Abstract
The faithful charging of amino acids to cognate tRNAs by aminoacyl-tRNA synthetases (AARSs) determines the fidelity of protein translation. Isoleucyl-tRNA synthetase (IleRS) distinguishes tRNAIle from tRNAMet solely based on the nucleotide at wobble position (N34), and a single substitution at N34 could exchange the aminoacylation specificity between two tRNAs. Here, we report the structural and biochemical mechanism of N34 recognition-based tRNA discrimination by Saccharomyces cerevisiae IleRS (ScIleRS). ScIleRS utilizes a eukaryotic/archaeal-specific arginine as the H-bond donor to recognize the common carbonyl group (H-bond acceptor) of various N34s of tRNAIle, which induces mutual structural adaptations between ScIleRS and tRNAIle to achieve a preferable editing state. C34 of unmodified tRNAIle(CAU) (behaves like tRNAMet) lacks a relevant H-bond acceptor, which disrupts key H-bonding interactions and structural adaptations and suspends the ScIleRS·tRNAIle(CAU) complex in an initial non-reactive state. This wobble nucleotide recognition-based structural adaptation provides mechanistic insights into selective tRNA aminoacylation by AARSs.
Similar content being viewed by others
Introduction
The genetic code is established by the faithful charging of transfer ribonucleic acids (tRNAs) with cognate amino acids, a process catalyzed by a family of ancient enzymes named aminoacyl-tRNA synthetases (AARSs)1,2. In the universal genetic code table, 61 codons encode 20 common proteingenic amino acids. Generally, two pyrimidine (Y)-ending codons (NNU and NNC) encode the same amino acids, as do the two purine (R)-ending codons (NNA and NNG). However, an essential exception is that two AUR codons separately encode two different amino acids: AUA for l-isoleucine (l-Ile) and AUG for l-methionine (l-Met). In addition to AUA, l-Ile is also encoded by two AUY codons (AUU and AUC), making l-Ile the only amino acid encoded by three codons. Like most AARSs, isoleucyl-tRNA synthetase (IleRS) recognizes anticodon triplets as the primary identity element of its tRNA substrates, and in particular, it discriminates tRNAIle from tRNAMet by relying exclusively on the first (wobble) anticodon nucleotide (N34 of tRNA)3,4,5. tRNAMet can be efficiently isoleucylated by Saccharomyces cerevisiae IleRS (ScIleRS) when its C34 is replaced by G344. Therefore, IleRS must strictly exclude C34 to avoid mis-aminoacylation of tRNAMet and accommodate N34 with different sizes and chemical structures in tRNAIle isoacceptors, which is a more challenging task than that encountered by all other AARSs.
Various post-transcriptional modifications have been developed on the N34s of tRNAIle isoacceptors to facilitate their recognition by IleRS (Supplementary Fig. 1). In eukaryotes, modifications of tRNAIle(AAU) and tRNAIle(UAU) generate tRNAIle(IAU) and tRNAIle(ΨAΨ), respectively (I: inosine, Ψ: pseudouridine)4,6,7. The I34 modification by tRNA adenosine deaminase increased the isoleucylation of the in vitro transcript of tRNAIle(AAU) by 16-fold, while the tRNAIle(ΨAΨ) extracted from yeast cells was 40-fold more active than the unmodified in vitro transcript of tRNAIle(UAU)4. Interestingly, prokaryotes encode a tRNAIle bearing the CAU anticodon, and the tRNAIle(CAU) lacking modification at C34 behaves like a tRNAMet 8,9. The post-transcriptional modifications of C34 to 2-lysylcytidine (L34) in bacteria8 and to 2-agmatinylcytidine (agm2C34) in archaea9 efficiently changed the amino acid-accepting and mRNA-decoding specificities of tRNAIle(CAU) from l-Met to l-Ile.
To facilitate recognition of the modified or unmodified N34, IleRS recruits new C-terminal domains in addition to the canonical anticodon binding ___domain (ABD)5. C-terminal truncated IleRS is active for l-Ile activation but inactive for l-Ile tRNA transfer10,11, validating the important role of the C-terminal domains in tRNA recognition. Interestingly, IleRSs from the three domains of life diverge sharply in their C-terminal domains (Fig. 1a)12. Eukaryotic and archaeal IleRSs contain C-terminal sequences that are at least twice as long as that of bacterial IleRS, and IleRS in higher eukaryotes has an additional unique ___domain (UNE-I) for multi-synthetase complex (MSC) assembly13. Notably, the zinc-binding ___domain (ZBD), which is essential for N34 recognition by bacterial IleRS5, does not exist in eukaryotic and archaeal IleRSs, suggesting that eukaryotic and archaeal IleRSs develop distinct N34 recognition mechanisms which may be related to eukaryotic/archaeal-specific N34 modifications.
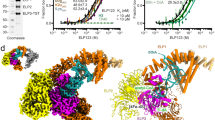
a Domain diagram of IleRSs from Saccharomyces cerevisiae, Homo sapiens, Pyrococcus horikoshii and Staphylococcus aureus. b Cloverleaf models of tRNAIle(GAU) and tRNAIle(CAU) from Escherichia coli. c Cartoon representation of the overall structure of ScIleRS in complex with tRNAIle(GAU) and l-Ile. ScIleRS is colored the same as the ___domain diagram, the substrate l-Ile is represented as spheres, and tRNAIle(GAU) is colored in orange with the nucleotides directly interacting with ScIleRS shown in filled rings. d Structural superposition of the ScIleRS·tRNAIle(GAU)·l-Ile complex and the Thermus thermophilus IleRS (TtIleRS) ED·Val-2AA complex (PDB ID: 1WNZ, green) confirms the protein‒tRNA interactions in the editing state revealed by the small-molecule probe Val-2AA. The H-bonds between residues of ScIleRS and nucleotide A76 of tRNAIle(GAU) are shown as black dashed lines. e tRNALeu was modeled to IleRS according to the EcLeuRS·tRNALeu·Leu-AMP complex structure in the aminoacylation state (PDB ID: 4AQ7). The CP core of ScIleRS in the editing state largely overlaps with the acceptor arm of tRNALeu. f In contrast, structural modeling revealed that there is no significant conflict between the acceptor arm of tRNALeu and the CP core of SaIleRS in the SaIleRS·tRNAIle(GAU)·mupirocin complex (PDB ID: 1FFY).
Moreover, how a small difference at the single nucleotide N34 could control whether eukaryotic/archaeal IleRS aminoacylates a tRNA remains a mystery. The local geometry difference resulting from a single substitution at the G3·U70 base pair of tRNAAla could be transmitted along the acceptor stem, finally causing the tRNA 3’ CCA end to fold back into a non-reactive route14. However, the distance between N34 and the CCA end of tRNAIle (approximately 70 Å) is significantly greater than that between G3·U70 and the CCA end of tRNAAla, and the transmission of N34 recognition information could be even more difficult.
Here, we report two parallel cocrystal structures of ScIleRS bound with tRNAIle(GAU) and unmodified tRNAIle(CAU) (Fig. 1b). The unique C-terminal domains of ScIleRS were found to employ a eukaryotic/archaeal-specific and more robust N34 recognition mechanism based on a conserved arginine. Importantly, because of the lack of the arginine-mediated N34 interactions, the ScIleRS·tRNAIle(CAU) complex cannot trigger necessary structural adaptations to reach the preferable conformation that the functional ScIleRS·tRNAIle(GAU) complex adopts.
