2o). A KEGG pathway analysis of these genes reveals them to be enriched in 'Signaling pathways regulating the pluripotency of stem cells' further implicating OLIG2 as playing a role in the stem cell function (Supplementary Fig. 2p).
Next, to validate the functional role of Olig2 in MB tumour cells, we used CRISPR-Cas9 to knock out Olig2 in cells derived from mouse medulloblastomas (PTCP53 304). We first validated a loss of Olig2 mRNA expression and protein abundance in the knockout (KO) cells (Supplementary Fig. 2f–j). We found Olig2-KO cells exhibited a deficiency in growth and in sphere-forming capacity compared with cells that had been transfected with a scrambled guide RNA (control), indicating an effect on stem cell function (Fig. 2f, g). We next tested the ability of these cells to initiate tumours in a subcutaneous allograft assay performed in immunocompromised mice. The Olig2-KO cells formed no tumours (0/6) compared with control cells (5/8) (chi-square test p-value = 0.0157) (Fig. 2h), suggesting an important role for Olig2 in supporting tumour initiation and progression.
Olig2 plays a key role in stem cell fate transition from quiescence
In order to determine if the tumorigenic defect in Olig2-KO cells is due to Olig2 regulating stem cell activation, we tested the ability of cells to exit from quiescence, following its induction using a modified culture medium containing BMP4 as previously described60,61,62. Dormancy is induced without an increase in cell death or differentiation, or a loss of proliferation or differentiation potential61. Following 28 days of induction of quiescence, the cells were replated in conditioned proliferative expansion media. Olig2-KO cells showed a diminished ability to re-initiate proliferation compared with control cells, implicating a role for Olig2 in exiting the quiescent cell state in vitro (Fig. 3a). Analysis of publicly available TF ChIPseq data in mouse neural stem cells revealed that OLIG2 binding on the chromatin overlapped sites bound by TFs involved in stem cell maintenance, as well as factors involved in proliferation and differentiation (Supplementary Fig. 3b), highlighting the role of Olig2 in regulating dormant and activated states. Further, Olig2-expressing cells are distributed across all phases of the cell cycle (Supplementary Fig. 3h). These data further support the role of Olig2 in the critical fate transition of stem cells. In vivo, we observed that a large majority of SOX2+ cells in the external granule layer of preneoplastic postnatal day (P14) Ptc mice cerebella co-expressed Olig2 (Fig. 3b, c; Supplementary Fig. 3d, e), suggesting that persister stem cells27 require expression of Olig2 to generate a clonal output and form a tumour.
a Phase object confluence of adherently grown Ptch1+/−;Trp53−/− CRISPR-Cas9 control or Olig2-KO mouse tumour cells measured across 9 days using live cell imaging. The cells were grown in either proliferation media, modified quiescence-inducing media containing BMP4 or modified media control for 3 days and then washed and re-exposed to proliferation media; data shown are representative of three independent experiments; error bars denote mean ± SEM; two-tailed unpaired t-test performed at final time point (212 h); two-tailed p-values; ns p ≥ 0.05, ****p < 0.0001; p = 0.4693 (Olig2-KO + veh vs. control + veh), p = 0.4967 (Olig2-KO + MM vs. control + MM), p < 0.0001 (Olig2-KO + MM + wash vs. control + MM + wash). b, c Expression of endogenous SOX2 and OLIG2 proteins detected by immunofluorescence in the external granule layer (EGL) at P14 in Ptc WT and Ptc mice (b) and quantification of the SOX2+/OLIG2+ cells as a fraction of the SOX2+ cells and OLIG2+ cells respectively (c); n = 3 biological replicates; error bars denote mean ± SEM; two-tailed unpaired t-test; *p < 0.05; p = 0.0177. d–k scRNAseq analysis on CGNP-like (SOX2+ or DCX+) Math1-Cre;SmoM2 mouse tumour cells at P7; unsupervised clustering using Uniform Manifold Approximation and Projection (UMAP) performed on ~5000 single cells (d), dot plot of the top genes most differentially expressed in each cluster (e), graphical demonstration of dimension separation strategy of neoplastic CGNP-like cells (f), construction of a neuronal differentiation trajectory by Monocle2 and the expression of Sox2, Olig2, Dcx, Neurod1, and Stmn2 across the trajectory (g), construction of a cell cycle pseudotime by slingshot and the expression of cell cycle markers across the cell cycle pseudotime (Pcna, Mki67, and Top2a) (h), heatmap of the differentially expressed genes across the neurogenesis trajectory and the cell cycle pseudotime (i), comparison of the neurogenesis and cell cycle trajectories; gene names associated with each heat map are listed in Supplementary Data 2 (j), violin plots of cell cycle pseudotime and Mki67 expression between OLIG2+ vs. OLIG2– cells; centre line, median; box bounds, upper and lower quartiles; whiskers, 1.5x interquartile range; points, outliers; n = 1096/3996 (OLIG2+/OLIG2−); two-tailed unpaired t-test (k). Source data are provided as a Source Data file.
To investigate whether this putative neoplastic role for Olig2 is also seen in normal development, we independently re-analysed scRNAseq data of the developing murine cerebellum from Vladoiu et al.16. We identified Olig2-expressing cells in normal cerebellar developmental context (Supplementary Fig. 3c); subsets of OLIG2+ cells overlap with markers of stem cells (Sox2) as well as of progenitors (Dcx) and proliferative cells (Mki67) (Supplementary Fig. 3f). This pattern was recapitulated in the scRNAseq data we generated above from Ptc mouse tumour cells (Supplementary Fig. 3g–i), further emphasising the potential role of OLIG2 in governing a transition between quiescent neural or cancer stem cells and the progenitor cell stage.
To determine if Olig2 had a similar function in an alternative SHH-constitutive MB model (Math1-Cre;SmoM2), that also contains similar fractions of SOX2+, DCX+ and NeuN+ cell populations24, we studied the role of Olig2 in the early stages of tumour development by performing scRNAseq pseudotime on cerebellar granule neuron progenitor (CGNP)-like SOX2+ or DCX+ cells extracted computationally from the transcriptomic profile of developing (P7) tumour cells (Fig. 3d, e; Supplementary Fig. 3j, k). Using Monocle and RNA Velocity software to order cells by their inherent biological processes, we constructed a neuronal differentiation trajectory of the cells (Fig. 3f, g, i; Supplementary Data 2;). Whereas SOX2+ cells are more widely distributed in the neuronal trajectory, the SOX2+/OLIG2+ cells represent the apex of the trajectory (Fig. 3g). A very similar pattern was also observed in an identical analysis of endpoint (P21) tumour cells from the same model63 (Supplementary Fig. 3q–u). To probe the proliferation potential of the OLIG2+ cells within the SOX2+ or DCX+ subset of tumour cells, we constructed a cell cycle pseudotime (Fig. 3f, h, i; Supplementary Data 2; Supplementary Fig. 3l–p). Combining the lineage and cell cycle trajectories reveals a clear deceleration in the cell cycle pseudotime, concomitant with the decrease in expression of Olig2 (Fig. 3j). Stratifying the cells based on Olig2 expression, we found that cell cycle pseudotime, as well as cell cycle marker Mki67 expression, were significantly higher in OLIG2+ cells compared to OLIG2– (Fig. 3k; Supplementary Fig. 3l–p). Together, these data support a role for Olig2 as a pioneer factor for SOX2+ MB stem cell activation in the setting of SHH-driven neoplasia.
A pharmacological inhibitor of OLIG2 blocks lineage output from SOX2+ MB cells
To test whether the role of Olig2 in governing MB stem cell activation could be exploited therapeutically, we obtained a specific pharmacological inhibitor of OLIG2, CT-17964 (Fig. 4a). This small-molecule inhibitor of OLIG2 impedes OLIG2 dimerization and its TF activity on the chromatin, but does not decrease OLIG2 protein abundance (Supplementary Fig. 4a–d). CT-179 was developed by Curtana Pharmaceuticals following an in-silico identification of small molecules inhibiting OLIG2 activity and was validated for OLIG2 selectivity and potent cytotoxicity in cell-based assays performed in glioblastoma65 and medulloblastoma cell lines66. A proliferation assay of CT-179 treatment in the CRISPR control and Olig2-KO cells revealed a significant difference in cell viability, suggesting on-target effect of the treatment (Supplementary Fig. 4e). Consistent with the genetic KO data, CT-179-treated cells showed a diminished ability to re-initiate proliferation compared with control cells, implicating a role for Olig2 in exiting the quiescent cell state in vitro (Supplementary Fig. 3a). Further, CT-179 treatment disrupted the growth of a variety of mouse tumour cells derived from the MB tumours of Ptch1+/−;Trp53−/− as well as Ptch1+/− mice irradiated at postnatal day 0 (P0)56,67 (Supplementary Fig. 4f). CT-179 also had a dose-dependent effect on the proliferation of human diffuse intrinsic pontine glioma (DIPG) cell lines68 and primary human MB samples (Supplementary Fig. 4g–i). OLIG2 protein is present in primary human SHH-MB samples in a range of frequencies (Supplementary Fig. 4k, l), as identified through immunohistochemistry, with the inhibitor effect being linked to OLIG2 abundance rather than specific cell hierarchies (Supplementary Fig. 4j, k). Taken together, these data demonstrate the effectiveness and relevance of targeting OLIG2+ cells in human SHH-MB. The fraction of CT-179-treated cells incorporating EdU (Fig. 4b, c), cycling through the G0/G1 and S phases of the cell cycle (Fig. 4d), and expressing Mki67 (Supplementary Fig. 4m, n) is significantly decreased compared with control. The CT-179-treated cell fraction showed a depletion of actively cycling cells and mitosis defects as seen by an accumulation in G2/M phase (Fig. 4d). Conversely, Olig2 overexpression using a DOX-inducible construct increased the rate of cellular proliferation (Fig. 4e). This effect was abrogated when the Olig2-overexpressing cells were treated with CT-179 (Fig. 4e), illustrating that the OLIG2 inhibitor CT-179 acts on target. Taken together, these data emphasise the importance of Olig2 in regulating SOX2+ stem cell proliferation.
