Abstract
Large foundation models show promise in biomedicine but face challenges in clinical use due to performance gaps, accessibility, cost, and lack of scalable evaluation. Here we show that open-source small multimodal models can bridge these gaps in radiology by generating free-text findings from chest X-ray images. Our data-centric approach leverages 697K curated radiology image-text pairs to train a specialized, ___domain-adapted chest X-ray encoder. We integrate this encoder with pre-trained language models via a lightweight adapter that aligns image and text modalities. To enable robust, clinically relevant evaluation, we develop and validate CheXprompt, a GPT-4-based metric for assessing factual accuracy aligned with radiologists’ evaluations. Benchmarked with CheXprompt and other standard factuality metrics, LLaVA-Rad (7B) achieves state-of-the-art performance, outperforming much larger models like GPT-4V and Med-PaLM M (84B). While not immediately ready for real-time clinical deployment, LLaVA-Rad is a scalable, privacy-preserving and cost-effective step towards clinically adaptable multimodal AI for radiology.
Similar content being viewed by others
Introduction
Foundation models, trained on massive amounts of unlabelled data using self-supervised learning, enable rapid adaptation to various downstream tasks with minimal requirement for task-specific labeled data1,2,3. Due to the high cost of annotating biomedical data4,5, foundation models are poised to become a new paradigm in biomedicine, achieving state-of-the-art results on many applications, including medical question answering2,6 and medical image classification7,8. Recently, multimodal generative artificial intelligence (AI) has emerged as an exciting frontier in the biomedical ___domain, expanding the application scope from single-modality to multi-modality (e.g., text and image), such as visual question answering and radiology report generation6,9,10. While existing models are still largely evaluated on artificial biomedical benchmarks, their promising performance demonstrates their potential in clinical applications.
However, there are still major bottlenecks hindering the use of foundation models in real-world clinical settings. First, sharing patient data with large foundation models hosted on the cloud is subject to privacy concerns11. Therefore, clinicians may prefer to run inference and fine-tuning locally. Second, existing state-of-the-art models are often very large and resource-intensive, which makes local deployment challenging. Smaller models incur smaller carbon footprint12 and offer reduced serving costs and latency, which is of particular importance in resource-constrained settings outside of data centers13. However, while small language models have shown success in text domains14,15,16, small multimodal models (SMMs) still have significant performance gaps compared to larger models6,10. Third, many state-of-the-art models are inaccessible13, necessitating the development of effective open-source models for biomedicine. Finally, even the best models are still subject to errors such as hallucination, and existing automated evaluation methods of factual correctness exhibit limited correlation with expert assessments17. Hence it is crucial to develop reliable methods to evaluate the correctness of model outputs at scale, especially in the specialized field of biomedicine18.
We focus our study on identifying key findings from chest X-ray (CXR) images, the most commonly performed medical imaging examination. Automatically drafting high-quality radiology reports is a challenging but clinically meaningful task that could directly increase radiologist productivity and potentially improve communication and decrease burnout19. Existing frontier models such as GPT-4V still have a large performance gap even on such a fundamental medical application. To bridge this gap between existing medical foundation models and real-world clinical applications, we have developed LLaVA-Rad, a SMM that attains state-of-the-art performance in standard radiology imaging tasks (Fig. 1), in addition to CheXprompt, an automated scoring metric for factual correctness. To develop LLaVA-Rad, we adopt a modular approach by incorporating state-of-the-art open-source pretrained models for image and text modalities, and focusing on training a lightweight adapter to ground each modality to the text embedding space.
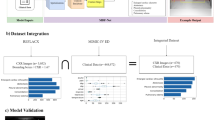
a To train LLaVA-Rad, we assemble a large dataset with over 697 thousand chest X-ray image-text pairs; GPT-4 is used to synthesize reports from labels, translate reports from Spanish, and process and structure the corresponding radiology reports. b We adopt a modular three-stage approach to train LLaVA-Rad, comprised of pre-training, alignment and fine-tuning. c A qualitative visualization of the model’s attention during its generative process. d For evaluation, we also propose a novel factual error scoring approach using GPT-4 and demonstrate its parity with expert evaluation. e LLaVA-Rad outperforms much larger generalist and specialized models like GPT-4V and Med-PaLM M on prior standard report evaluation metrics. MLP multi-layer perceptron. The example chest X-ray image in b is obtained from ref. 27 with permission for reproduction from the authors.
For training, we assemble a large dataset comprising 697,435 radiology image-report pairs from 7 diverse sources. Some data sources only contain structured labels of key findings, in which case we use GPT-4 to synthesize the report based on the ground-truth labels. For evaluation, we report standard metrics such as n-gram-based BLEU and ROUGE, as well as factuality checks based on CheXpert and RadGraph20,21. Additionally, we propose CheXprompt, a factuality evaluation method based on GPT-4. Compared to existing automated metrics, we show that CheXprompt is more consistent with error quantification by practicing radiologists, thus demonstrating the potential of using GPT-4 for evaluation in a manner that is both scalable and highly relevant to medical practice. To establish best practices for biomedical multimodal learning, we conduct a systematic ablation study on various choices in data engineering and multimodal training.
The resulting LLaVA-Rad (7B) model attains state-of-the-art results on standard radiology tasks such as report generation and cross-modal retrieval, even outperforming much larger models such as GPT-4V and Med-PaLM M (84B)6. LLaVA-Rad inference is fast and can be run on a single V100 GPU in private settings, offering a promising state-of-the-art tool for real-world clinical applications. In addition, LLaVA-Rad training is also very efficient, taking just one day on over 697 thousand image-text pairs using a standard 8-A100 cluster. This means that clinicians can further efficiently fine-tune the model as needed using their private data. By examining the model weights, we found that LLaVA-Rad can ground key regions of abnormalities to generated words in the output report, which signifies future opportunities to synergize with the latest progress in biomedical segmentation and grounded report generation.
In summary, we develop LLaVA-Rad, a lightweight yet high-performing radiology multimodal model for clinical applications. The promising performance of LLaVA-Rad shows that its underlying modular approach can effectively and efficiently bridge the multimodal performance gap in existing frontier models, enabling clinical access with limited computational resources.
Results
Overview of LLaVA-Rad
LLaVA-Rad represents an emerging paradigm in exploring SMMs, following the proliferation of small language models (Fig. 1). Our intuition for designing LLaVA-Rad is that a lightweight, specialized SMM can be efficiently developed by decomposing training into unimodal pretraining on individual modalities followed by lightweight cross-modal learning focusing on a small adapter to ground a non-text modality to the text embedding space.