Results
ScIleRS and tRNAIle(GAU) form an editing state complex
Our attempts to crystallize the complex of ScIleRS with S. cerevisiae tRNAIle(IAU) were unsuccessful. It has been reported that Escherichia coli tRNAIle(GAU) is recognized and isoleucylated by ScIleRS with an efficiency similar to that of SctRNAIle(IAU)4. Consequently, EctRNAIle(GAU) was utilized as a substitute for S. cerevisiae tRNAIle in the study of the eukaryotic tRNA recognition mechanism. The structure of full-length ScIleRS in complex with in vitro transcribed EctRNAIle(GAU) and l-isoleucine (l-Ile) (Fig. 1c) was determined by X-ray crystallography to 2.8 Å (Supplementary Fig. 2 and Supplementary Table 1). The crystallographic asymmetric unit contains two ScIleRS·tRNAIle(GAU)·l-Ile ternary complexes that adopt similar conformations to each other (Supplementary Fig. 2), and the complex consisting of chains A (ScIleRS) and T (tRNAIle) is discussed below owing to its superior electron density.
ScIleRS comprises three parts: the aminoacylation main body, the connective peptides (CPs), and the C-terminal appendant domains contributing to tRNA binding (Fig. 1a, c). The aminoacylation main body can be further divided into the Rossmann-fold catalytic ___domain (CD), stem-contact fold (SCF) and anticodon-binding ___domain (ABD). CPs contain the CP core, editing ___domain (ED, also known as CP1), CP2 and CP3. Unlike bacterial IleRS, whose C-terminal domains consist of a C-terminal junction ___domain and a ZBD, the C-terminal sequence of ScIleRS can be divided to three small domains, named the C-ter A, C-ter B and C-ter C domains (Fig. 1a, c). The C-ter A ___domain resembles the C-terminal junction ___domain of bacterial IleRS (Supplementary Fig. 3). The C-ter B ___domain is unique to eukaryotic and archaeal IleRSs and has no corresponding ___domain in bacterial IleRS. Although the C-ter C ___domain is the functional counterpart of the ZBD of bacterial IleRS, its structure is distinct from that of the ZBD, but despite the low sequence homology, it resembles the C-ter A ___domain (Supplementary Fig. 3).
The anticodon stem-loop of tRNAIle(GAU) is clamped between the aminoacylation main body and C-terminal domains (Fig. 1c). The acceptor stem of tRNAIle(GAU) forms only a few interactions with the CD, and the amino acid-accepting 3’ CCA end is directed into the ED, indicating that the complex was crystallized in the editing state (Fig. 1c). Owing to their similar sizes and physicochemical properties, l-valine (l-Val) and non-proteinogenic norvaline are incorrectly activated and charged to tRNAIle by IleRS at certain rates, and IleRS quickly hydrolyzes the mischarged tRNAIle with its ED to maintain protein translation fidelity15,16. The A76 of tRNAIle(GAU) interacts with the editing pocket in a manner similar to 2’-(l-valyl)amino-2’-deoxyadenosine (Val-2AA), a post-transfer editing substrate analog that mimics the 3’ end of the aminoacyl-2’-ester Val-tRNAIle17 (Fig. 1d). The 2’-OH of A76 of tRNAIle(GAU) is precisely oriented towards a subpocket for the l-Val (Fig. 1d). Our ScIleRS·tRNAIle(GAU)·l-Ile complex is the first structure that clearly shows how the entire 3’ CCA end interacts with the ED (Supplementary Fig. 4), and it strongly supports the editing substrate recognition mechanism of IleRS suggested previously by small-molecule probes17.
Class Ia AARSs containing EDs (IleRS, LeuRS and ValRS) prefer to bind substrate tRNAs in the editing state, and thus far, the aminoacylation state complex has been captured only for LeuRS5,18,19. In the editing state of the ScIleRS·tRNAIle(GAU)·l-Ile ternary complex, the CP core of ScIleRS packs over the l-Ile pocket, and structural comparison suggested that the CP core closes the cleft for the tRNAIle CCA end to enter the aminoacylation site (Fig. 1e). The closed conformation of the CP core has also been observed in tRNA-free Thermus thermophilus IleRS (TtIleRS, PDB ID: 1ILE)20, Candida albicans IleRS (CaIleRS, PDB ID: 6LDK)10 and Helicobacter pylori IleRS (HpIleRS, PDB ID: 8WNF)21 (Supplementary Fig. 5), as well as LeuRS and ValRS in tRNA-free and tRNA-editing states18,22,23. Translocation of the tRNA 3’ CCA end to CD requires a large rotation of the CP core and ED in these three AARSs19. In the SaIleRS·tRNAIle(GAU)·mupirocin complex (PDB ID: 1FFY), although the acceptor arm of tRNAIle(GAU) is orientated towards the ED, the ED and CP core of Staphylococcus aureus IleRS (SaIleRS) rotated by approximately 42° relative to those of ScIleRS in the ScIleRS·tRNAIle(GAU)·l-Ile complex (Supplementary Fig. 5), opening the conformation for translocating the tRNA 3’ CCA end between the CD and ED (Fig. 1f)5. Thus, the structure of the SaIleRS·tRNAIle(GAU)·mupirocin complex may represent an intermediate state from the editing state to the aminoacylation state, probably induced by the co-binding of the Ile-AMP-mimicking inhibitor mupirocin. In contrast, our structure provides the first unambiguous editing conformation for studying the catalytic process and possibly inhibitors of IleRS.
Productive tRNAIle binding induces C-terminal ___domain movements
The C-ter B ___domain, which is highly dynamic in tRNA-free ScIleRS (PDB ID: 7D5C)11, is stabilized upon tRNAIle binding through interactions with the tRNAIle elbow (consisting of D and T loops) (Fig. 1c). The C-ter B ___domain consists of three small α-helices, a short β-hairpin and an antiparallel two-stranded β-sheet, and the helix-turn-helix sequence from Trp900 to Ser920 is rich in basic residues (Supplementary Fig. 6). Residues Trp900, Pro901 and Lys915 form stacking, hydrophobic and hydrogen-bonding (H-bonding) interactions with nucleotides G19 and U20 of tRNAIle(GAU), respectively (Fig. 2a). As a result, U20 flips approximately 180° compared to that of tRNAIle(GAU) bound to SaIleRS (PDB ID: 1FFY) (Fig. 2b). When the C-ter B ___domain was deleted by replacing the sequence from Val897 to Asn948 with the linker of -GSGS-, ScIleRSΔCB could still activate amino acid (Fig. 2c) but completely lost aminoacylation activity against tRNAIle (Fig. 2d), indicating that C-ter B ___domain deletion does not affect ScIleRS folding but disrupts functional ScIleRS·tRNAIle binding. Consistently, the formation of ScIleRSΔCB·tRNAIle(GAU) complex is weaker than that of the wild-type ScIleRS·tRNAIle(GAU) complex as indicated by the electrophoretic mobility shift assay (EMSA) (Fig. 2e). We also introduced mutations at the G18·U55 and G19·C56 tertiary base pairs located in the elbow region of tRNAIle(GAU), and the results indicated that G19C, G18C&C56A and U55A mutants exhibited significant reductions in isoleucylation compared to the wild-type tRNAIle(GAU) (Fig. 2f). These tRNA mutants, along with ScIleRSΔCB, underscored the critical role of the C-ter B–elbow interaction in the isoleucylation of tRNAIle by ScIleRS. Notably, U20, which directly interacts with the C-ter B ___domain, as well as the tertiary base pairs G18·U55 and G19·C56, which are important for maintaining the elbow conformation, are conserved across all tRNAIle isoacceptors in both E. coli and S. cerevisiae, suggesting that the C-ter B ___domain of ScIleRS likely employs a similar mechanism to recognize the elbow of S. cerevisiae tRNAIle.