a Structure of the OLIG2 inhibitor, CT-17967. b, c Immunocytochemistry of Ptch1+/−;Trp53−/− mouse tumour cells (b), quantification of EdU incorporation in OLIG2+ cells after a 1-hour pulse (c); data shown are representative of three independent experiments; error bars denote mean ± SEM; two-tailed unpaired t-test; ***p < 0.001; p = 0.0001 (c). d Proportion of Ptch1+/−;Trp53−/− mouse tumour cells in each phase of the cell cycle as measured through FACS analysis of propidium iodide (PI) staining and analysed using FlowJo software; data shown are representative of three independent experiments; error bars denote mean ± SEM; two-tailed unpaired t-test; *p < 0.05; p = 0.0143 (G0/G1), p = 0.0157 (S), p = 0.0210 (G2/M). e Percentage confluence of Ptch1+/−;Trp53−/− mouse tumour cells transfected with a doxycycline (DOX)-inducible OLIG2 overexpression (O/E) construct or empty vector control upon treatment with CT-179 or vehicle for 10 days; data shown are representative of three independent experiments; error bars denote mean ± SEM; two-tailed unpaired t-test; *p < 0.05; p = 0.0404 (O/E vector control vs. OLIG2 O/E), p = 0.0484 (OLIG2 O/E vs. OLIG2 O/E + CT-179). f–h Immunocytochemistry of Ptch1+/−;Trp53−/− mouse tumour cells treated with IC90 of CT-179 (276.1nM) (f), quantification of SOX2+ cells (g), quantification of the fluorescence intensity of DAPI, SOX2 and OLIG2 measured using ImageJ software (h); data shown in are representative of three independent experiments; scale bar: 80µM (f); g, h: error bars denote mean ± SEM; two-tailed unpaired t-test; ns p ≥ 0.05, **p < 0.01, ***p < 0.001; p = 0.0001 (SOX2) (g); p = 0.4551 (DAPI), p = 0.0003 (SOX2), p = 0.0035 (OLIG2) (h). i, j Secondary limiting dilution analysis (LDA) performed on Ptch1+/−;Trp53−/− mouse tumour cells pre-treated with vehicle, IC10 (129.7 nM), IC50 (189.2 nM) or IC90 (276.1nM) dose of CT-179 for 24 hours; data show the percentage of sphere-forming capacity (i) and a quantification of the spheres size (j); data shown are representative of three independent experiments; line at the median; two-tailed unpaired t-test; ns p ≥ 0.05, *p < 0.05, ****p < 0.0001; p = 0.0724 (IC10 vs. vehicle), p = 0.0259 (IC50 vs. vehicle), p = 0.0259 (IC90 vs. vehicle) (i); p = 0.3791 (IC10 vs. vehicle), p < 0.0001 (IC50 vs. vehicle), p < 0.0001 (IC90 vs. vehicle) (j). k, l Cell confluence measured using high-throughput live-cell imaging of Ptch1+/−;Trp53−/− mouse tumour cells pre-treated with vehicle or IC50 (189.2 nM) CT-179 for 24 hours, immediately followed by a dose-response assay to AraC (k) and Vismodegib (GDC) (l). Source data are provided as a Source Data file.
To further interrogate the role Olig2 plays in stem cell state transition from quiescent to activated, we probed the functional potential of the residual cells following OLIG2 inhibition. Hypothesising that OLIG2 inhibition affects only activated stem cells and their output, but not quiescent, the residual cells may still remain potent to generate progeny in the absence of the CT-179 treatment constraint. CT-179 treatment induces cell death (Supplementary Fig. 4o), and increases cell doubling time (Supplementary Fig. 4p) in PTCP53 304 mouse tumour cells in a dose-dependent manner. Plating equal numbers of residual cells following CT-179 treatment, we found an increase in both the frequency of SOX2+ cells (Fig. 4f, g) and in SOX2 protein abundance as measured by fluorescence intensity (Fig. 4f, h). To determine if the post-treatment residual cells were enriched for stem cell activity, we treated mouse tumour cells with a range of doses of CT-179 (vehicle, IC10, IC50 and IC90) for 24 hours, after which they were washed and replated in equal numbers for downstream assays. A proliferation assay and a primary limiting dilution assay did not show any difference in rates of growth or sphere-forming capacity in the treated and untreated cells (Supplementary Fig. 4q, r). Remarkably, we observed a significant dose-dependent increase in both the sphere-forming capacity, (Fig. 4i) as well as the size of the spheres (Fig. 4j; Supplementary Fig. 4s) in a secondary sphere-forming assay. These data indicate an effect on stem cell function (more spheres) and in generation of progeny (larger spheres), supporting the idea that OLIG2 inhibition enriches a pool of cells with a greater potential for self-renewal. To further determine the role of Olig2 in potentiating stem cell activation, we tested the effect of OLIG2 inhibition in conjunction with conventional therapies that target proliferating cells. We measured CT-179-treated cells for sensitivity to the anti-mitotic Cytarabine (AraC) or the SHH pathway inhibitor Vismodegib (GDC-0449, GDC) (Fig. 4k, l). The cells pre-treated with CT-179 for 24 hours were more resistant to both AraC and Vismodegib. Taken together, this data suggests that inhibiting OLIG2 enriches for a more potent and primitive stem cell state. Though somewhat counterintuitive, this is consistent with its role in facilitating state transition from quiescence to activation, suggesting a chronic treatment paradigm may be needed in the clinical context to prevent emergence from quiescence.
To explore a downstream mechanism of CT-179 treatment, we performed RNA sequencing (RNAseq) in PTCP53 304 cells treated with vehicle or CT-179 for 1 and 6 days (Fig. 5a; Supplementary Fig. 5a; Supplementary Data 3). Markers of more primitive cells (Sox2 and Gfap) were significantly upregulated (Fig. 5b; Supplementary Fig. 5d) after 6 days of treatment, further highlighting the impact of OLIG2 inhibition in enriching for a more self-renewing stem cell state. Targets of Olig2 and notably, constituents of the SHH pathway (Ptch1 and Gli1), as well as active progenitor markers (Dcx and Egfr) show a significantly decreased expression upon 6 days of treatment (Fig. 5b). Among the most down-regulated pathways in the treated cells included 'DNA replication', 'regulation of mitotic cell cycle', and 'regulation of chromosome organization' (Supplementary Fig. 5b, c), demonstrating that CT-179 attenuates proliferation by decreasing SHH signalling and downregulating proliferative machinery.
a Volcano plot comparing fold change (x-axis) and false discovery rate (y-axis) obtained from EdgeR analysis of the differentially expressed genes between IC50 (189.2 nM) CT-179 or vehicle treatment of Ptch1+/−;Trp53−/− (PTCP53 304) mouse tumour cells for 6 days. b Relative fold change expression of select genes in vehicle and CT-179 treated cells from dataset shown in (a); n = 3 biological replicates, error bars denote mean ± SEM; two-tailed unpaired t-test; ns p ≥ 0.05, *p < 0.05, **p < 0.01. c, d Representative immunofluorescence (IF) images of endogenous SOX2, OLIG2, and EdU in early (c) or late (d) neoplastic lesions of mice sacrificed after 7 days of continuous EdU label in the presence of either vehicle (dH2O) or CT-179 daily injections. Scale bar: 50 µM for 20X and 25 µM for 40X. e Quantification of the fraction of SOX2 + cells and of EdU incorporation in (c) relative to DAPI+ cells. n = 4 for CT-179, n = 3 for vehicle; error bars denote mean ± SEM; two-tailed unpaired t-test. unpaired t-test; *p < 0.05, ****p < 0.0001; p = 0.0240 (SOX2+), p = 0.0081 (OLIG2+/EdU+). f Quantification of the fraction of SOX2+ or SOX2+ and OLIG2+ cells that have incorporated EdU in (d). n = 3 for CT-179, n = 3 for vehicle; error bars denote mean ± SEM; two-tailed unpaired t-test; **p < 0.01, ***p < 0.001; p = 0.0006 (EdU+), p < 0.0001 (EdU+). g Representative immunofluorescence (IF) images of endogenous SOX2, EdU, and BrdU in early neoplastic lesions in mice sacrificed after 4 days of EdU label followed by an 8-day chase period of which 7 days were treatment with either vehicle (dH2O) or CT-179 by daily IP injections, followed by a single BrdU pulse. Scale bar: 50 µM for 20X and 25µM for 40X. SOX2+/EdU+ cells are indicated with white arrows in the 40x image, while the yellow arrow indicates a triple positive cell (SOX2+/EdU+/BrdU+). h Quantification of the fraction of SOX2+ cells that have incorporated EdU or BrdU as represented in (g). For the quantification, 8 independent lesions were evaluated from 3 mouse brains for each treatment condition (vehicle or CT-179); error bars denote mean ± SEM; two-tailed unpaired t-test; ns p ≥ 0.05, **p < 0.01; p = 0.0034 (EdU+), p = 0.9926 (OLIG2+/BrdU+). Source data are provided as a Source Data file.
OLIG2 inhibition constrains SOX2+ MB cell activation in vivo
To test the role of Olig2 in governing MB stem cell activation in the context of developing tumours, we performed an in vivo chronic thymidine analogue label experiment to determine stem cell dynamics in the presence of OLIG2 inhibition. After determining that 20 mg/kg is an effective and well-tolerated CT-179 dose in Ptc mice (Supplementary Fig. 5e–g), 5-week-old mice were injected with CT-179 or vehicle daily for 8 days. 24 hours after the first dose of CT-179, labelling of the actively dividing cells was carried out by administering drinking water containing ethynyl-2′-deoxyuridine (EdU) for 7 days, following which the mice were sacrificed (Supplementary Fig. 5h). Various independent tumour lesions were observed in the mice (Supplementary Fig. 5i), ranging from focal thickening at the pial surface (early neoplastic lesion) to more widespread neoplastic regions (late neoplastic lesions or tumours) (Supplementary Fig. 5j). In early neoplastic lesions of vehicle-treated mice, we observed a frequency of ~5% SOX2+ tumour-initiating stem cells, and >90% EdU+ cells, as described previously24,27, indicative of a high degree of proliferation and cellular turnover (Fig. 5c, e). In comparison, the proportion of SOX2+ cells in early lesions of CT-179-treated mice was significantly enriched at 8-fold the vehicle-treated frequency (Fig. 5c, e). In contrast, the overall fraction of cells incorporating EdU, as well as SOX2+/EdU+ cells were significantly diminished in the early lesions of CT-179 treated mice, highlighting the crucial role of OLIG2 in the activation of SOX2+ stem cells (Fig. 5c, e; Supplementary Fig. 5l). In the late neoplastic lesions of control (vehicle-treated) mice, we observed that ~40% SOX2+ cells had taken up EdU, consistent with previous findings24 (Fig. 5d, f; Supplementary Fig. 5k). Remarkably, the proportion of EdU+/SOX2+ cells in CT-179-treated mice were drastically diminished, indicating a failure of SOX2+ cells to enter into the cell cycle under the constraint of OLIG2 inhibition (Fig. 5d, f). In addition, there was a decrease of both double-(EdU+/OLIG2+) and triple-(EdU+/OLIG2+/SOX2+) positive cells in CT-179-treated mice, indicating the action on cells undergoing the fate transition within the malignant hierarchy (Fig. 5f; Supplementary Fig. 5m).
As a corollary, we hypothesised that OLIG2 inhibition following an EdU label would impair the ability of SOX2+ stem cells to become activated and re-enter the cell cycle. To test this, we performed an independent labelling of the actively dividing cells by administering drinking water containing EdU for 4 days, followed by CT-179 or vehicle treatment for 7 days (Supplementary Fig. 5n). To trace stem cell proliferation dynamics subsequent to OLIG2 inhibition, both groups of mice were administered a single pulse of the synthesis nucleoside analogue 5-bromo-2'-deoxyuridine (BrdU) after the end of vehicle/CT-179 treatment, and sacrificed the following day (Supplementary Fig. 5n). We observed a significant enhancement of EdU label retention in SOX2+ cells in the early neoplastic lesions of CT-179-treated animals compared with vehicle-treated, indicating the diminished ability of stem cells to proliferate under OLIG2 inhibition (Fig. 5g, h). A similar trend has been observed in a Gfap-Cre; SmoM2 model of SHH-MB66. Contrastingly, the SOX2+ pool in the vehicle-treated mice undergoes asymmetric cell division at the expected rate (see Supplementary Information), which corresponds to the diminishment of SOX2+/EdU+ cells, since the DCX+ progeny loses their entire EdU label by the end of a 7-day chase24. Thus, the percentage of SOX2+/EdU+ double-positive cells in the vehicle and treatment groups follow the mathematical modelling of stem cell proliferation kinetics in a striking fashion (Fig. 1a).