LLaVA-Rad can generate radiology report findings given a CXR image. Its training comprises three stages: a pre-training stage, an alignment stage, and a fine-tuning stage. In the first stage of pre-training, we train a ___domain-specific vision encoder (BiomedCLIP-CXR) by using 697 thousand pairs of CXR images and associated radiology reports from 7 diverse datasets22,23,24,25,26,27,28. These de-identified image-text pairs were sourced from approximately 258,639 patients. Since CXR images are often published with a limited number of associated findings or image labels instead of a complete report, we used GPT-4 to synthesize a report based on annotated image labels. Alternatively, reports may be available in other languages, such as the PadChest reports, which are available in Spanish, for which we leverage GPT-4 to translate them into English. We also exploit GPT-4 to extract findings from reports when the finding section cannot be reliably extracted using existing rule-based heuristics27. For examinations where only the CXR images are available, we studied the quality of these synthetic reports by measuring their alignment with their corresponding image using CXR-specific vision language models (Supplementary Table 1). We found that GPT-4-generated synthetic reports show significantly higher alignment compared to random, particularly in datasets with more granular labels (i.e. PadChest, VinDR, CheXpert, and Brax). Alternatively, for CheXpert reports -released after our model training- we found that while synthetic reports exhibit high overall similarity to their reference (cosine similarity 0.73), they contain on average 3.78 total errors (Supplementary Table 2). Finally, when comparing GPT-4 extracted findings to rule-based extraction, we found that GPT-4 findings exhibit high similarity (cosine similarity 0.93) and minimal error counts (0.20 average total errors).
In the second stage of alignment, we align the pre-trained vision encoder BiomedCLIP-CXR with a language model. In this alignment stage, we train a conditional generative decoder model that generates radiology report findings given an input CXR. We provide a CXR as the input, without any associated contexts such as clinical instructions or patient information. As noted by other works9,29 and also demonstrated by our ablation studies, this strategy can substantially improve the alignment by forcing the decoder model to focus on the image alone. In the third stage, we fine-tune the model to generate the findings given both the indication for the exam and the image, more closely reflecting the real-world setting. LLaVA-Rad exploits an efficient technique LoRA30 for fine-tuning, thus substantially reducing the computational time required for this stage. We further reduce the computational time by only using MIMIC-CXR training data instead of the entire 697 thousand image-text pairs in the second and third stages, since reports in MIMIC-CXR are of higher quality. The three stages of LLaVA-Rad can be finished in 8 hours, 4 hours, and 16 hours, respectively, using 8 A100 GPUs. We studied the effects on generalizability of our fine-tuning process using MT-bench, a general ___domain evaluation of language model capabilities31. Overall, we found minimal impact of our training regime on underlying language model capabilities (Supplementary Fig. 1). LLaVA-Rad showed moderate improvements in reasoning, at the expense of slightly lower quality of text extraction.
Evaluating LLaVA-Rad using existing report generation benchmarks
We evaluated LLaVA-Rad on the test set of the widely used radiology report generation benchmark MIMIC-CXR using metrics assessing lexical similarity and factual accuracy (Fig. 2, Supplementary Fig. 2). In particular, lexical similarity metrics, such as BLEU and ROUGE, are used in traditional natural language processing to assess the model’s ability to produce contextually and stylistically aligned output. On the other hand, factual accuracy metrics, including CheXbert-based and RadGraph-based F1 scores20,21 are more clinically relevant because they gauge the extent to which the generated reports accurately reflect imaging findings.
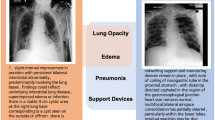
a Comparison between LLaVA-Rad and open-source models according to existing factual correctness (F1-CheXbert-14, F1-RadGraph) and lexical similarity (ROUGE-L) metrics. b Comparison between LLaVA-Rad and closed-source models according to existing factual correctness and lexical similarity metrics. c Comparison between model size and factual correctness shows that LLaVA-Rad is both smaller and more factually correct compared to existing approaches. d Illustration of a sample generated report from LLaVA-Rad compared with that of LLaVA and LLaVA-Med. LLaVA-Rad’s generations that match reference findings are highlighted. e Comparison of the performance on cross-modal retrieval demonstrated by LLaVA-Rad, LLaVA-Med and LLaVA. In a–e values correspond to mean statistic in MIMIC-CXR test-set (n = 2461 image-report pairs) with the exception of MAIRA-1 and Med-PaLM M which are derived from their original publications. In a, b error bars correspond to 95% bootstrap confidence intervals derived from 500 samples. Source data are provided as a Source Data file.
We found that LLaVA-Rad achieved superior performance on both groups of metrics (Fig. 2a–d). When compared to other models of equivalent size (7B parameters), such as LLaVA-Med9, CheXagent32 and MAIRA-133, LLaVA-Rad demonstrates significant advancements in performance across all evaluated metrics. Furthermore, LLaVA-Rad is more efficient than the current overall leading model, Med-PaLM M6, despite having an order of magnitude fewer parameters. This efficiency does not come at the cost of effectiveness; LLaVA-Rad outperforms Med-PaLM M in the most important existing lexical similarity and factual correctness metrics for radiology text (ROUGE-L and F1-RadGraph34, with a relative improvement of 12.1% and 10.1% respectively). A more detailed evaluation (Supplementary Table 3) shows that Med-PaLM M marginally surpasses LLaVA-Rad by F1-5 CheXbert metrics, which assess only a small subset of 5 potential abnormalities, and the performance gap is minimal (<1% relative improvement). Most of these competing models also use MIMIC-CXR for training (with the notable exception of LLaVA-Med). We attribute the promising performance of LLaVA-Rad to its modular design, which is more data efficient. The efficiency and the high degree of factual and lexical precision of LLaVA-Rad demonstrate its potential in real-world applications where large models are computationally too costly.
We studied the performance of LLaVA-Rad on other held out datasets: CheXpert22, Open-I35, and US-CXR, a private collection of 1,751 CXRs and reports sourced from various hospitals in the United States. Notably, only the CheXpert training images were used for pretraining the image encoder, while Open-I and US-CXR were entirely new to the model, allowing us to study its robustness and adaptability. We report summary statistics for each dataset in Supplementary Table 4, illustrating the varying prevalence of common findings in CXRs across datasets. Similar to the evaluation on MIMIC, We also employ CheXbert-14, F1-RadGraph, and ROUGE-L to assess the factual accuracy and lexical similarity of the reports on CheXpert, Open-I, and US-CXR. As illustrated in Fig. 3, LLaVA-Rad significantly outperforms LLaVA-Med, LLaVA, and GPT-4V across all metrics on these datasets, revealing that LLaVA-Rad’s superior performance is consistent across various settings.