a The C-ter B ___domain of ScIleRS is stabilized and binds to the elbow of tRNAIle(GAU). b Interactions with the C-ter B ___domain induce a conformation of the U20 nucleotide of ScIleRS-bound tRNAIle(GAU) opposite to that of SaIleRS-bound tRNAIle(GAU) (PDB ID: 1FFY). c ScIleRS proteins with mutations located far from the active site exhibited similar or comparable activity to that of the wild-type protein in the tRNA-independent pre-transfer editing assay. Data are presented as means ± SD (n = 3 independent experiments). d Most ScIleRS variants partially or completely lost the tRNAIle isoleucylation activity as measured by tRNA-dependent ATP consumption assay. EctRNAIle(GAU) overexpressed in E. coli cells was utilized in this assay. Data are presented as means ± SD (n = 3 independent experiments). e The EMSA assay result revealed that tRNAIle binding ability of ScIleRSΔCB is weaker than that of wild-type ScIleRS. The in vitro transcript of tRNAIle(GAU) was used in this assay. Similar results were observed in two independent experiments. f The aminoacylation activity of ScIleRS against in vitro transcribed tRNAIle(GAU) and its variants with G19C, G18C&C56A or U55A mutations. All the variants presented significantly lower isoleucylation than that of the wild-type tRNAIle(GAU). Data are presented as means ± SD (n = 3 independent experiments). g, h Structural comparison of the tRNAIle(GAU)-bound ScIleRS with the tRNA-free ScIleRS (PDB ID: 7D5C, colored in gray) (g) and apo ScIleRS (AlphaFold DB: AF-P09436-F1, colored in pink) (h) indicated the conformational changes in the C-terminal domains and ABD of ScIleRS upon tRNAIle binding.
Notably, the C-ter B ___domain exists only in eukaryotic and archaeal IleRSs among all class I AARSs, and its best structural homolog is the insertion 3 ___domain (Ins3) of eukaryotic α2 glycyl-tRNA synthetase (GlyRS, a class II AARS) (Supplementary Fig. 3), as revealed by DALI24,25. The Ins3 of α2 GlyRS as well as a similar B2 ___domain in bacterial α2β2 GlyRS were proposed to undergo a large conformational movement to interact with the tRNAGly elbow, which may contribute to protecting tRNAGly from undesired disassociation during aminoacylation25,26,27,28.
When tRNAIle(GAU)-bound ScIleRS was aligned with tRNA-free ScIleRS (PDB ID: 7D5C) based on the CD, the ABD and C-ter A ___domain underwent the rotations of approximately 7° and 25°, respectively (Fig. 2g). An apo structure of full-length ScIleRS was predicted by AlphaFold2 (AlphaFold DB: AF-P09436-F1)29, and its ABD and C-ter A ___domain were well aligned with those of tRNA-free ScIleRS, highlighting the reliability of the C-terminal conformation of the predicted structure. Compared to this predicted structure, in addition to the ABD and C-ter A ___domain, the C-ter B and C domains also rotated approximately 20–30° during tRNAIle binding (Fig. 2h). Thus, in the editing state of ScIleRS, the C-terminal domains must undergo conformational movements to clamp tRNAIle together with the aminoacylation main body. However, additional experimental data are needed to further elucidate the potential ___domain movements, which are currently inferred primarily from the AI-predicted structure.
The mechanism for recognition of anticodon A35/U36
The backbone of the anticodon loop sits in a positively charged cavity (Fig. 3a) and forms extensive electrostatic and H-bonding interactions with multiple residues from the C-ter A, C-ter C and ABD domains (Supplementary Fig. 7), including Asn660, whose mutation leads to the resistance of eukaryotic IleRS to the natural product inhibitor reveromycin A11,30. Notably, Arg739 from the ABD plays a key role in anticodon loop binding by inserting its side chain into the cavity between C32, U33, A35 and U36. It forms six H-bonds with these bases, three of which contribute to the direct recognition of A35 and U36, the second and third nucleotides of the anticodon triplet (Fig. 3b). Arg739 is well-conserved among all aligned eukaryotic IleRSs (Supplementary Fig. 6), and its mutation to alanine in ScIleRS caused complete failure of tRNAIle isoleucylation in vitro (Fig. 2d). The important role of Arg739 in isoleucylation may explain the clinical finding that the compound heterozygous variants of human cytoplasmic IleRS, R739C (corresponding to Arg739 in ScIleRS) and F556S (a mutation in the CD that impairs enzyme function), caused growth delay, hepatic dysfunction, and neurodevelopmental disabilities31. Notably, R739A and other site-directed mutations of ScIleRS discussed later all exhibited activity comparable to that of wild-type ScIleRS in the tRNA-independent pre-transfer editing assay (Fig. 2c), indicating that their inactivity in tRNAIle isoleucylation is due to deficiencies in appropriate tRNAIle binding for l-Ile transfer.
a Electrostatic surface potential of the positively charged cavity formed by the ABD, C-ter A, and C-ter C domains for binding the anticodon loop of tRNAIle(GAU). b Base-specific interactions between the anticodon loop of tRNAIle(GAU) and the ABD of ScIleRS. c As shown in the SaIleRS·tRNAIle(GAU)·mupirocin complex structure (PDB ID: 1FFY), the ABD of SaIleRS only forms base-specific interactions with A35 of tRNAIle(GAU). d Different conformations of the anticodon loop between SaIleRS-bound and ScIleRS-bound tRNAIle(GAU) molecules. The nucleotides U33, G34 and U36 in the SaIleRS-bound tRNAIle(GAU) clash with the C-ter C ___domain of ScIleRS.
The Arg739-based A35/U36 recognition mechanism is specific to eukaryotic IleRS but not to bacterial IleRS, which relies on both the unique binding conformations of the tRNAIle anticodon loop and the anticodon loop-binding residues of IleRS. In the ScIleRS·tRNAIle(GAU) complex, C32 stacks with U33, and they are both buried in a cavity of ScIleRS. Arg736 contributes to stabilizing this conformation by forming a cation‒π interaction with the cytosine ring of C32 (Fig. 3b), and its mutation to alanine abolished the tRNAIle isoleucylation activity of ScIleRS (Fig. 2d). In contrast, C32 and U33 are partially exposed to the solvent in the SaIleRS·tRNAIle(GAU)·mupirocin complex (PDB ID: 1FFY)5 (Fig. 3c, d). Moreover, although A35 stacks with U36, and they both point to the ABD in ScIleRS·tRNAIle(GAU) complex (Fig. 3b), U36 of SaIleRS-bound tRNAIle(GAU) is directed inside the anticodon loop (Fig. 3c, d). Thus, unlike that of ScIleRS, the ABD of SaIleRS only forms base-specific interactions with A35 but not with C32, U33 or U36 (Fig. 3c). The distinct conformations of U36 in tRNAIle(GAU) bound to ScIleRS and SaIleRS propagate to the following nucleotides A37 and A38 (Fig. 3d). When the ABDs of the two IleRSs were superimposed, the anticodon loop backbones of the two tRNAIle(GAU) molecules shifted by up to 6.5 Å (Fig. 3d).
Notably, the ABD of eukaryotic ScIleRS makes considerably more contacts with the anticodon loop than that of bacterial SaIleRS does, but it does not recognize the nucleotide N34 to discriminate tRNAIle from tRNAMet; this task must be performed with additional C-terminal domains.