Notably, the rate at which SOX2+ cells in both groups incorporate the BrdU pulse post-treatment is equivalent, providing evidence that stem cells are only constrained during OLIG2 inhibition, with no change to their rate of re-entry into the cell cycle after inhibition has been lifted (Fig. 5g, h). Finally, only the CT-179-treated animals have triple-positive (SOX2+/EdU+/BrdU+) cells in their early neoplastic lesions, indicating the presence of cells that had been quiescent during the treatment and only began cycling again following the end of the treatment (Supplementary Fig. 5o), adding further evidence that OLIG2 acts on the SOX2+ compartment to promote its activation in the malignant hierarchy. Importantly, we demonstrate that the SOX2+ stem cells are unable to exit quiescence and enter the proliferative state during OLIG2 inhibition. Taken together, our data show that OLIG2 inhibition constrains the ability of SOX2+ tumour-initiating stem cells to get activated and generate downstream proliferative tumour output at the crucial stages of tumour development.
Inhibiting OLIG2 prevents the formation of tumour growth and relapse in vivo
After determining that OLIG2 regulates SOX2+ stem cell activation in vivo, we next investigated whether CT-179 can alter MB growth. We first examined if CT-179 could inhibit early tumour formation, by testing its effect on Ptc mice from P3–P21 followed by sacrifice at P22 and assessment of pre-neoplastic lesion (PNL) formation. Vehicle-treated mice developed characteristic PNLs whereas CT-179 treated mice did not have any identifiable lesions (Fisher’s exact test p-value = 0.0286) (Fig. 6a; Supplementary Data 4). To test the longer-term effects of OLIG2 inhibition on early tumour progression, we assessed the survival of mice after treating them with vehicle or CT-179 during a critical and finite window (P3–P28) of tumour development. Notably, although treatment was limited, median survival was significantly extended from 160 to 216 days (Fig. 6b). To visualise the impact of CT-179 more definitively on early tumour development, we used magnetic resonance imaging (MRI) technology to track tumour growth in a second cohort of mice, treating tumours for a 4-week window from P65–P93. Images taken after treatment had ended reveal that vehicle-treated mice had developed significantly larger tumours with CT-179-treated mice (Fig. 6c; Supplementary Fig. 6a), consistent with previous results. Together, these multimodal data support the conclusion that without Olig2 function, SOX2+ stem cells are attenuated in their ability to generate the cellular output required for early tumour initiation and subsequent growth.
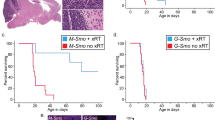
a Treatment strategy and H&E staining of Ptc mouse hindbrains treated with 20 mg/kg CT-179 or vehicle daily by IP injection from P3–P21 and sacrificed at P22. Lesion is indicated by the black arrow. n = 4 for CT-179, n = 4 for vehicle. b Treatment strategy and Kaplan–Meier survival curve of Ptc mice treated with 20 mg/kg CT-179 (Vehicle: n = 12; CT-179: n = 8). The dotted lines represent the duration of treatment of vehicle or CT-179 by daily IP injection (P3–P28). Significance was estimated using the log-rank (Mantel–Cox) test. Chi square = 5.425, p = 0.0198. c Sagittal MRI images of the brains of Ptc mice taken at age 3.5 months. The mice were treated with MCT (vehicle for GDC) from P56-P64 followed by CT-179 or vehicle from P65–P93. Tumours are indicated by the yellow arrows or yellow dotted lines. d Phase object confluence of adherently grown Ptch1+/−;Trp53−/− mouse tumour cells measured across 14 days using live cell imaging. Cells were treated with vehicle control, IC30 CT-179 (163.5 nM), 50 µM Vismodegib (GDC-0449), or a combination of CT-179 and Vismodegib; data shown are representative of three independent experiments; error bars denote mean ± SEM; two-tailed unpaired t-test; ns p ≥ 0.05, *p < 0.05, ***p < 0.001; p = 0.7291 (water vs. CT-179 at 172 h), p = 0.0365 (water vs. GDC at 172 h), p = 0.0277 (water vs. GDC + CT-179 at 172 h), p = 0.8717 (water vs. CT-179 at 184 h), p = 0.0614 (water vs. GDC at 184 h), p = 0.0101 (water vs. GDC + CT-179 at 184 h), p = 0.6868 (water vs. CT-179 at 220 h), p = 0.8109 (water vs. GDC at 220 h), p = 0.0002 (water vs. GDC + CT-179 at 220 h). e Treatment strategy (top) and H&E staining of Ptc mouse hindbrains treated with Vismodegib from P28–P35 followed by CT-179 or vehicle from P35–P61. Lesion is indicated by the black arrows. n = 4 for CT-179, n = 6 for vehicle. f Treatment strategy (below) and Kaplan–Meier survival curve of Ptc mice treated GDC followed by vehicle or 20 mg/kg CT-179. n = 16 for CT-179, n = 16 for vehicle. The dotted lines represent the durations of GDC debulking (daily gavage) (P56–P63) followed immediately by treatment with vehicle or CT-179 by daily IP injection (P64–P92). Significance was estimated using the log-rank (Mantel–Cox) test. Chi square = 5.452, p-value = 0.0195. Source data are provided as a Source Data file.
We next tested CT-179 in the more clinically relevant context of tumour relapse, hypothesising that Olig2 is required for tumour regrowth following conventional debulking therapy using the SHH pathway inhibitor Vismodegib. In an in vitro screen on PTCP53 304 cells to identify favourable drug combinations with CT-179, Vismodegib ranked among the top three hits, with the top interacting targets with OLIG2 inhibition overlaid being epigenetic and metabolic inhibitors (Supplementary Fig. 6b, c; Supplementary Data 5). Cellular proliferation was significantly diminished with a combined treatment compared to control and individual treatments (Fig. 6d). Further, a checkerboard assay at various dose combinations also showed favourable dose reduction indices between the two inhibitors, suggesting therapeutic potential (Supplementary Fig. 6d). In vivo, 5 days of Vismodegib treatment effectively eliminates small lesions in 4-week-old mice compared with vehicle (MCT) treatment (Supplementary Fig. 6e). After this debulking paradigm, mice were treated with CT-179 or vehicle for an additional 4 weeks. We observed that mice treated with CT-179 after debulking did not have recurrent lesions, whereas mice that had undergone debulking followed by vehicle did (Fisher’s exact test p-value = 0.0476) (Fig. 6e; Supplementary Data 4), demonstrating the critical function of Olig2 in tumour regrowth. To test the longer-term effects of OLIG2 inhibition on tumour relapse, we assessed the survival of 8-week-old mice after 8 days of debulking (Supplementary Fig. 6f) followed by treating them with vehicle or CT-179. Notably, median survival was significantly extended (p-value = 0.0195) with a visible difference in lesion formation tracked by MR imaging (Fig. 6f; Supplementary Fig. 6g, h). Therefore, in both early tumour initiation and tumour relapse contexts, we eliminated tumour output by inhibiting a key factor in the transition of quiescent stem cells to progenitors, offering proof of principle for the therapeutic targeting cell fate transitions within the MB stem cell hierarchy.
Discussion
The intricate balance between self-renewal and generation of progenitors by stem cells during normal development is disrupted in cancer, with genetic changes altering the functions of cells within the now corrupt neoplastic developmental lineage. Olig2 has been previously shown to be associated with both stem cell quiescence as well as with cycling progenitor populations53,69,70,71,72. Harnessing the power of transgenic mouse models to track and target tumour progression from the earliest stages of initiation, we demonstrate that in these MB tumours, Olig2’s particular role lies between these populations, enabling the transition of stem cells from a non-cycling inactive state to a proliferative one, which then subsequently leads to expansion of the neoplastic clone through progenitor proliferation and differentiation to comprise tumour bulk. Indeed, our EdU-label experiments revealed that inhibiting OLIG2 severely impairs the ability of SOX2+ cells to proliferate beginning from the earliest stages of tumour development. Olig2 is not widely expressed in the DCX+ population70 suggesting OLIG2 is not required for progenitor proliferation. Inhibiting OLIG2 blocked early tumour formation as well as tumour regrowth in a post-treatment recurrence model, demonstrating it is acting earlier within the MB hierarchy than the conventionally targeted progenitor stage. Though analyses to link these findings to human tumours would be imperative to ascertain clinical applicability, we propose a new clinical paradigm of targeting tumour growth at key earlier stages for more effective eradication.
The stem cell population marked by Sox2 expression in SHH-subgroup medulloblastoma has a distinct epigenomic and transcriptomic landscape compared to the tumour bulk. There are dynamic changes in chromatin accessibility at the level of distal regulatory elements, including at key transcription factors determining hierarchy and cell fate such as Sox2, between these functionally distinct subpopulations. This finding is suggestive of a larger mechanism by which enhancers show dynamism as cells progress down lineage hierarchies, fine-tuning and controlling target gene expression depending on cell context within a lineage.
SHH-MB arises due to dysregulated cerebellar development27,73. Although OLIG2 is found to be correlated with Sox2 expression in MB tumour-initiating cells24,70,74,75, its precise functional role in this neoplastic lineage was previously not clear. This study sheds light on cell fate events within the malignant hierarchy that lead to tumour initiation and relapse. Through pulse and label-chase experiments tracking stem cell proliferative dynamics prior to, during and after treatment with OLIG2 inhibition, we identified Olig2 as playing a crucial role in activating stem cells in the MB neural lineage to generate a tumour bulk. We propose a mechanism by which Olig2 functions as the molecular switch that triggers the cell fate transition from quiescent stem cell to an activated state that then gives rise to rapidly cycling progenitors that will form the tumour bulk. Inhibition of OLIG2 activity by treatment with CT-179 limits the ability of the quiescent stem cells to shift to a proliferative state, thereby preventing this lineage output (Fig. 7).
Inhibiting OLIG2 does lead to an enrichment of the stem cell population, resulting in heightened self-renewal capacity and resistance to anti-proliferative therapies including an antimitotic agent (AraC) and a smoothened inhibitor (Vismodegib), along with an increase in cell death. Remarkably, however, initial treatment of established tumour lesions with Vismodegib, mirroring a clinical paradigm, followed by inhibition of OLIG2 drastically reduces tumour relapse. These functional experiments support the notion that targeting OLIG2 may be a key to preventing eventual tumour relapse by constraining the more resistant, quiescent pool of stem cells, left behind after current conventional therapies.
Focusing on cell transitions in the stem cell hierarchy, we have defined a rare but critical transcription factor-orchestrated event of stem cell fate transition from quiescence to activation. This transition leads to the generation of rapidly proliferating tumour output in MB. Blocking this event constrains the stem cell pool in a non-proliferative state, and is sufficient to prevent the onset of tumour initiation and post-treatment relapse. In a clinical context, additional therapies for patients with SHH-MB are needed, particularly for high-risk patients with TP53 mutation. Although quiescence itself remains a cancer stem cell property that is very difficult to target, limiting the ability of the quiescent cells to undergo a proliferative transition can substantially affect tumour growth by acting at a specific point in the hierarchy. We propose that a targeted approach to block cancer stem cell activation and subsequent generation of proliferative progeny may find a role in combination therapy for MB. Unlocking TF-dependent regulated cell fate transitions suggest new treatment approaches for limiting the output of stem cells at the apex of a neoplastic hierarchy.