Open-I (a, b) CheXpert (c, d) and US-CXR (e, f). LLaVA-Rad outperforms baselines across all external validation datasets, as assessed by traditional factual correctness metrics (F1-CheXbert-14, F1-RadGraph) and lexical similarity (ROUGE-L). CheXprompt evaluation (b, d, f) further demonstrates that LLaVA-Rad produces fewer clinically significant and overall errors compared to baselines. Each dataset sample consists of image-report pairs (Open-I: n = 2163; CheXpert: n = 61; US-CXR: n = 1751). Values represent mean metric scores for each dataset, and error bars indicate 95% bootstrap confidence intervals derived from 500 resampling iterations. Source data are provided as a Source Data file.
Finally, to verify the effectiveness of our approach in generating aligned vision and language representations, we examined the learned image encoder in LLaVA-Rad by comparing the performance of using it for retrieval against the image encoders from LLaVA and LLaVA-Med. We observed that BiomedCLIP-CXR attained the best results on both image-to-text and text-to-image retrieval, indicating the high quality of its image encoder by training on 697 thousand text-image pairs (Fig. 2e). Moreover, LLaVA-Med performed better than LLaVA, suggesting that better performance can be gained as increasing ___domain specialization is performed.
Evaluating LLaVA-Rad using CheXprompt, a GPT-4-based evaluation system
It is well known that existing n-gram and findings-based automated report evaluation methods might be biased to pre-defined conditions and have limited correlation with expert assessments17. We thus explore the utility of an large language model-based evaluation system, which has shown success in other domains36,37,38. Specifically, we employ GPT-4 as an evaluator to count how often the generated report contains errors in each of the following six categories, as per a previous study17: false positive finding, omission of finding, incorrect ___location/position of finding, incorrect severity of finding, mention of comparison that is not present in the reference report, omission of comparison describing a change from a previous study. For each error type, we further instruct GPT-4 to distinguish clinically significant and clinically insignificant errors.
We first assessed the rigor of CheXprompt by examining its consistency with expert scoring. To this end, we exploited the ReXval dataset39, which contains annotations from 6 board-certified radiologists on 200 pairs of ground-truth reports from MIMIC-CXR and AI-generated reports. In ReXval, each radiologist counts each of the aforementioned errors in the generated report, also discriminating between clinically significant and insignificant errors. We found that GPT-4-based evaluations were highly correlated with expert scoring by achieving Kendall’s Tau-b correlations greater than 0.75 for total errors and greater than 0.70 for clinically significant errors (Fig. 4b). In contrast, none of the existing preferred metrics (ROUGE-L metric, RadGraph-F1, and RadCliQ) obtained a correlation greater than 0.57. Moreover, we found that a similar evaluation system using GPT-3.5 Turbo, a less capable model compared to GPT-4, attains a much lower association with expert scoring due to an overestimation of the number of total and clinically significant errors, demonstrating the difficulty of automatically scoring radiology reports.
a GPT-4 based CheXprompt is more similar to average left-in radiologists in total error quantification, compared to the left-out radiologist (mean absolute difference 0.55 vs 0.71). b Comparison between CheXprompt and existing metrics in terms of agreement with radiologist error quantification. c Comparison between LLaVA-Rad and competing methods using CheXprompt on the MIMIC-CXR test set. d Illustration of how CheXprompt can be used to evaluate a report generated by LLaVA-Rad, with errors highlighted. GPT-4T stands for GPT-4 Turbo. In a p values correspond to two-sided paired t-test. In b, c values represent mean metric scores and error bars correspond to 95% bootstrap confidence intervals. Source data are provided as a Source Data file.
Beyond correlation, we performed a head-to-head comparison of the calculation of total errors as determined by GPT-4 Turbo with manual radiologist ratings in a leave-one-rater-out fashion. Fig. 4a summarizes the results of this comparison, which on average shows a mean absolute difference (MAD) of 0.71 between the left-out human rater and the average of the remaining ones, whereas GPT-4 Turbo has on average 0.55 MAD. We find that the MAD between GPT-4 Turbo CheXprompt score and the left-in expert average is smaller compared to the left-out expert in 3 out of 6 cases (P < 0.001), and not significantly different (P > 0.05) in the remaining 3 out of 6 cases. Altogether, we find that GPT-4 Turbo is indistinguishable from expert raters in calculating the total number of errors, increasing our confidence in using this proposed automated metric as an evaluation method that directly aligns with expert opinions.
After assuring the effectiveness of the GPT-based metric, we evaluated the performance of LLaVA-Rad on the held-out MIMIC-CXR test set using CheXprompt(Fig. 4c, d). In line with our observation using existing metrics, LLaVA-Rad outperforms publicly available report generation models, generating fewer clinically significant and total errors compared to GPT-4V and CheXagent. Moreover, by comparing models within the LLaVA family (e.g., LLaVA-Rad, LLaVA-Med, LLaVA), we observed that fewer errors are made in the generated reports as increasing ___domain specialization is performed. In particular, LLaVA-Rad generates fewer errors than LLaVA-Med, a LLaVA model tailored to medicine, and LLaVA-Med generates fewer errors than the general-___domain model LLaVA. This suggests a trade-off between ___domain-specific performance and broad applicability, supporting our intuition of developing LLaVA-Rad by continual pretraining of a general model using large amounts of ___domain-specific data.
Finally, to determine the clinical utility of LLaVA-Rad, we explore using the percentage of error-free reports to track the overall performance of report-generation models. A higher percentage of error-free reports increases the utility of a report generation model, given that it directly reflects the number of reports that require little to no radiologist modification following automated generation. Notably, LLaVA-Rad has the highest percentage of error-free reports, with 6.79% reports free of clinically significant errors, and 2.58% free of errors (Fig. 3c). The same trend of improved performance of LLaVA-Rad was observed in the external validation datasets (CheXpert, Open-I, and US-CXR), where LLaVA-Rad demonstrated fewer clinically significant and total errors (Fig. 3b, d), with up to 26% error-free reports. While this stringent metric allows us to estimate overall proportion of error-free reports, we find that when studying the percentage of error-free reports as determined by each of the six CheXprompt error types, LLaVA-Rad can achieve >50% error-free reports across the majority of error types (Supplementary Table 5). To further understand types of errors made by LLaVA-Rad, we report sensitivity and specificity values for common CXR findings in Supplementary Table 6, which overall show that LLaVA-Rad favors high specificity for common findings, at the expense of varying levels of sensitivity. This observation applied to all four evaluation datasets, and is is in line with granular error counts as determined by CheXprompt, reported in Supplementary Table 5, which show that the most common form of error is clinically significant false negatives, followed by incorrect assessments of severity. Overall, CheXprompt illustrates that while there undoubtedly remains room for improvement in fully automated CXR radiology report generation, the improvement demonstrated by our model is promising.