A conserved arginine recognizes a common carbonyl of N34s
The nucleotide G34 of tRNAIle(GAU) flips out from the anticodon loop to form multiple polar interactions with the C-ter C ___domain (Fig. 4a): the N1 atom of G34 H-bonds with the backbone oxygen of Leu1004; the 6-carbonyl group forms two H-bonds with Arg999; and N7 forms a water-mediated H-bond with Gln996. While the l-Ile tRNA transfer is the rate-limiting step in tRNAIle isoleucylation compared to l-Ile activation32, ScIleRS bearing the Q996A mutation catalyzed tRNAIle(GAU) isoleucylation at a rate comparable to that of the wild-type ScIleRS, so the Q996A mutation was unlikely to significantly affect tRNAIle recognition by ScIleRS. In contrast, ScIleRS bearing the R999A mutation completely lost aminoacylation activity, indicating that Arg999 plays a more important role in G34 recognition (Fig. 2d).
a The binding of G34 to the C-ter C ___domain of ScIleRS. The top panel shows a zoomed-in view of the G34 binding site of the ScIleRS·tRNAIle(GAU)·l-Ile complex. Direct and water-mediated H-bonding interactions are shown as black dashed lines. The bottom panel is the 2D presentation of G34–residues interactions, and the direct H-bonds are shown as orange dashed lines. The water-mediated H-bonding interactions have been omitted in the 2D presentation. b 2D presentations of the modeling of A34 and I34 at the N34 binding site of ScIleRS. A34 is unable to interact with the N34 binding residues due to the lack of appropriate H-bond acceptors or donors, so it is poorly recognized by ScIleRS. In contrast, after deamination, I34 can bind to these residues like G34, and be well recognized by ScIleRS. c Modeling of U34 and Ψ34 at the N34 binding site of ScIleRS. d Modeling of C34 and agm2C34 at the N34 binding sites of ScIleRS and PhIleRS, respectively.
In addition to G34, ScIleRS also recognizes U34 as well as I34 and Ψ34 (the modified A34 and U34) but poorly accommodates unmodified A34 and C344. When G34 was mutated in silico to A34 in the structural model of the ScIleRS·tRNAIle(GAU)·l-Ile complex, A34 lost direct contacts with Arg999 and Leu1004 because its chemical groups at positions 1 and 6 have H-bond acceptor-donor properties opposite to those of G34 (Fig. 4b and Supplementary Fig. 8). After the deamination of A34, product I34 has the same chemical properties as G34 at positions 1 and 6, so I34 can bind to ScIleRS in a manner similar to G34 (Fig. 4b and Supplementary Fig. 8). Interestingly, the modified nucleotide I can also function as a mimic of G to pair with C in both translation and splicing of mRNA33,34, highlighting a similar recognition mechanism of I by both protein and RNA. U34 could H-bond with Arg999 via its 4-carbonyl group, and after modification, Ψ34 may form an additional H-bond with Gln996 through its N1 atom (Fig. 4c and Supplementary Fig. 8). This new H-bond can partially explain the 40-fold increase in the activity of fully modified tRNAIle(ΨAΨ) compared with the unmodified tRNAIle(UAU) transcribed in vitro4. Notably, compared with G34 and I34, U34 and Ψ34 are smaller in size and may not interact with the backbone of Leu1004 (Supplementary Fig. 8).
Thus, it is a unified N34 recognition mechanism of ScIleRS in which Arg999 utilizes its side chain as the H-bond donor to form specific H-bonds with the carbonyl group (H-bond acceptor) of various N34s of tRNAIle. In contrast, Arg999 cannot form this critical interaction with nucleotide C34 because C34 lacks an H-bond acceptor at the corresponding position (Fig. 4d and Supplementary Fig. 8), thereby preventing tRNAMet from mis-isoleucylation by ScIleRS. Thus, our structure highlights that the carbonyl group of N34 acts as a positive determinant of eukaryotic tRNAIle. Consistently, its reader, Arg999, is conserved among eukaryotic IleRSs we aligned, except in Drosophila melanogaster IleRS, where it is replaced with a similar lysine residue (Supplementary Fig. 6). Mutation of Arg999 to lysine in ScIleRS resulted in approximately a 70% reduction in aminoacylation activity (Fig. 2d), which is consistent with the fact that lysine is able to form only one H-bond with N34, whereas arginine can form two H-bonds. Archaeal IleRS has C-terminal domains similar to those of eukaryotic IleRS. In addition to G34, archaeal IleRS can also charge the tRNAIle isoacceptor containing agm2C34, a modified C34 (Supplementary Fig. 1)9. According to structural modeling, Pyrococcus horikoshii IleRS (PhIleRS) can also use the arginine to form H-bonds with the 4-imine group (also an H-bond acceptor) of agm2C34 (Fig. 4d and Supplementary Fig. 8), supporting the unified N34 recognition mechanism of eukaryotic/archaeal-type IleRS.
The C-ter A and C domains are structurally similar (Supplementary Fig. 3). The C-ter A ___domain also has an arginine residue (Arg838) at the position corresponding to Arg999 of the C-ter C ___domain (Supplementary Fig. 9). Although Arg838 does not directly interact with tRNAIle, both Arg838 and Arg999 may contribute to structural stability by interacting with the backbones of nearby loops (Supplementary Fig. 9). Mutations of Arg838 to lysine or alanine caused approximately a half-reduction or complete failure in tRNAIle isoleucylation by ScIleRS, respectively (Fig. 2d). We propose that the C-ter C ___domain was a duplication of the C-ter A ___domain during the evolution of eukaryotic/archaeal IleRS, and it was retained because its conserved arginine happened to provide an effective way, alternative to that of the ZBD in bacterial IleRS, to recognize N34. Considering the facts that 1) the overall structural similarity between the C-ter A and C domains is more significant than that between the C-ter A ___domain and its corresponding C-terminal junction ___domain in bacterial IleRS, and that 2) the important arginine is conserved only in the C-ter A and C domains but not in the C-terminal junction ___domain (Supplementary Figs. 3 and 9), the acquisition of the C-ter C ___domain likely occurred later than the separation of bacterial and eukaryotic/archaeal IleRSs.
The ScIleRS·tRNAIle(CAU) complex in non-reactive state
Consistent with previous reports8, the unmodified tRNAIle(CAU) (Fig. 1b) was unable to be isoleucylated by ScIleRS and was instead methionylated by MetRS, suggesting that it functions like a tRNAMet in the term of aminoacylation specificity (Fig. 5a). However, a C34G mutation was sufficient to restore the isoleucylation of tRNAIle(CAU) (Fig. 5a). EMSA showed that tRNAIle(CAU) could still interact with ScIleRS, although its interaction is weaker than that of tRNAIle(GAU) (Fig. 5b). To understand how the Arg999–N34 interaction determines whether a tRNA should be isoleucylated by ScIleRS, we solved a cocrystal structure of the ScIleRS·tRNAIle(CAU)·l-Ile ternary complex at 2.83 Å resolution with R/Rfree = 23.8%/27.8% (Fig. 5c, Supplementary Fig. 2 and Supplementary Table 1). tRNAIle(CAU) is still located between the aminoacylation main body and the C-terminal domains of ScIleRS, but it has fewer interactions with ScIleRS. The interface area of 513 Å2 is much smaller than that between tRNAIle(GAU) and ScIleRS (2,362 Å2) as measured by program PISA35. Notably, the entire anticodon loop of tRNAIle(CAU), including C34, loses interactions with ScIleRS and becomes too dynamic to be traced in the density map. To our surprise, the acceptor stem of tRNAIle(CAU) points to neither the editing site nor the aminoacylation cavity, but binds to the backside of the ED (Fig. 5d). In this non-reactive conformation, only the nucleotides G2, C72 and C73 of the tRNAIle(CAU) acceptor stem H-bond with the residues Gln393 and Asn201 of ScIleRS ED, and the 3’ CCA end of tRNAIle(CAU) is exposed to the solution and invisible.
a The aminoacylation activities of ScIleRS and SaMetRS against in vitro transcribed tRNAIle(GAU), tRNAIle(CAU) and tRNAIle(CAU) C34G variant. Data are presented as means ± SD (n = 3 independent experiments). b EMSA revealed that in vitro transcribed tRNAIle(CAU) can still form a complex with ScIleRS, although it is slightly weaker than in vitro transcribed tRNAIle(GAU). Similar results were observed in two independent experiments. c The overall structure of the ScIleRS·tRNAIle(CAU)·l-Ile complex. The anticodon loop of tRNAIle(CAU) is dynamic because of the lack of interactions with ScIleRS, and the acceptor stem of tRNAIle(CAU) binds to the back of the ED. d Binding of the acceptor stem of tRNAIle(CAU) to the back of ScIleRS ED. The 3’ CCA end was invisible in the electronic density map. e Structural comparison between the reactive and non-reactive ScIleRS·tRNA complexes revealed an approximately 25° rotation between tRNAIle(GAU) and tRNAIle(CAU). f The C-terminal domains of tRNAIle(CAU)-bound ScIleRS adopt a conformation generally similar to that of ScIleRS in the tRNA-free state except for the distal C-ter C ___domain, but not to that of tRNAIle(GAU)-bound ScIleRS.