Methods
Experimental model and subject details
Ethics statement
All experiments carried out on mice were approved by the Hospital for Sick Children’s Animal Care Committee (Animal Utilisation Protocol# 1000046813) and University Health Network (UHN) Animal Resources Centre (Animal Utilisation Protocol# 6454) in accordance with the Canadian Council on Animal Care Guidelines. Protocols were approved by the Research Ethics Board at the Hospital for Sick Children and UHN, Toronto, respectively.
Human tumour samples were obtained from patients during their operative procedure following informed consent. All experimental procedures were performed in accordance with the Research Ethics Board at The Hospital for Sick Children (Toronto, Canada). All primary MB tissues were obtained from The Hospital for Sick Children (Toronto, Canada). Research was performed on de-identified samples in accordance with the Hospital for Sick Children Research Ethics Board protocols (REB 0020010404 and REB 0020020238). All human DIPG lines68 were provided by Dr. Michelle Monje, Stanford University (Stanford, California, United States).
Mice
Mice had free access to rodent chow and water in a 14-hour light, 10-hour dark cycle room with ambient temperature at 22–24 °C and 45–50% humidity (Laboratory Animal Services (LAS) Facility, SickKids); and in a 12-hour light, 12-hour dark cycle room with ambient temperature at 21–22 °C and 45–60% humidity (Spatio-Temporal Targeting and Amplification of Radiation Response (STTARR) Facility, UHN). All mice were healthy with no apparent behavioural phenotypes. For the subcutaneous engraftment assay, female NSG mice were used. For the transgenic mouse studies, mice of either sex were used. Ages of all mice used in experiments are indicated in the figure legends. The transgenic mouse strain Ptc1LacZ (Ptch1tm1Mps/J)30 was provided by Dr. C.C. Hui, Toronto, Hospital for Sick Children and authenticated as described30. Sox2eGFP (Sox2tm1Lpev) mice32 were provided by Dr. Freda Miller, Toronto, Hospital for Sick Children and were genotyped as described32. Experimental Ptc mice were administered 3 Gy γ-radiation from a caesium 137 source on the day of birth (P0). For OLIG2 inhibition treatment, Ptc mice were administered 20 mg/kg of CT-179 (Curtana Pharmaceuticals) dissolved in dH2O daily by intraperitoneal (IP) injection. For the 5-ethynyl-2'-deoxyuridine (EdU)-labelling, 0.82mg/mL EdU (Invitrogen, Cat# E10187) was dissolved into the mouse drinking water and continuously administered for 7 days by water bottle. Fresh EdU water top up solution was prepared every other day. For the pulse, BrdU (Invitrogen, Cat# 000103) was reconstituted in DMSO at a concentration of 200 mg/mL, then diluted in PBS to a working concentration of 50 mg/mL, and administered to the mice by a single intraperitoneal injection at 100 mg/kg. For the debulking experiment, 50mg/kg Vismodegib (GDC-0449) (Selleck Chemicals Cat# S1082) was administered once daily for 5 days in 0.5% methylcellulose 0.2% Tween 80 buffer (MCT vehicle) by gastric gavage. For each experiment, a power analysis was used to conduct the sample size and the mice were randomised into treatment and control groups ensuring a fairly equal distribution of males and females.
Cell culture
Mouse MB tumour cells (PTCP53 304 and PTCP53 302 derived from the tumours of Ptch1+/−;Trp53−/− mice, and IPM130 and IPM131 derived from the tumours of Ptch1+/− mice irradiated at P0 were grown adherently in serum-free medium as described previously23,50. All cell lines were regularly tested for mycoplasma contamination. Briefly, cells were grown on PRIMARIATM culture plates (Corning) coated with poly-L-ornithine (Sigma) and laminin (Sigma) and maintained in Neurocult NS-A basal medium (Mouse & Rat) (StemCell Technologies) containing 2 mM L-glutamine (Wisent), 75 µg/mL bovine serum albumin (Life Technologies), in-house hormone mix equivalent to N2 (home-made), B27 supplement (Life Technologies), 10 ng/mL recombinant human epidermal growth factor (rhEGF; Sigma), 10 ng/mL basic fibroblast growth factor (bFGF; StemCell Technologies), and 2 µg/mL heparin (Sigma).
For the quiescence-induction assay, media was modified as described53,59, briefly, as follows: 100–150 cells per well were plated into normal proliferation medium (EGF and FGF2, both at 10 ng/mL) in a 96-well plate (Corning) and, after 16 h, fresh medium was added without EGF and with 10 ng/mL BMP4 (R&D Systems) and 20 ng/mL FGF2. For reactivation, after 5–7 days in BMP4-containing medium, cells were passaged with Accutase (Sigma) and plated into proliferation medium.
For the human sample dose-response assays, primary human MB samples were cultured in serum-free medium as described previously23,50. Briefly, cells were grown on PRIMARIATM culture plates (Corning) coated with poly-L-ornithine (Sigma) and laminin (Sigma) and maintained in Neurocult NS-A basal medium (Human) (StemCell Technologies) containing 2 mM L-glutamine (Wisent), 75 µg/mL bovine serum albumin (Life Technologies), in-house hormone mix equivalent to N2 (home-made), B27 supplement (Life Technologies), 10 ng/mL recombinant human epidermal growth factor (rhEGF; Sigma), 10 ng/mL basic fibroblast growth factor (bFGF; StemCell Technologies), and 2 µg/mL heparin (Sigma). DIPG cell lines were cultured as described previously68. Briefly, cells were grown in vented T75 tissue culture flasks (Thermo Fisher Scientific) and maintained in Neurobasal-A Medium (Invitrogen) and D-MEM/F-12 1:1 (Invitrogen), HEPES Buffer Solution (Invitrogen), MEM Sodium Pyruvate Solution (Invitrogen), MEM Non-Essential Amino Acids Solution 10mM (Invitrogen), GlutaMAX-I Supplement (Invitrogen), 1x Antibiotic-Antimycotic (Wisent Bioproducts), and supplemented with B-27 Supplement Minus Vitamin A (Invitrogen), 20 ng/mL rhEGF (Sigma), 20 ng/mL bFGF (StemCell Technologies), 10 ng/mL h-PDGF-AA (Shenandoah Biotechnology), 10 ng/mL h-PDGF-BB (Shenandoah Biotechnology), and 2 µg/mL heparin (Sigma).
Experimental details
CRISPR knockout
An Olig2 CRISPR/Cas9 KO plasmid with guide RNAs targeting Olig2 (Santa Cruz, sc-424530) and a control CRISPR/Cas9 plasmid (Santa Cruz, sc-418922) was purchased from Santa Cruz Biotechnologies. Ptch1+/−;Trp53−/− MB cells (PTCP53 304) were cultured as described (“Cell lines”). PTCP53 304 cells (1–2 million) were nucleofected using the Lonza Nucleofector Kit for Mouse Neural Stem Cells (as per the manufacturer’s guidelines). Cells were nucleofected with 5 μg of either Olig2 KO or control CRISPR plasmid. Media was changed after 24 hours. GFP+ cells were sorted using a MoFlo XDP Cell Sorter after 48–72 hours. For verification of the KO, RNA was extracted using the Qiagen RNeasy kit as per the manufacturer’s protocol. Reverse transcription was performed using the Roche Transcriptor two-step Reverse Transcriptase kit. Quantitative RT-PCR (qRT-PCR) analysis was performed using Life Technologies SsoFast EvaGreen kit (SYBR green) and samples were run on the Bio-Rad CFX Connect Real-Time PCR detection system. Primers used for qRT-PCR are: mOlig2_F_1 CCCTCCTGTTGTCTCTCCTG; mOlig2_R_1 ATGTGTGTTGCGTGAGTGTG; mOlig2_F_2 GTCATCTTCCTCCAGCACCT; mOlig2_R_2 GATGTTGAGGTCGTGCATGC; mGapdh_1_F AACTTTGGC ATTGTGGAAGG; mGapdh_1_R ACACATTGGGGGTAGGAACA.
Dose-response assay
Mouse MB cell lines (PTCP53 302, PTCP53 304, IPM130, and IPM131) or primary human MB samples were plated in 96-well PrimariaTM plates, with three technical replicates per dose, and cultured across nine concentrations (ranging from 5 nM to 5 µM) of OLIG2 inhibitor (CT-179) for 6 days. The response to OLIG2 inhibition was estimated from the confluency data normalised to the dH2O control, measured using the IncuCyte ZOOM™ Live Cell Analysis System. The response of the cell lines to OLIG2 inhibition was estimated from cell viability normalised to the dH2O control, assessed by Alamar Blue assay (Thermo Fisher Scientific) using a Gemini EM Fluorescence Microplate Reader (Molecular Devices), after 6 hours of incubation with the active ingredient. Cell viability confluence values were normalised to the control endpoint. GraphPad Prism software was used to generate dose–response curves (GraphPad Software, San Diego, CA, USA). Inhibiting concentration (IC) values IC10, IC50 and IC90 were generated by GraphPad Prism using the sigmoidal dose–response (variable slope) equation.
Cell proliferation assay
PTCP53 304 cells were plated in triplicate in 96-well PRIMARIATM plates (Corning). Cells were treated with the IC50 dose of OLIG2 inhibitor (189.2nM CT-179) or vehicle (dH2O) for 6 days. Cell growth was monitored using the IncuCyte ZOOM™ Live Cell Analysis System. Confluency was calculated from cell images taken every 8 hours using the IncuCyte ZOOM software.
Limiting dilution assay
PTCP53 304 mouse tumour cells pre-treated with vehicle or CT-179 were plated in serial dilutions on non-adherent 96-well plates with six technical replicates per dilution in the media described above for adherent cells (see 'Cell culture'). LDAs were performed by plating cells in suspension in 1:2 serial dilution as follows: 2000, 1000, 500, 250, 125, 63, 31, 16, 8, 4 cells per well. Each cell dilution was plated as six technical replicates, all on the same plate. Each well was replenished with 50 μL of fresh media after 1 week. 7 and 14 days after plating, wells from each plate were scored blindly for the presence of spheres. For the secondary limiting dilution assay, spheres collected from the primary assay after 14 days. They were mechanically and enzymatically (0.05% Accutase) dissociated followed by filtering and visualisation with Trypan Blue to ensure a single cell suspension. They were then counted and replated for the subsequent limiting dilution assay experiment. The readout at the assay endpoint was the presence or absence of spheres in each well. Sphere size was imaged and measured using a DS-Fi3 camera (Nikon) and NIS Elements Imaging software. The fraction of technical replicates positive for the presence of spheres at each dilution was entered into the extreme limiting dilution analysis (ELDA) software (http://bioinf.wehi.edu.au/software/elda/) to determine the estimated sphere-forming frequency along with upper and lower limits (denoted by the error bars) with a 95% confidence interval. Data were tested for inequality in frequency between multiple groups and for adequacy of the single-hit model using the ELDA software76.