Analyzing components of LLaVA-Rad using ablation and case studies
Conducting thorough ablation studies for large language and multimodal models is often intractable due to the costly training of multiple variants. In contrast, the small size of LLaVA-Rad enables us to efficiently conduct ablation studies that explain the promising performance of LLaVA-Rad and potentially inform design choices for larger models. We compared LLaVA-Rad with 8 variants described in Supplementary Table 8. In particular, we investigate two key technical ideas used in LLaVA-Rad: the effect of pre-training a ___domain-specific image encoder using 697 thousand diverse CXR image-text pairs (Fig. 5a) and the effect of using GPT-4 to augment and organize the data (Fig. 5b). First, to understand the effect of pre-training an image encoder, we compare LLaVA-Rad with three increasingly ___domain-specific variants: an image encoder from OpenAI CLIP, an image encoder using BiomedCLIP, and an image encoder from BiomedCLIP but continually pre-trained using the only the training split of MIMIC-CXR (Fig. 5a). We did not find noticeable overlap between MIMIC-CXR training split and the PubMed data used to pre-train BiomedCLIP. We found that the MIMIC-CXR-based image encoder (BiomedCLIP-MIMIC-CXR) outperforms the other two variants, indicating the effectiveness of training a ___domain-specific image encoder. In addition, BiomedCLIP-CXR outperforms BiomedCLIP-MIMIC-CXR, illustrating the advantage in pre-training using more diverse CXR image datasets and their corresponding synthetic reports. Second, we studied the effect of using GPT-4 to process and augment the MIMIC-CXR report data (Fig. 5b). The data used to train our model in the second and third stages is a combination of rule-based and GPT-4 structured data, as summarized in Supplementary Table 7. We compare LLaVA-Rad with a variant that only uses rule-based data and a variant that only uses GPT-4-structured data. We found that LLaVA-Rad attains a better performance than both variants, indicating the effectiveness of GPT-4 data augmentation. The variant that only uses GPT-4-structured data outperforms the one that only uses rule-based data on factual accuracy metrics, confirming the effectiveness of GPT-4-based structuring in generating clinically precise reports (Supplementary Table 8). Finally, it is expected that rule-based variant outperforms GPT-4-structured variant on n-gram lexical metrics, because the test data is also from rule-based data. These ablation studies support our intuition that ___domain-specific data can help us build a small but effective ___domain-specific model, and help inform best practice in training larger models.
a Comparison of using different image encoders (BiomedCLIP-CXR from LLaVA-Rad, BiomedCLIP continually pre-trained on MIMIC-CXR, BiomedCLIP, and OpenAI CLIP) to start the alignment and fine-tuning stages. b Ablation study on only using rule-processed MIMIC-CXR training data or GPT-4 processed training data in alignment and fine-tuning stages. c Attention visualization qualitatively demonstrates the appropriate grounding of LLaVA-Rad in-specific image regions when generating a word (bold text) as part of a specific finding (bottom row). In a, b values represent mean metric scores and error bars indicate 95% bootstrap confidence intervals derived from 500 resampling iterations. Source data are provided as a Source Data file.
We also developed a method to investigate how LLaVA-Rad’s attention map on the input image correlates with a given generated word in the report (Fig. 5c), which demonstrates the model’s ability to focus on relevant image regions for the generation. A detailed examination reveals a significant variability in attention across different layers and attention heads, with different configurations gravitating towards distinct regions of the image (Supplementary Figs. 3, 4, 5, 6, 7). Our evaluation also identifies that the aggregation of attention, particularly through averaging the outputs of all heads within the 20th layer, generally yields the most coherent and relevant focal points across a wide array of scenarios. However, this approach does not uniformly apply, as deviations in alignment were observed in certain instances. Conversely, an alternative strategy of taking the maximum across all layers, coupled with an average across heads, demonstrates a consistently high correlation with pertinent image regions. Our proposed attention visualization indicates a strong alignment between the model’s attention and the specific image regions relevant to the generated words. This alignment underscores the model’s efficacy in synthesizing contextual information from visual cues to ground its linguistic output.
Discussion
To address the significant challenges of developing foundation models for real-world clinical settings, our work introduces LLaVA-Rad, a lightweight radiology SMM that offers open-source accessibility while attaining state-of-the-art results in the ___domain of CXR report generation. By curating a dataset of 697 thousand CXR images paired with radiology reports from diverse sources, using GPT-4 for generating synthetic data, coupled with a modular three-stage curriculum training method, we have developed a model that outperforms its larger counterparts, such as GPT-4V and Med-PaLM M, and demonstrates exceptional proficiency in generating accurate and lexically similar radiology reports on the evaluation datasets. Through our attention visualization techniques, LLaVA-Rad offers deep insights into how it prioritizes key regions in chest X-rays, correlating them with specific findings in the generated reports. Furthermore, our work introduces CheXprompt, which successfully resolves a major bottleneck in the automated evaluation of factual accuracy of generated radiology reports, by not only demonstrating a closer alignment with manual expert scoring compared to traditional metrics, but also exhibiting performance on par with expert radiologist annotators. This improved evaluation further illustrates LLaVA-Rad’s superiority in clinical report generation.
The landscape of AI-driven radiology report generation has evolved significantly with the advent of transformers and large multimodal models, ushering in a new era of more sophisticated and accurate models33,40,41,42,43,44,45. R2Gen stands out as a pioneering effort in leveraging memory-efficient transformers for report generation46. A notable leap forward is CheXagent32, which leverages an instruction fine-tuned foundation model trained across 28 publicly available datasets, demonstrating an enhanced capability for analyzing and summarizing CXR images. Concurrently, Flamingo-CXR fine-tuned the Flamingo vision language model47 and incorporated regularization and adaptation techniques to tailor their applications to the nuances of radiology report generation10. Med-PaLM M pushed the boundaries by creating a versatile 84-billion-parameter biomedical AI system capable of addressing multiple tasks across various medical modalities6. In contrast to these advancements, our method, LLaVA-Rad, distinguishes itself by not only achieving superior performance across several benchmark metrics but also by being comparatively lightweight. We refer to LLaVA-Rad as a highly performant SMM due to its small size compared to other large language model approaches with tens of billions of parameters and its ability to run on local hardware such as V100 GPUs. This attribute is particularly important, as it offers a more accessible and efficient solution for scaling radiology report generation, addressing both the need for factual correctness and the practicality of deployment in clinical settings. Furthermore, the data-centric focus of our work distinguishes our approach from prior models.