Two ScIleRS·tRNA complex structures were aligned based on the CD of ScIleRS (Fig. 5e, f), and a difference of approximately 25° was observed between tRNAIle(CAU) and tRNAIle(GAU) (Fig. 5e). The C-ter A and C-ter B domains of tRNAIle(CAU)-bound ScIleRS exhibited conformations quite similar to those of tRNA-free ScIleRS (PDB ID: 7D5C and AlphaFold DB: AF-P09436-F1) but not to those of tRNAIle(GAU)-bound ScIleRS (Fig. 5f). In contrast, the conformation of the distal C-ter C ___domain of tRNAIle(CAU)-bound ScIleRS is dramatically different from those of both tRNA-free and tRNAIle(GAU)-bound ScIleRS (Fig. 5f). Thus, the dynamic anticodon loop of tRNAIle(CAU) did not attract but probably even kicked away the C-ter C ___domain in the ScIleRS·tRNAIle(CAU) complex. As a result, the C-ter C ___domain of tRNAIle(CAU)-bound ScIleRS adopts a more open conformation, and it must rotate approximately 60° and 40° to align with the C-ter C ___domain of tRNAIle(GAU)-bound and tRNA-free ScIleRS, respectively (Fig. 5f).
Based on the above structural observations and the biochemical results, we propose a possible route for the discriminative aminoacylation of tRNAIle (Supplementary Fig. 10): both tRNAIle and tRNAMet could initially dock to ScIleRS based on their rough shape and charge complementarity; ScIleRS–tRNAIle binding is then strengthened by the formation of multiple stacking and bonding interactions between the anticodon triplets of tRNAIle and ScIleRS, in which the H-bonds between the N34 of tRNAIle and Arg999 of ScIleRS contribute an indispensable part of the binding energy; in the meantime, mutual structural adaptation between tRNAIle and ScIleRS, including the rotation of tRNAIle along with the C-terminal domains of ScIleRS, stabilizes the complex in an editing conformation; tRNAIle can then move its acceptor arm to the aminoacylation site to get charged and subsequently move it back to the editing site for proofreading, followed by its release from ScIleRS for protein translation. Because C34 cannot contribute to H-bonding with Arg999, the binding of tRNAMet to ScIleRS will be suspended at the first step, after which tRNAMet will dissociates from ScIleRS without being mis-isoleucylated.
Discussion
In this study, the cocrystal structures showed that the complex of ScIleRS with the cognate tRNAIle(GAU) adopts an editing conformation, while the complex of ScIleRS with the unmodified tRNAIle(CAU) remains in a non-reactive state. Many structural studies have indicated that class Ia AARSs with ED (IleRS, LeuRS and ValRS) bind their substrate tRNAs mostly stably in the editing state5,18,19. The ED is known to ensure the fidelity of tRNA charging through amino acid proofreading. This fidelity check occurs prior to the release of aminoacyl-tRNA and is performed using the ED’s amino acid pocket and catalytic activity. Here, the fact that only the cognate tRNAIle(GAU) but not the unmodified tRNAIle(CAU) can bind to the ED of ScIleRS at the correct editing conformation suggests that the ED likely also facilitates the fidelity check of tRNA. This tRNA check is performed upon tRNA entry using the ED in conjunction with other tRNA binding domains. Thus, the ED likely plays fidelity check roles for both amino acids and tRNAs, but in two distinctive ways.
The accurate recognition of N34 by IleRS plays the most important role in the discrimination of tRNAIle from tRNAMet and subsequently maintains the fidelity of protein translation. Bacterial and eukaryotic/archaeal IleRSs recruit kingdom-specific domains to recognize cognate N34s. Bacterial SaIleRS was shown to recognize the guanine ring of G34 of tRNAIle(GAU) by stacking and H-bonding interactions with its Trp890 and Arg888 from the ZBD (PDB ID: 1FFY)5 (Supplementary Fig. 11). Interestingly, although both the ZBD and the C-ter C ___domain H-bond with G34 via an arginine residue, the ZBD utilizes the main chain carbonyl group of arginine (Arg888 in SaIleRS) as an H-bond acceptor to interact with N1 and 2-NH2 of G34, while the C-ter C ___domain utilizes the side chain guanidyl group of arginine (Arg999 in ScIleRS) as an H-bond donor to interact with the 6-carbonyl group of G34. Moreover, the orientations of G34 are completely opposite in SaIleRS- and ScIleRS-bound tRNAIle (Fig. 3d). Therefore, the recognition mechanisms of G34 by bacterial and eukaryotic/archaeal IleRSs are completely independent of each other in both the recognition ___domain and the binding mode.
Eukaryotic/archaeal IleRS utilizes a unified mechanism to recognize all the N34s of their cognate tRNAIle substrates (Fig. 4 and Supplementary Fig. 8). Mupirocin, a natural product that selectively inhibits bacterial IleRS, is widely used to treat skin infections. However, some bacteria acquire mupA or mupB genes from the environment, which express mupirocin-resistant IleRS (MupA or MupB), resulting in the ineffectiveness of mupirocin in clinic36,37. In addition, some bacteria, such as the mupirocin-producing Pseudomonas fluorescens, contain an endogenous mupirocin-resistant IleRS (IleRS2)38,39. These mupirocin-resistant IleRSs contain eukaryotic/archaeal-type C-terminal domains40, and can isoleucylate bacterial tRNAIle with L34 to decode the AUA codon on mRNA41. Structural modeling suggested that the C-ter C ___domain of PfIleRS2 also employs a conserved arginine to H-bond with the 4-imine group of L34 of bacterial tRNAIle (Supplementary Fig. 8). Thus, the arginine-mediated N34 recognition mechanism of eukaryotic/archaeal-type IleRS could also apply to the recognition of N34 modifications of bacterial tRNAIle, but bacterial IleRS is unable to cross-charge eukaryotic tRNAIle 12, suggesting that eukaryotic/archael-type IleRS likely has developed a more robust way for N34 discrimination than its bacterial counterpart.
The unusual non-reactive complex of ScIleRS with C34-unmodified tRNAIle(CAU) highlights the important roles of N34 modifications in IleRS–tRNA recognition. Bacterial, archaeal and eukaryotic tRNAIle molecules have developed their own modifications on N34 (such as Ψ34, I34, L34 and agm2C34)4,8,9. Thus, the N34 recognition domains and key residues on IleRS as well as the anticodon loop conformations and N34 modifications of tRNAIle are different between bacteria, archaea and eukaryotes, presenting an example of coevolution of AARS–tRNA pairs and also providing a valuable opportunity for drugging aminoacylation with lineage specificity.