Probe screen
We used 75 well-characterised chemical probes from the Structural Genomics Consortium at the University of Toronto (SGC – Toronto)77. Each compound selectively and potently inhibits a specific epigenetic or growth signalling pathway protein and has significant cellular activity at ≤10 µM. Five hundred MB tumour cells (PTCP53 304, and IPM131) were plated adherently in 384-well CELLBINDTM plates (Corning) and imaged using the IncuCyte® ZOOM Live-Cell Analysis System (Essen Biosciences). Cells were imaged with a ×10 objective using phase-contrast every 8 hours until the experimental endpoint. Culture media was refreshed every 5 days with ×1 chemical probe concentration. Cell confluency or fluorescence was analysed using the IncuCyte Live Cell Analysis System (Essen Biosciences), in which an algorithm was used to approximate the cell confluency of each well. The processing definition was tailored for each cell type, allowing for reproducible and robust monitoring of cell confluency over time. The data represented in the heatmap is log2 of the average confluency of triplicates in the probe wells normalised against the average confluency in the DMSO control wells.
Fluorescence-activated cell sorting (FACS) analysis and cell cycle assay
To PTCP53 304 mouse tumour cells pre-treated with vehicle or CT-179, 50 gm/mL propidium iodide (PI) solution was added. Cell sorting was performed on either a Beckman Coulter MoFlo or Beckman Coulter MoFlo-XDP cell sorter. Data were analysed using FloJo software.
Immunocytochemistry
PTCP53 304 cells were plated onto poly-L-ornithine/laminin-coated coverslips in culture media as described ('Cell culture') with an additional 1:1000 spike of laminin and then fixed with 4% paraformaldehyde at confluence. Fixed cells were washed 3x in PBS and 0.1 % Triton-X (PBS- T). Cells were then incubated in a blocking buffer containing PBS-T and 10% horse serum for 1 hour at room temperature. Cells were incubated with the primary antibody in a humidified chamber at 4 °C overnight. The primary antibody was washed off and replaced with secondary antibody and the nuclear marker DAPI (1:1000) in blocking solution and incubated for one hour at room temperature. Cells were again washed in PBS before mounting and imaging. Primary antibodies used were: goat anti-Sox2 at 1:100 (R&D Systems Cat# AF2018); mouse anti-Ki67 at 1:100 (BD Pharmingen Cat# 550609); rabbit anti-Olig2 at 1:500 (EMD Millipore Cat# AB9610). Secondary antibodies were used at 1:500 (Alexa Fluor 488 Goat anti-Rb, Invitrogen Cat# A-11034; Alexa Fluor 568 Goat anti-Ms, Invitrogen Cat# A-11004).
Protein extraction and western blot
Lysis buffer containing 10 mM Tris-HCl, 0.5% Triton X-100, 150 mM NaCl, 1 mM EDTA, 10 mM MgCl2, 1% sodium dodecyl sulphate, protease inhibitors, and benzonase was used to harvest total protein for western blot. In all, 30 µg of protein was loaded per well. PTCP53 304 cell lysates were run on 10% Bis-Tris SDS-PAGE protein gels and transferred onto polyvinylidene difluoride membranes. Primary antibodies used were anti-Olig2 1:2500 (EMD Millipore Cat# AB9610), and anti-B-actin 1:10,000 (Sigma, Cat #A5441). Secondary antibodies used were anti-mouse-HRP 1:10,000 (Millipore Cat# 2774) and IRDye® 800CW Donkey anti-Mouse 1:15,000 (Licor Cat# P/N 925-32212). Membranes were visualized on an Odyssey® CLx Imaging System (LI-COR).
Single-cell RNA sequencing and analysis
Human medulloblastoma tumour sample analysis
The SHH MB tumour data set downloaded from Hovestadt et al.17 (N = 5) was merged and processed using Seurat. Cells with less than 200 expressed genes, greater than 8000 genes, greater than 18,000 transcripts were removed resulting in 1400 cells for further analysis. The gene expression values were further log normalized and corrected for biases from cell cycle states, transcript distribution and mitochondrial gene distribution. Top variable genes were identified using variance stabilising transformation method incorporated in Seurat followed by principal component analysis and graph-based clustering. The clusters were then visualised using Uniform Manifold Approximation and Projection for Dimension Reduction (UMAP)78. Top marker genes of each cluster were identified using FindAllMarkers function from Seurat.
Developmental mouse cerebellum sample analysis
Filtered gene matrices from Vladoiu et al.16 for scRNAseq produced at 9 different developmental stages (E10,12,14,16,18 and P0,5,7,14) were loaded and filtered independently using Seurat79 to a minimum of 3 cells expressing each gene and 200 genes expressed in each cell. Normalisation was carried out using SCTransform, regressing out the percentage of mitochondrial reads in each sample. SingleR was used to filter out immune cells. Finally, Seurat was used to cluster cells and carry out differential expression analysis between fractions of cells expressing or not expressing Olig2 within the SOX2+ fraction.
Ptc MB sample single-cell RNA sequencing and analysis
Sample preparation and sequencing
Viably-frozen single-cell suspensions (−80 °C) from primary Ptc end-point MB were thawed in DMEM supplemented with DNAse I. Cells were pelleted at 200 RCF and resuspended in 0.1% BSA/PBS, supplemented with DNAse I, and filtered through a 35 µM nylon filter. Cells were stained with propidium iodide or DAPI before live-cell sorting using either: MoFlo Astrios, MoFloXDP, or AriaII-SC. Live cells were collected into 0.1% BSA/PBS before proceeding with single-cell mRNA-seq using the 10x Genomics platform, as per the manufacturer’s guidelines. Target cell number ranged from 2000 to 6000 cells with post- sort sample viability ranging from 71 to 92%, as assessed by Trypan Blue. Samples were sequenced with an Illumina HiSeq 2500 machine as paired-end reads (read 1: 26 bases; read 2: 98 bases; index 1: 8 bases) at The Princess Margaret Genomics Centre (PMGC).
Analysis
The four libraries were aggregated using CellRanger aggr80 from 10X Genomics using default parameters. Quality check, filtering and downstream processing of the aggregated sample was done by adapting the pipeline from Seurat79,81 which is briefly outlined as follows. In order to avoid low-quality cells and doublets from further analysis we excluded the cells with less than 200 expressed genes, greater than 7500 genes, greater than 60,000 transcripts or mitochondrial gene expression greater than 10% of the total gene expression of the cell. After filtering we obtained 6739 cells. The gene expression values were further log normalised and corrected for biases from cell cycle states, transcript distribution and mitochondrial gene distribution. Top 4000 variable genes were identified using the variance stabilising transformation method incorporated in Seurat followed by principal component analysis and graph-based clustering. The clusters were then visualised using Uniform Manifold Approximation and Projection for Dimension Reduction (UMAP)78. Top marker genes of each cluster were identified using FindAllMarkers function in Seurat and the genes were then used to annotate the clusters through literature search. In order to identify differentially expressed genes between SOX2+ and SOX2– cells, differential expression analysis was performed using the Wilcoxon rank sum test implemented in FindMarkers function of Seurat.
Math1-Cre;SmoM2 MB sample single-cell RNA sequencing and analysis
Sample preparation and sequencing
Math1-Cre; SmoM2 MB from P7 mice was harvested followed by mechanical and enzymatic dissociation using the Papain Dissociation System (Worthington Biochemical Corporation). Single cell suspension was assessed by Trypan blue. Approximately 5000 cells at a concentration of ~1000 cells/μl were loaded onto the Chromium Controller. Gel Bead-In EMulsions (GEMs) and library generation were performed following standard 10X Genomics protocols using the Chromium Single Cell 3ʹ Reagents Kit. 10X libraries were sequenced using the Illumina NovaSeq 6000 sequencing platform to achieve ~40,000 reads per cell. Single-cell RNA sequencing was performed by the Princess Margaret Genomics Centre.
Pre-processing of scRNAseq dataset
We used cell ranger mkfastq to demultiplex Illumina sequencer’s base call files (BCls) into FASTQ files. The Cell Ranger count was adopted for aligning FASTQ files to GRCm38 (mm10) mouse genome reference to generate barcode-labelled sparse matrices. To identify expression dynamics of mRNAs, we passed the outputs of Cell Ranger to Velocyto run10x pipeline. The aligned .loom files, containing qualified 'unspliced', 'spliced' and 'ambiguous' molecules, were imported by SeurateWrappers package.
Clustering and cell type identification
The pre-processed barcode-labelled sparse matrices were passed to Seurat v4.0 for data filtering. Cells with high mitochondria gene percentage (percent.mt > 5–10%) and low gene saturation (nFeatures < 200) were filtered. Data normalisation was performed by NormalizeData() with the 'LogNormalize' method. Top 2000 highly variable feature genes were selected by FindVariableFeatures() for principal component analysis. Significant principal components (PCs) dimensionalities were identified by the ElbowPlot method. The significant PCs were used for Uniform Manifold Approximation and Projection (UMAP)78 to reduce to 2 dimensions by RunMAP() function. Cell clustering was generated by FindNeighbors() and FindCluster() functions. Marker genes of each cluster were determined by FindAllMarkers() with 'roc' test, min.pct at 0.25 and logfc.threshold at 0.5. We categorised MB cells into six subgroups according to the gene expression pattern of each cluster. Non-tumoral cell types were identified by referring to the CellMarker database.
Pseudotime reconstruction of SOX2+ or DCX+ MB cells
SOX2+ or DCX+ MB cells were identified using the WhichCells() function from the Seurat R package. To reconstruct neuronal differentiation trajectory of SOX2+ or DCX+ MB cells, we first reduced the cells into 2 dimensions on UMAP by selecting the feature genes in GO term neurogenesis gene list (http://www.informatics.jax.org/go/term/GO:0022008). Then, the trajectory and pseudotime of neuronal differentiation were inferred by the infer_trajectories() function from the dyno package. Component 1 method was selected in infer_trajectories() for neuronal differentiation pseudotime estimation. For cell cycling pseudotime reconstruction, we first identified cell cycle phases of SOX2+ or DCX+ MB cells by the CellCycleScoring() function. Then, we embedded these cells into two-dimensional distribution according to S and G2M scoring. Thirdly, we used the dyno pipeline to generate a cell cycling trajectory. To reconstruct a linear-like trajectory, tscan was selected from the dyno pipeline. Expression matrix was transformed by the wrap_expression() function from the Seurat object. The cell cycling trajectory was inferred by infer_trajectorires().The neuronal differentiation and cell cycling trajectories were then confirmed by RNA velocity analysis.
RNA velocity of SOX2+ or DCX+ MB cells
We analysed RNA velocity of SOX2+ or DCX+ MB cells by velocyto.R and SeuratWrappers packages. Seurat objects with RNA velocity quantifications were generated by the WhichCells() function. We renormalized the matrices and selected the top 2000 highly variable feature genes for PCA. SOX2+ or DCX+ MB cells were embedded into the neuronal differentiation and cell cycling embeddings. mRNA velocity was estimated using a gene-relative model via RunVelocity(), which passed parameters to gene.relative.velocity.estimates(). We combined cell k nearest neighbours (kNN) pooling with gamma fit with min/max quantile fit at 0.02. The number of kNN in slope calculation smoothing was set at 7. Minimum average expression count for spliced and unspliced expression matrices was set as default at 0.2 and 0.05. We visualised mRNA velocity in embeddings by the show.velocity.on.embedding.cor() function. Fitting of individual genes was visualised by the gene.relative.velocity.estimates() function via 'show.gene'.