In line with the growing recognition of the importance of data-centric AI, our study emphasizes the systematic engineering of high-quality data as a key element in building robust AI systems. Specifically, we introduce: (1) a paradigm for creating synthetic radiology reports for datasets lacking publicly available reports, addressing a major limitation in publicly available CXR data, and (2) the use of a large language model (GPT-4) to automate the parsing of radiology reports into standardized sections while removing references to prior images. These data-centric contributions enable us to effectively train BiomedCLIP-CXR and LLaVA-Rad, building on the successful frameworks of Biomed-CLIP and LLaVA. Our ablation study (Fig. 5, Table 8, variants 1, 6–8) validated the effectiveness of this pre-training strategy, showing a 5-10% relative improvement in performance across multiple evaluation metrics compared to various baselines. Our analysis of the quality of generated reports (Supplementary Table 1 and 2) indicates that GPT-4-generated synthetic reports exhibit significantly higher quality than random reports, particularly in datasets with granular labels such as PadChest, VinDR, CheXpert, and BraX. However, comparison of our synthetic reports to ground-truth reports on the CheXpert dataset, showed that while the synthetic reports have high similarity, they still may contain errors, likely due to the non-exhaustive label availability and inherent limitations of synthetic label-based descriptions that lack localization and other nuanced details. These evaluations illustrate the added value and limitations of synthetic GPT-4 generated reports, which improve overall model performance, particularly in identifying common findings in CXRs. By utilizing models like GPT-4 to synthesize high-quality large-scale pre-training data, our approach advances automated CXR report generation and may be useful in other medical domains.
Generative models are subject to producing inaccurate statements, an important concern in a factuality-focused ___domain such as radiology. Our work proposes methods to identify and mitigate potential errors in generated radiology reports. For identification, we develop CheXprompt, which can identify various kinds of inaccuracies, such as false positive or false negative findings. While CheXprompt can quantify such errors, its flexibility also allows us to understand the types of errors most frequently observed (Supplementary Table 5). Furthermore, to mitigate the ocurrence of such errors in the first place, we leverage the GPT-4 processing of reports, in which we remove references to prior examinations. Despite this removal, since our training datasets comprise both GPT-4 processed and rule-based processed reports (which include references to prior images), LLaVA-Rad may still produce such comparison errors. Our results show these occur relatively infrequently when compared to the rule-based ground truth (for significant and insignificant errors, on average 0.01 and 0.05 false positive comparisons per report, corresponding to 99.3% and 95.1% of reports without false positive comparisons). Altogether, our synthesis of training data and CheXprompt enable error minimization and detection.
Overall, LLaVA-Rad and CheXprompt provide opportunities to enhance automated generation of draft CXR findings. We anticipate that models such as LLaVA-Rad can help draft the bulk of the report, and such drafts can be quickly edited by specialists, or alternatively compared with draft radiologist reports to identify potential discrepancies. LLaVA-Rad favors specificity over sensitivity, suggesting its potential use as a confirmatory diagnostic tool during CXR report generation. Furthermore, CheXprompt can be used to uncover and quantify potential errors in draft reports, allowing the development of interfaces that highlight errors, facilitating both model development and human-in-the-loop applications.
While LLaVA-Rad represents a substantial advancement in radiology multimodal models, our research acknowledges several areas for future exploration and improvement. First, the current scope of LLaVA-Rad is limited to CXRs. While CXR is the most common medical image examination, future iterations should evaluate the feasibility of our method on alternative anatomies (e.g., abdomen or extremities) and modalities (e.g., computed tomography or ultrasound) to enhance the model’s applicability and utility across diverse application scenarios. In addition, while our ablation studies enable us to understand the importance of training a specialized image encoder, and examine the impact of various types of training data, image encoders, and input resolutions, it is possible that other language model sizes or pre-training strategies may further benefit ___domain-specific performance. While text-only evaluations have studied the strengths and limitations of varying language model sizes and pre-training data14,48, further exploration of SMMs appropriately controlling for language model pre-training data, number of parameters, and compute could further clarify the role of the underlying language model in multimodal performance. Regarding model interpretability, attention-based attribution methods have been found to be more effective at explaining model decisions and to be more useful by radiologists49, and our attention visualization technique does appear to highlight sensible patterns. However, alternative saliency-based methods for CXR interpretation algorithms such as Grad-CAM have been shown to have limited correlation with expert assessments and limited robustness to input perturbations50,51. There is a pressing need for a more exhaustive evaluation of such grounding strategies. These would further improve the model’s explainability and interpretability, making it more transparent and trustworthy for clinical use. Another consideration is the inherently multimodal nature of modern medical practice, which integrates various patient information streams, including historical medical images, medical records, lab tests, and vital signs. Integrating these diverse and longitudinal data sources into medical multimodal models like LLaVA-Rad could significantly enrich the model’s understanding and analysis, leading to more nuanced and holistic patient assessments.
LLaVA-Rad illustrates the potential of SMMs in significantly enhancing CXR interpretation by improving diagnostic accuracy and potentially providing radiological expertise in underserved regions. However, its use raises important concerns around biases and limitations. Biases present in the training data, such as underrepresentation of specific demographics, may result in disparities in performance across different population groups. To address this, we included diverse, publicly available datasets during model pre-training. Additionally, the model’s generalizability may vary when applied to populations outside its training scope, as indicated by our external validation results. An important contributing factor to the observed variability in the number of errors across datasets is the varying prevalence of disease in underlying evaluation populations (critically ill or hospitalized patients in MIMIC-CXR and CheXpert, compared to other populations including outpatients in OPEN-I and US-CXR). This further enhances the need to consider the potential differences in performance of LLaVA-Rad based on the deployment population. Another consideration is the risk of automation bias, where over-reliance on AI systems could reduce critical oversight by human clinicians. Therefore, ethical considerations–including transparency, accountability, and adherence to regulatory standards–are essential for responsible deployment. LLaVA-Rad is designed as a supportive tool to assist clinicians by generating draft CXR findings, and is not meant for replacing human expertise.
LLaVA-Rad exemplifies a significant leap toward making advanced diagnostic capabilities accessible with limited computational resources, thus paving the way for broader clinical applications and impact. The pursuit of open-source, lightweight, high-performing models that not only extends to various medical imaging types but also incorporates multimodality and interpretability, embodies the next frontier in medical multimodal model development. Such advancements will bridge the gap between current technological capabilities and the real-world demands in clinical applications, moving us closer to achieving meaningful improvements in patient outcomes.
Methods
Ethics Statement
This study utilized both public datasets and private de-identified data sources. The research was exempt from institutional review board oversight under relevant institutional and federal guidelines. All data were handled in accordance with rigorous ethical standards, including full adherence to applicable data use agreements and licensing requirements. As this study did not involve direct human participant recruitment, participant compensation and informed consent were not applicable.