Methods
Protein preparation
The wild-type ScIleRS (UniProtKB ID: P09436) was expressed and purified as described11. Briefly, the DNA sequence encoding ScIleRS was cloned into the pET20b(+) plasmid (Novagen) with a C-terminal hexahistidine tag. The E. coli BL21(DE3) cells transformed with IleRS-pET20b(+) were grown in Luria-Bertani (LB) medium supplemented with 0.1 mg/mL ampicillin at 37 °C until the OD600 reached approximately 0.6, and then 0.2 mM isopropyl-β-D-thiogalactoside (IPTG) was added to induce protein overexpression at 20 °C for 16 h. The cells were harvested by centrifugation, resuspended in washing buffer (400 mM NaCl, 50 mM Tris-HCl pH 8.0, 10% v/v glycerol, 5 mM β-ME and 10 mM imidazole) and lysed by sonication. The lysate was centrifuged at 4000 × g for 30 min to remove cell debris. The supernatant was loaded onto a Ni-NTA column (Qiagen) and washed with 20 column volumes of washing buffer. The target protein was eluted with 5 column volumes of elution buffer (400 mM NaCl, 50 mM Tris-HCl pH 8.0, 10% v/v glycerol, 5 mM β-ME and 100 mM imidazole). The elution was concentrated to 2 mL and injected into a HiLoad 16/60 Superdex 200 pg (GE healthcare) column. The peak fractions eluted using gel filtration buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 5 mM β-ME and 5% v/v glycerol) were collected and concentrated to ~30 mg/mL. The plasmids for overexpressing ScIleRS mutants were constructed by amplifying the whole plasmid using PCR. For each mutation, a pair of primers that partially overlap and both contain the targeted mutation were used (Supplementary Table 2). The expression and purification of all ScIleRS variants were performed in the same manner as the wild-type protein.
The DNA sequence encoding SaMetRS (Uniprot ID: V7IMS7) was inserted into pET15b, and full-length SaMetRS protein was overexpressed in BL21(DE3) and purified by Ni-NTA column (Qiagen) and HiLoad 16/60 Superdex 200 pg column (GE Healthcare). The detailed methods for producing SaMetRS were described previously42.
In vitro transcription and purification of tRNA
The E. coli tRNAs were produced using in vitro T7 RNA polymerase transcription assay as described11. The DNA templates for tRNAIle(GAU) transcription were generated by PCR using Primer1 (5’-TAATACGACTCACTATAGGGCTTGTAGCTCAGGTGGTTAGAGCGCACCCCTGATAAG-3’) and Primer2 (5’-TGGTGGGCCTGAGTGGACTTGAACCACCGACCTCACCCTTATCAGGGGTGCGCTCTAAC-3’). This two primers cover the full-length of tRNAIle(GAU) gene and are partially complementary to each other (the underlined nucleotides), and Primer1 contains T7 promoter sequence (nucleotides in bold). The wild-type base pair A1·U72 of E. coli tRNAIle(GAU) was replaced by the G1·C72 pair to increase transcription3. The PCR product was further amplified by the second round of PCR using primer3 (5’-TAATACGACTCACTATAGGGCTTGT-3’) and primer4 (5’- TGGTGGGCCTGAGTGGACTTGAAC-3’). The first two nucleotides in italics and bold at the 5’ end of primer4 were methylated at their 2’-hydroxyl groups to reduce the non-templated addition of nucleotides to the 3’ end of tRNA by T7 RNA polymerase. The DNA templates for tRNAIle(CAU) and mutants of tRNAIle(CAU) and tRNAIle(GAU) are prepared by the same way, and the primers used are listed in Supplementary Table 3.
The product of second PCR was then used as the DNA template for in vitro transcription assay without additional purification. In a 15 mL centrifugal tube, 2 mL of the PCR product was mixed with 2 mL of 5 × transcription buffer (1 M Tris pH 8.0, 10 mM spermidine and 50 mM DTT), 1 mL of 40 mM NTPs (each), 0.2 mL of 1 M MgCl2, 0.2 mL of 10 mg/mL T7 polymerase and 3.6 mL of DECP water, and incubated at 37 °C for 3 to 4 h. The transcripts were purified using 12% polyacrylamide gel electrophoresis supplemented with 8 M urea, extracted from gels by 0.5 M ammonium acetate, and precipitated by ethanol at -20 °C overnight. The tRNA pellets were collected by centrifugation, washed by 70% v/v ethanol, and redissolved in a buffer consisting of 20 mM Tris pH 8.0 and 1 mM EDTA to 1 mg/mL. The redissolved tRNA was heated at 65 °C for 5 min, and then refolded by slowly cooling to room temperature after the addition of 10 mM MgCl2. The refolded tRNA was concentrated to ~10 mg/mL using a 3 kDa Ultra-4 centrifugal filter device (Millipore), aliquoted and stored at -80 °C for further use.
Overexpression and purification of tRNA
The E. coli tRNAIle(GAU) gene with the native base pair A1·U72 substituted with G1·C72 was inserted between the T7 promoter and terminator of pET29b(+) by homologous recombination. The transformed E. coli BL21(DE3) cells were cultured in LB medium supplemented with 50 μg/mL kanamycin until the OD600 reached approximately 0.6, and then 1 mM IPTG was added to induce the overexpression of tRNAIle(GAU) at 30 °C for 16 h. The tRNA transcript was extracted from the cell pellets using RNAiso Plus (Cat. No. 9109, TakaRa) and chloroform, and precipitated from aqueous fractions by isopropanol. The tRNA pellets collected by centrifugation were washed with 70% v/v ethanol and redissolved in a buffer containing 20 mM Tris pH 8.0 and 10 mM MgCl2. The sample was loaded onto a HiTrap Q XL (GE healthcare) column and the elution fractions between 0.55 and 0.70 M NaCl were collected. Finally, the tRNA was concentrated to ~20 mg/mL and stored in a buffer containing 10 mM HEPES pH 7.5 and 10 mM MgCl2.
Crystallography
The sitting-drop vapour-diffusion method was employed to crystallize the ScIleRS·tRNA complex. The full-length ScIleRS (10 mg/mL) was preincubated with tRNAIle(GAU) or tRNAIle(CAU) (transcribed in vitro, 2.5 mg/mL), together with 5 mM l-Ile, at room temperature for 30 min. Each drop containing 1 μL of protein–tRNA mixture, 0.5 μL of reservoir solution and 0.5 μL of seed stocked in reservoir solution was equilibrated against 100 μL of reservoir solution at 8 °C for 3–7 days to allow crystals to grow. For ScIleRS·tRNAIle(GAU) complex, the reservoir solution contains 0.2 M ammonium sulfate, 0.1 M BIS-TRIS pH 5.5, 25% PEG3350 and 0.06 M sodium citrate. For ScIleRS·tRNAIle(CAU) complex, the reservoir solution contains 2% Tacsimate pH 6.0, 0.1 M BIS-TRIS pH 6.5 and 20% PEG3350. Large crystals were immersed in a reservoir solution supplemented with 20% ethylene glycol for a few seconds and then flash frozen in liquid nitrogen. The diffraction data were collected using a single crystal at 100 K with a wavelength of 0.979 Å at the BL19U1 beamline at National Facility for Protein Sciences Shanghai (NFPS) and Shanghai Synchrotron Radiation Facility (SSRF) and were indexed, integrated and scaled using XDS43 and Aimless44. The structure was solved by molecular replacement using the ScIleRS structure (PDB ID: 7D5C)11 as the search model in the program Molrep45. Iterative refinements of the structure model were carried out using Coot46 and Refmac547. The stereochemical quality of the final model was assessed using MolProbity48. The statistics of the data collection and structural refinement are listed in Supplementary Table 1. Final Ramachandran statistics were as follows: 94.4% favored, 5.5% allowed and 0.1% outliers for the ScIleRS·tRNAIle(GAU)·l-Ile complex; 93.6% favored, 6.3% allowed and 0.1% outliers for the ScIleRS·tRNAIle(CAU)·l-Ile complex. The coordinate and structural factors of the ScIleRS·tRNAIle(GAU)·l-Ile and ScIleRS·tRNAIle(CAU)·l-Ile complex have been deposited in the Protein Data Bank (PDB) under the accession code 8WND and 8Z1P respectively.