ATAC sequencing
Tumour samples (n = 4) from primary Ptc; Sox2eGFP/+ and (n = 2) from primary Ptc; Sox2eGFP/+; Ki67RFP/+ end-point MB tumours were dissociated. Fresh or viable-frozen single-cell suspensions (−80 °C) were used. If frozen, cells were thawed in DMEM supplemented with DNase I (Thermo Fisher Cat# 89836). Cells were pelleted at 200 RCF and resuspended in 0.1% BSA/PBS, supplemented with DNaseI, and filtered through a 35 μM nylon filter. Cells were stained with DAPI before live-cell sorting using a MoFlo Astrios and sorted by FACS into GFP+ and GFP– or GFP+/RFP+ and GFP+/RFP– components. Live cells were collected into 0.1%BSA/PBS before proceeding with ATACseq, which was performed as previously described (Buenrostro et al., 2013). Briefly, 35,000–50,000 cells were collected, depending on sample viability. Equal numbers of GFP+ and GFP– or RFP+ and RFP– cells were used for downstream ATAC protocol. Each sample was washed in PBS and centrifuged at 600 × g for 10 minutes. Cold lysis buffer (10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2 and 0.1% IGEPAL CA-630, 0.1% tween) was used to lyse cells and the nuclei were immediately spun down at 600 × g for 10 minutes at 4 °C. Following nuclei prep, the pellets were resuspended in a transposase reaction mix (25 μL 2× TD buffer, 2.5 μL transposase (Illumina) and 22.5 μL nuclease-free water) for 30 minutes at 37 °C. Following transposition, the samples were purified using Qiagen MinElute Kit. Next, library fragments were amplified using the NEBNext High Fidelity 2X PCR Master MixEndFragment (NEB) and the libraries were purified using a Qiagen MinElute Kit kit. Following purification, the libraries were selected for a size range of 100–600bp using Pippin Prep (Sage Science). The resulting libraries were controlled for quality by qPCR using the following primers: mGapdh_2_F GGACTGGATAAGCAGGGCG; mGapdh_2_R GGAACAGGGAGGAGCAGAGA; mKat6b_F GAACACTTAGGGCCAACCGT; mKat6b_R GAGATGACCGGCAGGAACTT (positive controls); mQml5_F CAAGGGCGGAGACCAGTAAG; mQml5_R GGTCAGCGCAAGGGATGATA; mQml7_F CACATTGCCAGTCTGCCCTA mQml7_R CTTGGACCTTCTAAGGGACTTT (negative controls). Libraries passing QC were sequenced on an Illumina HiSeq 2500 platform, using a Rapid Run mode flow cell to generate 50 bp single-end reads which were mapped to mm10 using the Bowtie 2 aligner82. Duplicate reads, unmapped or poor quality (Q < 30) reads, mitochondrial reads, chrY reads, and regions documented to show artificially high signals (blacklisted regions) were filtered out. Following alignment, open chromatin regions/peaks were called using Macs2.083. Default parameters were used except for the following: --keep-dup all -B --nomodel --SPMR -q 0.05 --slocal 6250 --llocal 6250. The Macs2.0 bdgcmp function was used to calculate the signal intensity as the fold enrichment of the signal per million reads in a sample over a modelled local background. A composite of unique peaks for GFP+ and GFP–, and RFP+ and RFP– subpopulations were generated. The signal intensity was calculated as the fold enrichment of the signal per million reads in a sample over a modelled local background using the bdgcmp function in MACS2. The distribution of genomic features across the ATAC peaks was determined using cis-regulatory element annotation system (CEAS)84. Interactions between regions of open chromatin were predicted using Cross-cell-type correlation in DNase I hypersensitivity (C3D). To identify gene promoters associated with the GFP+ and GFP– and the RFP+ and RFP– subpopulations, the unique open chromatin regions or peaks associated with each group were overlapped with a 3 kb window around the Gencode v19 transcription start site (2.5 kb upstream, 0.5 kb downstream). From the obtained gene list, Gene Ontology Biological Process pathway analysis was performed85,86 using the WebGestalt web tool87. The enrichment map was generated via Cytoscape88 using the application Enrichment Map89. Overlaps with ChIPseq datasets from Mateo et al.53 were performed using IntersectBed (bedtools2)90. The case example was generated using Integrative Genomics Viewer (IGV). First, ATACseq output bedgraph files were converted into tiled data files (TDF) format by using the 'igvtools' package (toTDF command) and then loaded into the IG viewer (genome assembly mm10) along with RefSeq gene track.
Identification of enhancer-promoter interactions using C3D
We took advantage of the known relationship between the Cross-Cell type Correlation in DNaseI hypersensitivity signals (C3D) and chromatin interactions41 to identify gene promoters associated with chromatin-accessible regions identified in primary Ptc tumours (see 'ATACseq'). We used the uniformly processed DNaseI hypersensitivity sequencing signal files for 230 mouse cell lines and tissues downloaded from the ENCODE project, filtered for alignment to mm10 genome36. We performed the correlation of DNaseI signal intensities in a cell type-specific manner by interrogating the catalogue of ATACseq peaks identified in the primary Ptc tumours (see 'ATACseq').
Motif enrichment analysis
Enrichment for known motifs was screened using Homer v4.9.1 software using the unique ATAC peaks of the contrasting sample as a background91. The enrichment score was calculated as: % target sequences with motif/% background motifs with motif.
RNA sequencing
In vitro samples: PTCP53 304 cells were cultured adherently for 1 and 6 days with either IC50 (189.2nM) of CT-179 or vehicle, dH2O. In vivo samples: Ptc mice treated with 20 mg/kg of CT-179, administered through IP injection daily for 14 days, and sacrificed at P15. Transcardiac perfusion with phosphate-buffered saline was performed, after which whole brains were harvested.
For both in vitro and in vivo samples, RNA was extracted using the RNeasy Plus Mini Kit (Qiagen Cat# 74134) with on-column DNase digestion using the gDNA Eliminator Columns. RNAseq libraries were constructed using the NEB Ultra II Directional polyA mRNA Library Prep Kit (NEB Cat# E7765), following the manufacturer’s instructions. Quantitative PCR (qPCR) was performed on select genes and all the primers and expected amplicon sizes are listed in Supplementary Data 6. qPCR reactions were performed using 10 ng cDNA in the presence of 0.2 mM each dNTP, 1.5 mM MgCl2, 0.6 μM each primer, and 0.2 units of Platinum Taq DNA polymerase (Thermo Scientific) in a final volume 10 μL. An initial denaturation step of 2 minutes at 95 °C was followed by 35 cycles at 94 °C 30 s, 55 °C 30 s, and 72 °C 60 s. The amplification was completed by a 2-minute elongation step at 72 °C. PCR reactions were performed on SimpliAmp thermocyclers PCR System (Applied Biosystems, Life Technologies).
RNA was sequenced using the NovaSeq 6000 System (Illumina) on the SP flow cell. Sequencing was done as paired-end reads with a read length of 100 bases. Raw sequencing information was collected in the form of FASTQ files. The quality of the FASTQ files was assessed using FastQC v.0.11.5. Adaptors are trimmed using Trim Galore v. 0.5.0. The quality of the trimmed reads was re-assessed with FastQC. The trimmed reads were also screened for presence of rRNA and mtRNA sequences using FastQ-Screen v.0.10.0. To assess the read distribution, positional read duplication and to confirm the strandedness of the alignments the RSeQC package v. 2.6.2 was used. The raw trimmed reads were aligned to the reference genome using the STAR aligner, v.2.6.0c. The STAR alignments were processed to extract raw read counts for genes using htseq-count v.0.6.1p2 (HTSeq). Assigning reads to genes by htseq-count was done in the mode 'intersection_nonempty', and only uniquely mapping reads were counted. In the 144-hour time point dataset sample CT-179_treated_B was an outlier and was removed for the identification of differentially expressed genes. To identify the set of differentially expressed genes between vehicle and CT-179 treatment conditions, two-condition differential expression was done with the edgeR R package v.3.28.1, using R v.3.6. The data set was filtered to retain only genes whose FPKM > 2 in at least 2/1 samples to remove genes that are not expressed, or expressed at a very low level. The method used for normalising the data was TMM, implemented by the calcNormFactors(y) function. All samples were normalised together. The glmQLF functionality in edgeR was used for the differential expression tests. From the obtained gene list, Gene Ontology Biological Process pathway analysis was performed85,86 using the WebGestalt web tool87. The enrichment map was generated via Cytoscape88 using the application Enrichment Map89.
In vivo transplantation for subcutaneous allografts
For subcutaneous allograft experiments, 50,000 viable PTCP53 304 CRISPR scrambled control or Olig2-KO cells were resuspended in 100 μL of 1:1 PBS/Matrigel (BD Biosciences) and injected subcutaneously into the flanks of female NSG mice. Mice were monitored for health twice a day for 3 days after surgery. Tumours were measured twice per week in two dimensions using callipers and tumour volume was calculated using the formula: (length * width * width)/2. The maximum allowable size of subcutaneously engrafted tumours by the SickKids Lab Animals Service is 20 mm in diameter. All animals were sacrificed when the largest tumour reached end-point dimensions.
Tolerability study of CT-179
Ptc mice were administered 100, 80, 60, 40, 30 and 20 mg/kg of CT-179 dissolved in dH2O daily by intraperitoneal (IP) injection. Weight and condition of mice were recorded every day. Mice who had lost >20% of their body weight were sacrificed. Plasma and brain samples from mice treated with the two lowest maximum tolerated doses (20 and 30 mg/kg) were submitted for pharmacokinetic analyses.
Pharmacokinetics (PK) of CT-179
Ptc mice treated with 20 and 30 mg/kg of CT-179, administered through IP injection daily for 14 days, and sacrificed at P15, following a maximum tolerated dose assessment. Blood was collected in heparinized tubes by cardiac puncture and plasma was separated by centrifugation. Transcardiac perfusion with phosphate-buffered saline was performed, after which whole brains were harvested. Perfused brains were flash-frozen in liquid nitrogen and stored at −80 °C. Samples were submitted to Quintara Discovery Inc. for the determination of concentration of CT-179 in plasma and brain. Plasma and brain tissue concentration measurements were done by LC-MS/MS. 6 mouse plasma samples + 10 calibrations for CT-179 untreated mouse plasma (3) were used to generate the calibration curves.
Survival analysis
Mice in the Ptc vehicle or CT-179-treated experiment presented in Fig. 6 were monitored daily following CT-179 daily treatments from P3–P28 (Fig. 6b) and from P64–P92 followed MCT/GDC treatment from P56–63 (Fig. 6f). The mice were sacrificed at their ethical endpoint; the first onset of neurological symptoms. Mice that died with no neurological symptoms were censored from the study. Survival analysis was performed using the GraphPad Prism 9 software. Kaplan–Meier curves show time elapsed (in days) from treatment until death. Statistical analysis was performed using the log-rank (Mantel–Cox) test in Prism 9 (GraphPad).
Tissue processing
Tissue from postnatal mice was dissected after transcardial perfusion of PBS followed by 4% PFA and subsequently fixed for 24–72 h at room temperature immersed in 4% PFA. For immunofluorescence (IF), tissue was cryoprotected in 30% sucrose/PBS for 24–48 h prior to snap freezing in 100% OCT. Sagittal frozen sections were cut at a 16 μm on a cryostat and kept at −20 °C until staining. For immunohistochemistry (IHC) and histological staining tissue was dehydrated in ethanol, wax embedded and cut sagittally at thickness of 5 μm on a microtome.