Sex or gender information are available in the corresponding demographic reporting of each of the MIMIC-CXR, CheXpert, and Open-I datasets. However, these data were not used in the study design or analysis, as the primary objective was to evaluate the overall quality of model-generated findings across diverse datasets, rather than to assess subgroup differences. The US-CXR dataset does not contain sex or gender information, as it has been fully de-identified in accordance with privacy regulations.
Details of the dataset
CXR-697K: We compiled a comprehensive dataset comprising 697 thousand pairs of CXR images, each accompanied by its corresponding radiology report, for pre-training the image encoder of LLaVA-Rad. This dataset amalgamates data from seven publicly available datasets as summarized in Supplementary Table 9. To maintain transparency and reproducibility, we adhere to the original train/val/test splits provided by each contributing public dataset, using only the train split for pre-training the image encoder.
The CheXpert dataset22 consists of retrospectively collected chest radiographic studies conducted between October 2002 and July 2017, encompassing both inpatient and outpatient centers at Stanford Hospital. BraX23, obtained from chest radiography studies at Hospital Israelita Albert Einstein in \({{{\rm{S}}}}\tilde{a}{{{\rm{o}}}}\) Paulo, Brazil, was labeled for 14 radiological findings using the CheXpert Label Extraction Algorithm22, which was adapted to detect findings in Portuguese for this dataset. CandidPTX24 encompasses data acquired between January 2010 and April 2020 from Dunedin Hospital in New Zealand. This dataset’s chest radiographs were manually annotated by radiology trainees and radiologists with respect to pneumothoraces, acute rib fractures, and intercostal chest tubes. VinDR25 was gathered from HMUH and H108 hospitals in Vietnam between 2018 and 2020, with images labeled for six diagnoses by multiple experienced radiologists from these institutions. JF Healthcare26 data was collected from approximately 300 township hospitals in China and manually annotated by multiple radiologists to identify foreign objects within the lung field on CXRs. The aforementioned datasets are comprised of images and associated binary labels that indicate whether common disease entities such as pneumonia, or pneumothorax are present in the image. However, they lack free-text reports. Thus, to enable pre-training of our image encoder using image and text methods, we create synthetic reports grounded on the labels provided. Detailed templates used for this synthetic rule-based generation can be found in Supplementary Table 10.
PadChest28 encompasses CXRs interpreted and reported by 18 radiologists at the Hospital Universitario de San Juan, Alicante (Spain), covering the period from January 2009 to December 2017, alongside their corresponding reports in Spanish. For this data, we harness the capabilities of GPT-4 to translate these reports into English, ensuring linguistic consistency. MIMIC-CXR comprises images and their corresponding radiology reports sourced from radiographic studies conducted at the Beth Israel Deaconess Medical Center in Boston, MA, spanning the years 2011 to 201627.
MIMIC-CXR free-text reports are utilized for training the text-generation component of LLaVA-Rad. For each report, we extract the Indication, Findings, and Impression sections. To do so, we employ rule-based heuristics as supported by the official MIMIC code repository.
Extracting reports in this rule-based manner poses two challenges. First, report structure varies within the dataset, with use of different section headers, merging of findings and impression into the same section, etc., which limits the availability of reports with findings available. Second, reports often contain references to prior examinations, such as “heart size remains unchanged". This poses a challenge for training report generation systems which often hallucinate references to prior examinations that are not available at inference time52. To mitigate these challenges, we leverage GPT-4 to extract the reason for exam, findings, and impression sections in the free-text reports from MIMIC-CXR. Prompt templates used to instruct GPT-4 for the organization are elaborated in Supplementary Table 11. Compared to the standard MIMIC-CXR rule-based extraction method, GPT-4 demonstrates proficiency in enhancing report quality by addressing issues like grammar errors, broken words, and synonymous section headers, while at the same time eliminating redundant phrases and references to previous exams. Supplementary Table 12 showcases examples of sections structured by GPT and those extracted through rule-based methods. The use of GPT to extract sections augments rule-based data by an additional 237, 073 image-text pairs for the training split and 1, 952 for the validation split, as summarized in Supplementary Table 7.
Modeling approach
Image encoder
The first stage of training LLaVA-Rad consists of pre-training the image encoder. Within the LLaVA framework, the image encoder plays a pivotal role in extracting complex image representations, crucial for tasks such as automated medical report generation where standard vision transformer models often do not capture the necessary detail and nuanced representations (Supplementary Table 13). To overcome this, we pretrain a ___domain-specific vision encoder, named BiomedCLIP-CXR, and integrate it into LLaVA to bolster its medical image analysis capabilities. Our method includes several key enhancements: firstly, we increase the image input resolution to 518px, substantially higher than the 224px or 336px resolutions typically used in LLaVA-Med, to capture more detailed image features. Secondly, we compile the CXR-697K dataset, an extensive collection of over 697 thousand CXR images from various sources, providing a rich foundation for pretraining. Lastly, we employ the BiomedCLIP recipe for training BiomedCLIP-CXR, which involves contrastive vision-language training with PubMedBERT, a text encoder specialized for the medical ___domain53. The initialization of our vision encoder uses a DINOv2 model checkpoint, benefiting from its extensive training on a diverse set of 142 million general-___domain images54.
Small multimodal model
LLaVA-Rad leverages the capabilities of a pre-trained image encoder and a pre-trained language model to create a SMM. We choose BiomedCLIP-CXR as our image encoder and Vicuna-7B-v1.555 as our language model. A multi-layer perceptron (MLP) is introduced to project image features extracted by the image encoder into the word embedding space of the language model. Conditioned on the projected image features (visual tokens) and textual tokens, LLaVA-Rad generates text in an autoregressive manner. We refer the reader to LLaVA29,56 for a more in-depth description of the model architecture.
Training strategy
Due to the introduction of our ___domain-specific image encoder, BiomedCLIP-CXR, LLaVA-Rad is not initialized with the pre-trained LLaVA weights. Instead, we initialize LLaVA-Rad with the pre-trained image encoder BiomedCLIP-CXR, the pre-trained language model Vicuna-7B-v1.5, and a randomly initialized MLP.
The second and third stages for training LLaVA-Rad are carried out similarly to LLaVA and LLaVA-Med, consisting of feature alignment and end-to-end fine-tuning, respectively. Given a set of training examples, where each example consists of a CXR Xv and the corresponding indication section Xi and finding section Xf from the processed report, the training procedure is described as follows:
In stage two (feature alignment), we freeze the image encoder and the language model, and only update the MLP projection layer. Given a CXR Xv, we train LLaVA-Rad to generate the corresponding findings section Xf. Note that the indication section Xi is not used in this stage. No text prompt is used, and the image is the only input. Our goal is to align the image features with word embeddings of the language model via the learning of the projection layer. In this stage, we train LLaVA-Rad on the training split of MIMIC-CXR for 1 epoch.