ATP consumption assay
ATP consumption assay was employed to evaluate the aminoacylation activities of ScIleRS and its variants on tRNAIle. The 60 μL of reactions contained 40 nM ScIleRS (wild type or variants), 200 μM ATP, 1 mM l-isoleucine, 1 mg/mL E. coli tRNAIle(GAU) (overexpressed in E. coli) in the reaction buffer (30 mM HEPES pH 7.5, 150 mM NaCl, 30 mM KCl, 40 mM MgCl2, 1 mM DTT and 0.1% BSA). The reaction was incubated at room temperature. Aliquots of 5 μL at various time points (2, 5, 10, 20 and 30 min) were transferred to a 384 well plate and mixed with 5 μL of Kinase-Glo® Max Reagent (Cat. No. V6071, Promega) to stop the reaction. After 20 min, the luminescence (L) which reflects the concentration of the remaining ATP, was read on a Synergy H1 microplate reader (BioTek). The reactions without the addition of tRNA were used as controls (Lc). The ATP consumption (μM) = 200 × (1-L/Lc). Each reaction was repeated three times, and the results are expressed as the means ± SD (n = 3). Statistical analyzes were performed using GraphPad Prism 7 software, and a one-phase association equation was used to fit the time response curves (ATP consumption vs reaction time).
To evaluate the aminoacylation activities and specificities of in vitro tRNA transcripts, 60 μL of reactions containing 50 nM ScIleRS or SaMetRS, 4 μM ATP, 1 mM l-isoleucine or l-methionine and 1 mg/mL in vitro transcribed tRNA were incubated at room temperature. Aliquots of 5 μL at various time points (2, 5, 10, 20 and 30 min) were transferred to a 384 well plate and mixed with 5 μL of Kinase-Glo® Reagent (Cat. No. V6711, Promega) to stop the reaction. After 10 min, the luminescence (L) was read and the ATP consumption was calculated. The time response curves (ATP consumption vs reaction time) were fitted.
tRNA-independent Pre-transfer editing assay
Assays were performed in 80 μL of reaction mixtures consisting of 80 nM wild-type ScIleRS or its mutants, 6 mM l-cysteine, 250 μM ATP, 50 μg/mL PPiase, 30 mM HEPES pH 7.5, 150 mM NaCl, 30 mM KCl, 40 mM MgCl2 and 1 mM DTT. After incubation at room temperature for 30 min, 20 μL of malachite green reagent (2.45 M sulfuric acid, 0.1% malachite green, 1.5% ammonium molybdate tetrahydrate and 0.2% tween-20) was added to the mixtures. After incubation for 10 min, absorbance (A) was measured at 620 nm. The reactions without the addition of ScIleRS were used as controls. The absorbance differences between the wells with ScIleRS and those without ScIleRS reflected the tRNA-independent pre-transfer editing activity of ScIleRS. The tRNA-independent pre-transfer editing activity of the wild-type ScIleRS was normalized to 100%. Each reaction was repeated three times, and the results are expressed as the means ± SD (n = 3).
Electrophoresis mobility shift assay (EMSA)
The 20 μL reaction mixtures containing 1 μM in vitro transcript of tRNAIle(GAU) or tRNAIle(CAU) and ScIleRS at different concentrations (2, 5 and 10 μM) in binding buffer (30 mM sodium cacodylate pH 6.5, 20 mM MgCl2, 150 mM NaCl, 2 mM DTT and 10% v/v glycerol) were incubated at 4 °C for 10 min. The samples were loaded to 5% native polyacrylamide gels and electrophoresed at a voltage of 80 V for 2 h on a ice-bath. The gel was stained with Gel-Red nucleic acid dye (BBI Life Sciences).
Data analysis and figure preparation
Multiple protein sequence alignment and conservation score calculation were performed using Clustal Omega program49 and Jalview program50. All data were analyzed using GraphPad Prism 8.0 software and are expressed as the means ± SD (n = 3). All protein structure figures were prepared using PyMOL (PyMOL v.2.5.0).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data supporting the findings of this study are available from the corresponding authors upon request. The coordinates and structural factors of ScIleRS·tRNAIle(GAU)·l-Ile and ScIleRS·tRNAIle(CAU)·l-Ile complexes have been deposited in the Protein Data Bank (PDB) under the accession codes 8WND and 8Z1P. The structures used for molecular replacement or structural analyses are publicly available in PDB under accession codes 1FFY, 1WNZ, 4AQ7, 7D5C, 1ILE, 6LDK, 8WNF and 4QEI. The source data for Figs. 2c–f and 5a, b are provided as a Source data file. Source data are provided with this paper.
References
Carter, C. W. Jr. Cognition, mechanism, and evolutionary relationships in aminoacyl-tRNA synthetases. Annu. Rev. Biochem. 62, 715–748 (1993).
Ibba, M. & Soll, D. Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 69, 617–650 (2000).
Nureki, O. et al. Molecular recognition of the identity-determinant set of isoleucine transfer RNA from Escherichia coli. J. Mol. Biol. 236, 710–724 (1994).
Senger, B., Auxilien, S., Englisch, U., Cramer, F. & Fasiolo, F. The modified wobble base inosine in yeast tRNAIle is a positive determinant for aminoacylation by isoleucyl-tRNA synthetase. Biochemistry 36, 8269–8275 (1997).
Silvian, L. F., Wang, J. & Steitz, T. A. Insights into editing from an Ile-tRNA synthetase structure with tRNAIle and mupirocin. Science 285, 1074–1077 (1999).
Pixa, G., Dirheimer, G. & Keith, G. Sequence of tRNAIleIAU from brewer’s yeast. Biochem. Biophys. Res. Commun. 119, 905–912 (1984).
Szweykowska-Kulinska, Z., Senger, B., Keith, G., Fasiolo, F. & Grosjean, H. Intron-dependent formation of pseudouridines in the anticodon of Saccharomyces cerevisiae minor tRNAIle. EMBO J. 13, 4636–4644 (1994).
Muramatsu, T. et al. Codon and amino-acid specificities of a transfer RNA are both converted by a single post-transcriptional modification. Nature 336, 179–181 (1988).
Ikeuchi, Y. et al. Agmatine-conjugated cytidine in a tRNA anticodon is essential for AUA decoding in archaea. Nat. Chem. Biol. 6, 277–282 (2010).
Chung, S., Kim, S., Ryu, S. H., Hwang, K. Y. & Cho, Y. Structural basis for the antibiotic resistance of eukaryotic isoleucyl-tRNA synthetase. Mol. Cells 43, 350–359 (2020).
Chen, B. et al. Inhibitory mechanism of reveromycin A at the tRNA binding site of a class I synthetase. Nat. Commun. 12, 1616 (2021).
Shiba, K. et al. Human cytoplasmic isoleucyl-tRNA synthetase: selective divergence of the anticodon-binding ___domain and acquisition of a new structural unit. Proc. Natl Acad. Sci. Usa. 91, 7435–7439 (1994).
Chung, S. et al. Regulation of BRCA1 stability through the tandem UBX domains of isoleucyl-tRNA synthetase 1. Nat. Commun. 13, 6732 (2022).
Naganuma, M. et al. The selective tRNA aminoacylation mechanism based on a single G•U pair. Nature 510, 507–511 (2014).