Immunostaining and histology
For IF, frozen sections were thawed, washed in PBS and blocked in a solution of 10% normal goat serum/0.1% Triton-X / PBS. Primary antibodies were incubated overnight at 4 °C at the specified concentrations: mouse anti-Sox2 1:100 (Abcam Cat# AB79351); goat polyclonal anti-Sox2 1:200 (R&D Systems Cat# AF2018) (for the EdU label experiment); rabbit anti-Olig2 1:200 (EMD Millipore Cat# AB9610); mouse monoclonal (Mobu-1) anti-BrdU (Ab-3) 1:50 (Millipore Sigma Cat# NA61) in the blocking solution. Goat anti-rabbit/mouse 488/568/647 fluorophore-conjugated secondary antibodies (Molecular Probes) were incubated 1:200 with DAPI for one hour at room temperature (RT). For the EdU label experiment, EdU was detected using the Invitrogen Click-iT Plus EdU Alexa Fluor 647 imaging kit (Invitrogen Cat# C10640), and the secondary antibodies used were Alexa Fluor 488 donkey anti-rabbit 1:500 (Thermo Fisher Cat# A-21206) and biotinylated horse anti-goat 1:200 (Vectolabs Cat# BA-9500) and biotinylated goat anti-mouse 1:200 (Vectolabs Cat# BA-9200). An amplification step was performed using Streptavidin DyLight 549 (BioLynx Cat# VECTSA5549), and the Vector® TrueVIEW® Autofluorescence Quenching Kit (BioLynx Cat# VECTSP8400). Images were acquired using a Leica DMi8 Inverted LED Fluorescence Motorized Microscope and analysed using the Leica Application Suite software.
For paraffin-embedded tissues, sections were deparaffinized and rehydrated prior to performing heat-induced antigen retrieval using 10 mM sodium citrate buffer (pH 6.0) in a microwave pressure cooker. Endogenous peroxide activity was blocked with 3% (v/v) peroxide in methanol for 15 minutes at RT and nonspecific binding was blocked with 2% (v/v) normal goat or horse serum (Vector Labs) in PBS with 2% (w/v) BSA for 30 minutes. Sections were incubated with primary antibody at 4 °C overnight: rabbit anti-Olig2 1:500 (EMD Millipore Cat# AB9610). Goat anti-rabbit/mouse ImmPRESS HRP (Vector Labs) were used to detect binding of the primary antibody. Normal goat serum was used for control sections. Hematoxylin and Eosin staining for histology were performed using standard methods. In all cases, analyses were performed on at least three biological replicates. Images were acquired using a 3DHistech Pannoramic 250 Flash II Slide Scanner and analysed using the Pannoramic Viewer software (3DHISTECH). For human primary tumour samples, the subgroups were confirmed by a nanoString analysis92.
Magnetic resonance imaging
High-resolution MRI was performed in the Ptc mice treated with either Vismodegib debulking agent or MCT vehicle, followed by CT-179 or vehicle treatment every 2–6 weeks. All MR imaging used a preclinical small bore 7T system (Biospec 70/30, Bruker Corporation, Ettlingen, DE), equipped with the BGA-12 gradient coil insert, 7.2 cm inner diameter linearly polarised RF coil for RF transmission, and a dedicated murine brain receive-only coil for RF signal reception. Mice were anaesthetised and maintained at 1.8% isoflurane throughout experimentation, on the provided slider bed. Respiration was monitored via a pneumatic pillow taped above the mouse (Model 1030 monitoring and gating system, SA Instruments, Stony Brook, NY). Tumours and brain anatomy were visualised via 2D-T2-weighted imaging in the horizontal plane of the mouse brain, as follows (2D-T2w-RARE technique; echo time 72 ms; repetition time 3800 ms; RARE factor 16; 160 × 160 matrix over 16 × 16 mm field-of-view for 0.1 × 0.1 image resolution; at least 15 0.5 mm contiguous slices providing whole-brain coverage; 3 averages; 3 min 48 s). Data processing and visualisation were performed using the National Institutes of Health MIPAV (Medical Image Processing, Analysis, and Visualization) application. Image quantification was performed by measuring # of voxels of the identified lesion regions by contour and slice using the MIPAV statistics generator function.
Quantification and statistical analysis
Statistical analyses of the limiting dilution assays were performed using the extreme limiting dilution analysis (ELDA) software (http://bioinf.wehi.edu.au/software/elda/). All other statistical analyses were performed in GraphPad Prism version 9.3.1. Unless otherwise stated, in all figures: ns = p ≥ 0.05; ∗ = p < 0.05; ∗∗ = p < 0.01; ∗∗∗ = p < 0.001; ∗∗∗∗ = p < 0.0001. Details of specific tests and sample numbers are reported in figure legends.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The accession numbers for the scRNAseq data reported in this paper are GSE234107 (Ptc) and GSE233652 and GSE197402 (Math1-Cre;SmoM2 P7 and P21 respectively). The accession numbers for the ATACseq data and RNAseq data reported in this paper are GSE233993 and GSE234105 respectively. Source data are provided with this paper.
Code availability
The following software and packages were used for data analysis: Cell Ranger v 3.0, Velocyto.py v0.17, R v3.6, Python v3.6.3, Seurat v3.0, gProfileR v0.6.7, Cytoscape v3.9.1, Enrichment Map v3.3.3, bedtools v2.29.0, IGV v2.16.1, and MIPAV v11.1.0. Additional analyses were performed using packages including Seurat v4.0.6, SeuratWrappers v0.3.0, Velocyto.R v0.6, dyno v0.1.2, dynwrap v1.2.2, dynmethods v1.0.5, dynguidelines v1.0.1, dynfeature v1.0.0, tidyverse v1.3.1.
References
Northcott, P. A. et al. Medulloblastoma comprises four distinct molecular variants. J. Clin. Oncol. 29, 1408–1414 (2011).
Northcott, P. A., Korshunov, A., Pfister, S. M. & Taylor, M. D. The clinical implications of medulloblastoma subgroups. Nat. Rev. Neurol. 8, 340–351 (2012).
Wechsler-Reya, R. & Scott, M. P. The developmental biology of brain tumors. Annu. Rev. Neurosci. 24, 385–428 (2001).
Gajjar, A. et al. Outcomes by clinical and molecular features in children with medulloblastoma treated with risk- adapted therapy: results of an international Phase III trial (SJMB03). J. Clin. Oncol. 39, 822–835 (2021).
Leary, S. E. S. et al. Efficacy of carboplatin and isotretinoin in children with high-risk medulloblastoma: a randomized clinical trial from the children’s oncology group. JAMA Oncol. 7, 1313–1321 (2021).
Ramaswamy, V. et al. Risk stratification of childhood medulloblastoma in the molecular era: the current consensus. Acta Neuropathol. 131, 821–831 (2016).
Gottardo, N. G. et al. Medulloblastoma Down Under 2013: a report from the third annual meeting of the International Medulloblastoma Working Group. Acta Neuropathol. 127, 189–201 (2014).
Tabori, U. et al. Universal poor survival in children with medulloblastoma harboring somatic TP53 mutations. J. Clin. Oncol. 28, 1345–1350 (2010).
Zhukova, N. et al. Subgroup-specific prognostic implications of TP53 mutation in medulloblastoma. J. Clin. Oncol. 31, 2927–2935 (2013).
Morrissy, A. S. et al. Divergent clonal selection dominates medulloblastoma at recurrence. Nature 529, 351–357 (2016).
Northcott, P. A. et al. The whole-genome landscape of medulloblastoma subtypes. Nature 547, 311–317 (2017).
Lin, C. Y. et al. Active medulloblastoma enhancers reveal subgroup-specific cellular origins. Nature 530, 57–62 (2016).
Hovestadt, V. et al. Decoding the regulatory landscape of medulloblastoma using DNA methylation sequencing. Nature 510, 537–541 (2014).
Northcott, P. A. et al. Subgroup-specific structural variation across 1,000 medulloblastoma genomes. Nature 488, 49–56 (2012).
Northcott, P. A. et al. Medulloblastomics: the end of the beginning. Nat. Rev. Cancer 12, 818–834 (2012).
Vladoiu, M. C. et al. Childhood cerebellar tumours mirror conserved fetal transcriptional programs. Nature 572, 67–73 (2019).
Hovestadt, V. et al. Resolving medulloblastoma cellular architecture by single-cell genomics. Nature 572, 74–79 (2019).
Cavalli, F. M. G. et al. Intertumoral Heterogeneity within Medulloblastoma Subgroups. Cancer Cell 31, 737–754.e6 (2017).
Ramaswamy, V. et al. Recurrence patterns across medulloblastoma subgroups: an integrated clinical and molecular analysis. Lancet Oncol. 14, 1200–1207 (2013).
Ocasio, J. K. et al. scRNA-seq in medulloblastoma shows cellular heterogeneity and lineage expansion support resistance to SHH inhibitor therapy. Nat. Commun. 10, 5829 (2019).
Robinson, G. W. et al. Irreversible growth plate fusions in children with medulloblastoma treated with a targeted hedgehog pathway inhibitor. Oncotarget 8, 69295–69302 (2017).
Singh, S. K. et al. Identification of human brain tumour initiating cells. Nature 432, 396–401 (2004).
Ward, R. J. et al. Multipotent CD15+ cancer stem cells in patched-1-deficient mouse medulloblastoma. Cancer Res. 69, 4682–4690 (2009).
Vanner, R. J. et al. Quiescent sox2(+) cells drive hierarchical growth and relapse in sonic hedgehog subgroup medulloblastoma. Cancer Cell 26, 33–47 (2014).
Schüller, U. et al. Acquisition of granule neuron precursor identity is a critical determinant of progenitor cell competence to form Shh-induced medulloblastoma. Cancer Cell 14, 123–134 (2008).
Luo, W. et al. Single-cell spatial transcriptomic analysis reveals common and divergent features of developing postnatal granule cerebellar cells and medulloblastoma. BMC Biol. 19, 135 (2021).
Selvadurai, H. J. et al. Medulloblastoma arises from the persistence of a rare and transient Sox2 granule neuron precursor. Cell Rep 31, 107511 (2020).
Roussel, M. F. & Stripay, J. L. Modeling pediatric medulloblastoma. Brain Pathol. 30, 703–712 (2020).
Ivanov, D. P., Coyle, B., Walker, D. A. & Grabowska, A. M. In vitro models of medulloblastoma: Choosing the right tool for the job. J. Biotechnol. 236, 10–25 (2016).
Goodrich, L. V., Milenković, L., Higgins, K. M. & Scott, M. P. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science 277, 1109–1113 (1997).
Shao, X. et al. scCATCH: Automatic annotation on cell types of clusters from single-cell RNA sequencing data. iScience 23, 100882 (2020).
Ellis, P. et al. SOX2, a persistent marker for multipotential neural stem cells derived from embryonic stem cells, the embryo or the adult. Dev. Neurosci. 26, 148–165 (2004).
Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y. & Greenleaf, W. J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013).
Stergachis, A. B. et al. Developmental fate and cellular maturity encoded in human regulatory DNA landscapes. Cell 154, 888–903 (2013).
Takayama, N. et al. The Transition from Quiescent to Activated States in Human Hematopoietic Stem Cells Is Governed by Dynamic 3D Genome Reorganization. Cell Stem Cell 28, 488–501.e10 (2021).