In the third stage (end-to-end fine-tuning), we train both the MLP projection layer and the language model. However, unlike the majority of existing work that fully fine-tunes the language model, we apply the parameter-efficient fine-tuning method LoRA30, which has recently been shown to achieve comparable performance to full fine-tuning while significantly reducing the training cost57,58. Given a CXR Xv and the corresponding indication section Xi, we train LLaVA-Rad to generate the finding section Xf, using the training split of MIMIC-CXR. Similar to the approaches taken by LLaVA and LLaVA-Med, our training process utilizes cross-entropy loss, applied in an auto-regressive manner, to optimize the generation of reports.
Model evaluation
Our model evaluation consists of cross-modal retrieval evaluation, where we evaluate the quality of alignment between LLaVA-Rad’s CXR their corresponding reports, attention visualization, which illustrates the level of grounding the model’s text predictions with regions of the input image, and the automated report evaluation which studied factual correctness and lexical similarity metrics and their alignment with radiologist error quantification. To ensure a thorough evaluation, the model is tested not only on a held-out subset of the MIMIC-CXR dataset, but also on a held-out subset of the CheXpert (n = 61)59, Open-I (n = 2163)35, and US-CXR (n = 1757) datasets. US-CXR corresponds to a private collection of CXRs and de-identified reports sourced from a mixture of inpatient and outpatients from a variety of hospitals across the United States. Notably, CheXpert CXRs from the training set were used alongside synthetic label-derived reports to train the image encoder, but the held-out evaluation set, derived from the CheXpert validation split, contains CXRs and radiologist reports that were not available during training. Alternatively, the Open-I and US-CXR datasets were fully held out during model development. Altogether. the inclusion of CheXpert, Open-I, and US-CXR tests the external generalizability and adaptability of the model across different datasets with varying degrees of familiarity and complexity. We compare LLaVA-Rad with both open and closed-source vision language models, including those specialized for CXR findings generation32,33,60 and other more general ones9,56,61,62,63,64.
Image-text alignment
Cross-modal retrieval evaluation: This task consists of matching radiology reports to their corresponding radiology images (text-to-image) and the reverse (image-to-text), thus evaluating the model’s ability to identify corresponding CXRs and reports by calculating similarity scores between images and text. We compared the performance of LLaVA-Rad, which uses the specialized BiomedCLIP-CXR, with more general image encoders used for LLaVA-Med and LLaVA, namely BiomedCLIP and OpenAI CLIP models. We used the official MIMIC-CXR test set for evaluation, quantifying performance using recall at K, a commonly used retrieval evaluation metric that measures the share of relevant items captured within the top K positions.
Attention Visualization: To qualitatively examine how well LLaVA-Rad’s image and text align, we develop a method to visualize the model’s attention mechanisms during its generative process. Specifically, we focus on analyzing LLaVA-Rad’s attention to each image token while generating words. This analysis enables us to understand how well each generated word aligns with relevant regions within the image. To achieve this, we conduct an in-depth examination of a fully trained LLaVA-Rad model across all its 32 layers and 32 attention heads. Furthermore, to provide a clearer insight into the model’s attention patterns, we calculate either the mean or maximum values (or both) across all layers and heads. For visualization purposes, we reformat the attention matrices into a 37 × 37 grid to mirror the original spatial dimensions of the image tokens.
A protocol was not prepared for this study, nor was it registered. No patients of the public were involved in the design, conduct, reporting, interpretation or dissemination of this study.
Quality of generated reports
Existing evaluation metrics
We employ a suite of automated evaluation metrics to determine the quality of generated reports. We report commonly used lexical similarity metrics (ROUGE-L, BLEU-4) for the sake of comparison with prior methods. However, we focus our model development and analysis on factual correctness metrics, employing commonly used metrics such as F1-CheXbert and F1-RadGraph, as well as proposing an automated language model scoring-based metric, CheXprompt. The F1-CheXbert metric65 corresponds to the F1 score of extracted disease labels of a generated report compared to the reference, as determined by the CheXbert labeler20. In line with prior work, we report F1-CheXbert for all 14 CheXbert classes, in addition to 5 classes that represent the most common findings in real-world CXR reports (atelectasis, cardiomegaly, consolidation, edema, and pleural effusion). The F1-RadGraph metric66 broadens the scope of the factual correctness evaluation by comparing the agreement of anatomy and observation entities extracted from the candidate report with that of the reference. Prior to this work, the F1-RadGraph metric was considered as a reference for the evaluation of factual correctness in radiology reports. However, it has limited correlation with manual error scoring as performed by radiologists, which has led to the proposal of composite metrics such as RadCliQ that aim to better reflect human evaluation of factual correctness17.
CheXprompt
Given the limitations of existing report evaluation methods and the challenge of accurately evaluating generated reports at scale, we explore the utility of a language model-based scoring system, which has shown success in other domains36,37,38. Specifically, we employ a large language model as an evaluator that quantifies the presence of the following six error types, as per17: false prediction of finding (false positive), omission of finding (false negative), incorrect ___location/position of finding, incorrect severity of finding, mention of comparison that is not present in the reference (false positive comparison), omission of comparison describing a change from a previous study (false negative comparison). We instruct the model to quantify the number of errors of each of the six error types, keeping a separate count for clinically insignificant and significant errors. We refer to a clinically significant error as one that likely affects treatment, management, or outcomes, and include this distinction in line with the RADPEER system, the most widely accepted peer evaluation framework in radiology67. In each rating prompt, we include a fixed set of five example report evaluation pairs alongside mean error counts for each type to enable the model to leverage in-context examples that quantify errors as requested. We evaluate the validity of the proposed CheXprompt using the ReXval dataset39, which is comprised of error annotations from 6 board-certified radiologists on 200 pairs of candidate and ground-truth reports, where each radiologist provides counts of each of the 6 aforementioned error types, also discriminating between clinically significant and insignificant errors.
For comparison, we evaluate the performance of three types of GPT models: GPT-3.5 Turbo (GPT-3.5T), GPT-4, and GPT-4 Turbo (GPT-4T, i.e. GPT-4-1106-preview). We quantify the alignment between errors quantified by radiologists with that of existing report evaluation methods, in addition to CheXprompt, using Kendall’s Tau b rank correlation coefficient. Further, we directly compare the performance of CheXprompt based on GPT-4T with that of each radiologist in a leave-one-rater-out fashion. For each comparison with a rater, the mean of the remaining left-in radiologist raters was calculated. The paired interobserver difference between the held-out radiologist rater and the mean was compared to the paired interobserver difference between CheXprompt and the mean. The mean absolute interobserver difference (MAD) for each left-out radiologist was compared with that of CheXprompt.