Fersht, A. R. Editing mechanisms in protein synthesis. Rejection of valine by the isoleucyl-tRNA synthetase. Biochemistry 16, 1025–1030 (1977).
Bilus, M. et al. On the mechanism and origin of isoleucyl-tRNA synthetase editing against norvaline. J. Mol. Biol. 431, 1284–1297 (2019).
Fukunaga, R. & Yokoyama, S. Structural basis for substrate recognition by the editing ___domain of isoleucyl-tRNA synthetase. J. Mol. Biol. 359, 901–912 (2006).
Fukai, S. et al. Structural basis for double-sieve discrimination of L-valine from L-isoleucine and L-threonine by the complex of tRNAVal and valyl-tRNA synthetase. Cell 103, 793–803 (2000).
Palencia, A. et al. Structural dynamics of the aminoacylation and proofreading functional cycle of bacterial leucyl-tRNA synthetase. Nat. Struct. Mol. Biol. 19, 677–684 (2012).
Nureki, O. et al. Enzyme structure with two catalytic sites for double-sieve selection of substrate. Science 280, 578–582 (1998).
Chen, X. et al. Structural basis for substrate and antibiotic recognition by Helicobacter pylori isoleucyl-tRNA synthetase. FEBS Lett. 598, 521–536 (2024).
Tukalo, M., Yaremchuk, A., Fukunaga, R., Yokoyama, S. & Cusack, S. The crystal structure of leucyl-tRNA synthetase complexed with tRNALeu in the post-transfer-editing conformation. Nat. Struct. Mol. Biol. 12, 923–930 (2005).
Liu, R. J. et al. Molecular basis of the multifaceted functions of human leucyl-tRNA synthetase in protein synthesis and beyond. Nucleic Acids Res. 48, 4946–4959 (2020).
Holm, L. & Rosenström, P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 (2010).
Deng, X. et al. Large conformational changes of insertion 3 in human glycyl-tRNA Synthetase (hGlyRS) during catalysis. J. Biol. Chem. 291, 5740–5752 (2016).
Ju, Y. et al. X-shaped structure of bacterial heterotetrameric tRNA synthetase suggests cryptic prokaryote functions and a rationale for synthetase classifications. Nucleic Acids Res. 49, 10106–10119 (2021).
Han, L. et al. The binding mode of orphan glycyl-tRNA synthetase with tRNA supports the synthetase classification and reveals large ___domain movements. Sci. Adv. 9, eadf1027 (2023).
Yu, Z. et al. Structural basis of a two-step tRNA recognition mechanism for plastid glycyl-tRNA synthetase. Nucleic Acids Res. 51, 4000–4011 (2023).
Varadi, M. et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 50, D439–D444 (2022).
Miyamoto, Y. et al. Identification of Saccharomyces cerevisiae isoleucyl-tRNA synthetase as a target of the G1-specific inhibitor Reveromycin A. J. Biol. Chem. 277, 28810–28814 (2002).
Orenstein, N. et al. Bi-allelic IARS mutations in a child with intra-uterine growth retardation, neonatal cholestasis, and mild developmental delay. Clin. Genet. 91, 913–917 (2017).
Fersht, A. R. & Kaethner, M. M. Mechanism of aminoacylation of tRNA. Proof of the aminoacyl adenylate pathway for the isoleucyl- and tyrosyl-tRNA synthetases from Escherichia coli K12. Biochemistry 15, 818–823 (1976).
Rueter, S. M., Dawson, T. R. & Emeson, R. B. Regulation of alternative splicing by RNA editing. Nature 399, 75–80 (1999).
Mendoza, H. G. & Beal, P. A. Structural and functional effects of inosine modification in mRNA. RNA 30, 512–520 (2024).
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007).
Hodgson, J. E. et al. Molecular characterization of the gene encoding high-level mupirocin resistance in Staphylococcus aureus J2870. Antimicrob. Agents Chemother. 38, 1205–1208 (1994).
Seah, C. et al. MupB, a new high-level mupirocin resistance mechanism in Staphylococcus aureus. Antimicrob. Agents Chemother. 56, 1916–1920 (2012).
Yanagisawa, T. & Kawakami, M. How does Pseudomonas fluorescens avoid suicide from its antibiotic pseudomonic acid?: Evidence for two evolutionarily distinct isoleucyl-tRNA synthetases conferring self-defense. J. Biol. Chem. 278, 25887–25894 (2003).
Zanki, V., Bozic, B., Mocibob, M., Ban, N. & Gruic-Sovulj, I. A pair of isoleucyl-tRNA synthetases in Bacilli fulfills complementary roles to keep fast translation and provide antibiotic resistance. Protein Sci. 31, e4418 (2022).
Brkic, A. et al. Antibiotic hyper-resistance in a class I aminoacyl-tRNA synthetase with altered active site signature motif. Nat. Commun. 14, 5498 (2023).
Suzuki, T. & Numata, T. Convergent evolution of AUA decoding in bacteria and archaea. RNA Biol. 11, 1586–1596 (2014).
Yi, J. et al. Fragment screening and structural analyses highlight the ATP-assisted ligand binding for inhibitor discovery against type 1 methionyl-tRNA synthetase. Nucleic Acids Res. 50, 4755–4768 (2022).
Kabsch, W. XDS. Acta Crystallogr. D. Biol. Crystallogr. 66, 125–132 (2010).
Evans, P. R. & Murshudov, G. N. How good are my data and what is the resolution? Acta Crystallogr. D. Biol. Crystallogr. 69, 1204–1214 (2013).
Vagin, A. & Teplyakov, A. Molecular replacement with MOLREP. Acta Crystallogr. D. Biol. Crystallogr. 66, 22–25 (2010).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 66, 486–501 (2010).
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D. Biol. Crystallogr. 53, 240–255 (1997).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. Biol. Crystallogr. 66, 12–21 (2010).
Madeira, F. et al. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res. 50, W276–W279 (2022).
Waterhouse, A. M., Procter, J. B., Martin, D. M., Clamp, M. & Barton, G. J. Jalview Version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 (2009).
Acknowledgements
We thank the staff of BL19U1 beamline (https://cstr.cn/31129.02.NFPS.BL19U1) at the National Facility for Protein Science in Shanghai (NFPS), Shanghai Advanced Research Institute, Chinese Academy of Sciences, for providing technical support in data collection and analysis. We also thank Prof. Xiang-Lei Yang from Scripps Research Institute for her helpful discussion on this research. This research was supported by National Natural Science Foundation of China (22177140 to H.Z. and 22207133 to B.C.), Guangdong Basic and Applied Basic Research Foundation (2023A1515012453, 2023B1515040006 and 2022B1515130008 to H.Z., 2023A1515012936 and 2021A1515110117 to B.C.), Guangdong Provincial Key Laboratory of Construction Foundation (2023B1212060022 to H.Z.) and Open Research Funds of the Affiliated Qingyuan Hospital (Qingyuan People’s Hospital), Guangzhou Medical University (202301-303 to H.Z. and H.L.).
Author information
Authors and Affiliations
Contributions
B.C. grew the crystals, solved the structures and performed the functional experiments. F.Y., Z.L., F.L. and S.L. contributed to the biochemical experiments. H.L. and Q.G. contributed to structural analysis and experiment design. B.C. and H.Z. wrote the manuscript. H.Z. supervised this research. All the authors have read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Bernhard Kuhle, Jinwei Zhang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Chen, B., Yi, F., Luo, Z. et al. The mechanism of discriminative aminoacylation by isoleucyl-tRNA synthetase based on wobble nucleotide recognition. Nat Commun 15, 10817 (2024). https://doi.org/10.1038/s41467-024-55183-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-55183-0