ENCODE Project Consortium. et al. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 583, 699–710 (2020).
Gorkin, D. U. et al. An atlas of dynamic chromatin landscapes in mouse fetal development. Nature 583, 744–751 (2020).
Ernst, J. & Kellis, M. ChromHMM: automating chromatin-state discovery and characterization. Nat. Methods 9, 215–216 (2012).
Sloan, C. A. et al. ENCODE data at the ENCODE portal. Nucleic Acids Res. 44, D726–32 (2016).
Shen, Y. et al. A map of the cis-regulatory sequences in the mouse genome. Nature 488, 116–120 (2012).
Thurman, R. E. et al. The accessible chromatin landscape of the human genome. Nature 489, 75–82 (2012).
Bailey, S. D. et al. ZNF143 provides sequence specificity to secure chromatin interactions at gene promoters. Nat. Commun. 2, 6186 (2015).
Mehdi, T., Bailey, S. D., Guilhamon, P. & Lupien, M. C3D: a tool to predict 3D genomic interactions between cisregulatory elements. Bioinformatics 35, 877–879 (2019).
Ong, C.-T. & Corces, V. G. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat. Rev. Genet. 12, 283–293 (2011).
Basak, O. et al. Mapping early fate determination in Lgr5+ crypt stem cells using a novel Ki67-RFP allele. EMBO J 33, 2057–2068 (2014).
Swartling, F. J. et al. Distinct neural stem cell populations give rise to disparate brain tumors in response to NMYC. Cancer Cell 21, 601–613 (2012).
Guo, D. et al. Tumor cells generate astrocyte-like cells that contribute to SHH-driven medulloblastoma relapse. J. Exp. Med. 218, (2021).
Sherwood, R. I. et al. Discovery of directional and nondirectional pioneer transcription factors by modeling DNase profile magnitude and shape. Nat. Biotechnol. 32, 171–178 (2014).
Zaret, K. S. & Carroll, J. S. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 25, 2227–2241 (2011).
Park, N. I. et al. ASCL1 reorganizes chromatin to direct neuronal fate and suppress tumorigenicity of glioblastoma stem cells. Cell Stem Cell 21, 209–224.e7 (2017).
Parras, C., Marie, C., Zhao, C. & Lu, Q. R. Chromatin remodelers in oligodendroglia. Glia 68, 1604–1618 (2020).
Yu, Y. et al. Olig2 targets chromatin remodelers to enhancers to initiate oligodendrocyte differentiation. Cell 152, 248–261 (2013).
Mateo, J. L. et al. Characterization of the neural stem cell gene regulatory network identifies OLIG2 as a multifunctional regulator of self-renewal. Genome Res. 25, 41–56 (2015).
Mizuguchi, R. et al. Combinatorial roles of olig2 and neurogenin2 in the coordinated induction of pan-neuronal and subtype-specific properties of motoneurons. Neuron 31, 757–771 (2001).
Novitch, B. G., Chen, A. I. & Jessell, T. M. Coordinate regulation of motor neuron subtype identity and panneuronal properties by the bHLH repressor Olig2. Neuron 31, 773–789 (2001).
Wetmore, C., Eberhart, D. E. & Curran, T. Loss of p53 but not ARF accelerates medulloblastoma in mice heterozygous for patched. Cancer Res. 61, 513–516 (2001).
Dulken, B. W., Leeman, D. S., Boutet, S. C., Hebestreit, K. & Brunet, A. Single-cell transcriptomic analysis defines heterogeneity and transcriptional dynamics in the adult neural stem cell lineage. Cell Rep. 18, 777–790 (2017).
Leeman, D. S. et al. Lysosome activation clears aggregates and enhances quiescent neural stem cell activation during aging. Science 359, 1277–1283 (2018).
Martynoga, B. et al. Epigenomic enhancer annotation reveals a key role for NFIX in neural stem cell quiescence. Genes Dev. 27, 1769–1786 (2013).
Conti, L. et al. Niche-independent symmetrical self-renewal of a mammalian tissue stem cell. PLoS Biol. 3, e283 (2005).
Mira, H. et al. Signaling through BMPR-IA regulates quiescence and long-term activity of neural stem cells in the adult hippocampus. Cell Stem Cell 7, 78–89 (2010).
Sun, Y., Hu, J., Zhou, L., Pollard, S. M. & Smith, A. Interplay between FGF2 and BMP controls the self-renewal, dormancy and differentiation of rat neural stem cells. J. Cell Sci. 124, 1867–1877 (2011).
Chen, X. et al. Mechanosensitive brain tumor cells construct blood-tumor barrier to mask chemosensitivity. Neuron 111, 30–48.e14 (2023).
PubChem. 1-(2-(3-(3,4-Dichlorophenyl)ureido)-6-methyl-pyrimidin-4-yl)-amino-3(dimethylamino)propane. https://pubchem.ncbi.nlm.nih.gov/compound/122520266.
Tsigelny, I. F. et al. Multiple spatially related pharmacophores define small molecule inhibitors of OLIG2 in glioblastoma. Oncotarget 8, 22370–22384 (2017).
Li, Y. et al. Suppressing recurrence in Sonic Hedgehog subgroup medulloblastoma using the OLIG2 inhibitor CT-179. Nat. Commun. https://doi.org/10.1038/s41467-024-54861-3 (2025).
Pazzaglia, S. et al. High incidence of medulloblastoma following X-ray-irradiation of newborn Ptc1 heterozygous mice. Oncogene 21, 7580–7584 (2002).
Grasso, C. S. et al. Functionally defined therapeutic targets in diffuse intrinsic pontine glioma. Nat. Med. 21, 555–559 (2015).
Ligon, K. L. et al. Olig2-regulated lineage-restricted pathway controls replication competence in neural stem cells and malignant glioma. Neuron 53, 503–517 (2007).
Zhang, L. et al. Single-cell transcriptomics in medulloblastoma reveals tumor-initiating progenitors and oncogenic cascades during tumorigenesis and relapse. Cancer Cell 36, 302–318.e7 (2019).
Sun, Y. et al. Phosphorylation state of Olig2 regulates proliferation of neural progenitors. Neuron 69, 906–917 (2011).
Mehta, S. et al. The central nervous system-restricted transcription factor Olig2 opposes p53 responses to genotoxic damage in neural progenitors and malignant glioma. Cancer Cell 19, 359–371 (2011).
Lu, Q. R. et al. Sonic hedgehog–regulated oligodendrocyte lineage genes encoding bHLH proteins in the mammalian central nervous system. Neuron 25, 317–329 (2000).
Xu, Z. et al. OLIG2 Is a Determinant for the Relapse of MYC-Amplified medulloblastoma. Clin. Cancer Res. 28, 4278–4291 (2022).
Tao, R. et al. Drives Group 3 medulloblastoma through transformation of Sox2 astrocyte progenitor cells. Cancer Res. 79, 1967–1980 (2019).
Hu, Y. & Smyth, G. K. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods 347, 70–78 (2009).
Sachamitr, P. et al. PRMT5 inhibition disrupts splicing and stemness in glioblastoma. Nat. Commun. 12, 979 (2021).
McInnes, L., Healy, J. & Melvilles, J. UMAP: Uniform manifold approximation and projection. J. Open Source Softw. 3, 861 (2018)
Satija, R., Farrell, J. A., Gennert, D., Schier, A. F. & Regev, A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502 (2015).
Zheng, G. X. Y. et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Shin, H., Liu, T., Manrai, A. K. & Liu, X. S. CEAS: cis-regulatory element annotation system. Bioinformatics 25, 2605–2606 (2009).
Ashburner, M. et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29 (2000).
The Gene Ontology Consortium. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 47, D330–D338 (2018).
Wang, J., Vasaikar, S., Shi, Z., Greer, M. & Zhang, B. WebGestalt 2017: a more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 45, W130–W137 (2017).
Cline, M. S. et al. Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2, 2366–2382 (2007).
Merico, D., Isserlin, R., Stueker, O., Emili, A. & Bader, G. D. Enrichment map: a network-based method for geneset enrichment visualization and interpretation. PLoS One 5, e13984 (2010).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
D’Arcy, C. E. et al. Immunohistochemical and nanoString-Based subgrouping of clinical medulloblastoma samples. J. Neuropathol. Exp. Neurol. 79, 437–447 (2020).
Acknowledgements
We would like to thank the SickKids Imaging Facility, SickKids Laboratory Animal Services, the UHN Spatio-Temporal Targeting and Amplification of Radiation Response (STTARR) Facility and the SickKids-UHN Flow Cytometry Facility for contributions to this work. We would also like to thank Dr. Warren Foltz at the STTARR facility for acquiring the mouse magnetic resonance images (MRI) and advice on MRI analysis. We would like to thank Dr. Michelle Monje, Pamelyn Woo, Alexis Ivec, and Ann-Helen Lijensten for generously sharing the Monje lab DIPG cell line resources with us. We would like to thank Dr. Patty Sachamitr and the Structural Genomics Consortium at the University of Toronto (SGC-Toronto) for generously sharing the resource of chemical probes for our CT-179 overlaid probe screen. We thank present and former members of the Dirks lab for all their support; including Dr. Charles Chesnelong and Dr. Sonam Dolma for valuable discussions on experimental design and analysis respectively, and Sophia Cusimano for assistance with tissue culture; we also thank Drs. Charles Chesnelong, Fiona Coutinho, Sonam Dolma, Morgane Morabito, and Matthaeus Ware for their constructive feedback on the manuscript. Research was supported by the Canadian Institutes of Health Research (FRN 133625). P.B.D. is additionally supported by the Ontario Institute for Cancer Research, the Terry Fox Research Institute, the Canadian Cancer Society, Cancer Research UK, Stand Up to Cancer, the Hospital for Sick Children Foundation, the Garron Family Cancer Centre, the Bresler family, Jessica’s Footprint Foundation, the Hopeful Minds Foundation, b.r.a.i.n.child, and Meagan’s Walk. P.B.D. holds a Harold Hoffman/Shopper Drug Mart Chair in Pediatric Neurosurgery at The Hospital for Sick Children.
Author information
Authors and Affiliations
Contributions
K.D. and P.B.D conceptualised and designed the study; K.D., E.L., R.J.V., H.S., and L.L. developed methodology; K.D., E.L., J.E.J., C.Y., H.W., L.L., M.K. and J.F. performed experiments; K.D., S.W., B.D.S., J.V., and P.G. analysed the data; G.S., S.K., X.H., and X.L. provided resources; K.D. and P.B.D. wrote the paper with inputs from all co-authors; P.B.D. provided funding and supervision.
Corresponding author
Ethics declarations
Competing interests
G.S. is the CEO of Curtana Pharmaceuticals, and S.K. is on its board of directors. They generously provided us with CT-179 and were given the opportunity to read the manuscript prior to submission. They were not involved in the experimental design, nor in the interpretation of the results. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Luca Tiberi, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Desai, K., Wanggou, S., Luis, E. et al. OLIG2 mediates a rare targetable stem cell fate transition in sonic hedgehog medulloblastoma. Nat Commun 16, 1092 (2025). https://doi.org/10.1038/s41467-024-54858-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-54858-y
This article is cited by
-
Suppressing recurrence in Sonic Hedgehog subgroup medulloblastoma using the OLIG2 inhibitor CT-179
Nature Communications (2025)