Finally, we use the GPT-4T version of CheXprompt to quantify the total number of clinically significant and overall errors in each generated report in the evaluation datasets. We quantify these totals in reports from publicly accessible models, enabling us to compare LLaVA-Rad with LLaVA-Med, LLaVA, CheXagent, and GPT-4V. Further, we study the overall proportion of error-free reports in the evaluation datasets, reflecting the potential of each model in directly impacting radiology workflows.
Study of language model generalizability
While our primary goal is to optimize LLaVA-Rad for CXR report generation, we perform an evaluation to examine whether model specialization affects the general language capabilities of the underlying language model (Vicuna-7b-v1.5). To do so, we utilize the MT-bench evaluation, a widely recognized benchmark for assessing language models across multiple dimensions in multi-turn, open-ended question answering31. In MT-bench, response quality is rated on a 1–9 point-based scale, with GPT-4 serving as an evaluator, as previously validated in MT-bench to be closely aligned with crowd annotator quality.
Synthetic report evaluation
To evaluate the quality of synthetic reports, we employ different strategies depending on the availability of ground truth reports. For synthetic reports without reference ground truths, we use a similarity comparison (cosine similarity) between the embeddings of corresponding CXR images and synthetic reports, as determined by BiomedCLIP-CXR and BioViL-T68. For each of the datasets described in CXR-697k, we compare the similarity between corresponding pairs with that of randomly assigned pairs from the same dataset. For datasets with available reference ground truth reports, such as MIMIC-CXR and the CheXpert Plus reports, we assess the similarity between synthetic and original reports using GatorTron69, and identify potential errors in synthetic reports using CheXprompt. We report results for similarity and CheXprompt metrics for the MIMIC-CXR test set, and a subset of 2000 reports with findings sections sampled from the training split of the CheXpert Plus dataset.
Statistics and reproducibility
No formal sample size calculations were carried out for this study. Instead, publicly available datasets containing CXR images, labels, and reports (when available) were curated to assemble the CXR-697K dataset for model training. For evaluation, we used held-out test sets as described in Model Evaluation. All images meeting the inclusion criteria (frontal view, presence of a Findings section) were included in the evaluation, while studies were excluded if they did not meet either of these criteria. Performance metrics for report generation were evaluated using existing standard metrics and CheXprompt, with 95% bootstrap confidence calculated using 500 resampling interations. MAD comparison for CheXprompt and left-out radiologist ratings using left-in radiologists as reference was carried out using two-sided paired t-tests. Synthetic report quality cosine similarity values were compared with random similarity scores using a permutation test with 1000 permutations. A significance threshold of 0.05 was applied to all statistical comparisons.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The GPT-4 and rule-based pre-processed MIMIC-CXR reports are publicly available as the LLaVA-Rad MIMIC-CXR Annotations at https://doi.org/10.13026/6ey5-df78, subject to credentialing and completion of an appropriate course for handling human participant data. The Open-I dataset35 is publicly accessible at https://doi.org/10.1093/jamia/ocv080. The CheXpert CXR images and reports are publicly accessible at https://doi.org/10.71718/6nvz-pm34. The US-CXR dataset is a private collection of images and reports and cannot be made publicly available due to privacy restrictions. Interested parties should contact Segmed, Inc (https://segmed.ai) to inquire about access to the dataset, subject to Segmed’s applicable ethical and legal requirements. Source data are provided with this paper.
Code availability
LLaVA-Rad is fully available at https://aka.ms/llava-rad, including the model weights and relevant source code for training, inference, and evaluation. A permanent version of the code including detailed methods and implementation steps to facilitate independent replication is available at https://doi.org/10.5281/zenodo.14897681. CheXprompt is publicly available at https://github.com/microsoft/chexprompt with a permanent version available at https://doi.org/10.5281/zenodo.14861615.
References
Bommasani, R. et al. On the opportunities and risks of foundation models. Preprint at https://arxiv.org/abs/2108.07258 (2021).
Moor, M. et al. Foundation models for generalist medical artificial intelligence. Nature 616, 259–265 (2023).
Xu, H. et al. A whole-slide foundation model for digital pathology from real-world data. Nature 630, 1–8 (2024).
Huang, S.-C. et al. Self-supervised learning for medical image classification: a systematic review and implementation guidelines. NPJ Digit. Med. 6, 74 (2023).
Huang, S.-C. et al. Developing medical imaging ai for emerging infectious diseases. Nat. Commun. 13, 7060 (2022).
Tu, T. et al. Towards generalist biomedical AI. NEJM AI 1, AIoa2300138 (2024).
Azizi, S. et al. Big self-supervised models advance medical image classification. In 2021 IEEE/CVF International Conference on Computer Vision (ICCV), 3458–3468 (2021).
Azizi, S., et al. Robust and data-efficient generalization of self-supervised machine learning for diagnostic imaging. Nat. Biomed. Eng 7, 756–779 (2023).
Li, C. et al. LLaVA-med: Training a large language-and-vision assistant for biomedicine in one day. In 37th Conference on Neural Information Processing Systems Datasets and Benchmarks Track. https://openreview.net/forum?id=GSuP99u2kR (2023).
Tanno, R. et al. Collaboration between clinicians and vision–language models in radiology report generation. Nat. Med. 31 1–10 (2024).
Thirunavukarasu, A. J. et al. Large language models in medicine. Nat. Med. 29, 1930–1940 (2023).
Truhn, D., Müller-Franzes, G. & Kather, J. N. The ecological footprint of medical AI. Eur. Radiol. 34, 1–3 (2023).
Wornow, M. et al. The shaky foundations of large language models and foundation models for electronic health records. npj Digital Med. 6, 135 (2023).
Gunasekar, S. et al. Textbooks are all you need. Preprint at https://arxiv.org/abs/2306.11644 (2023).
Mitra, A. et al. Orca 2: Teaching small language models how to reason. Preprint at https://arxiv.org/abs/2311.11045 (2023).
Lehman, E. et al. Do we still need clinical language models? In Proc Conference on Health, Inference, and Learning, vol. 209 of Proceedings of Machine Learning Research, (eds Mortazavi, B. J. et al.) 578–597 (PMLR, 2023).
Yu, F. et al. Evaluating progress in automatic chest x-ray radiology report generation. Patterns 4, 100802 (2023).
Clusmann, J. et al. The future landscape of large language models in medicine. Commun. Med. 3, 141 (2023).
Langlotz, C. P. The future of ai and informatics in radiology: 10 predictions. Radiology 309, e231114 (2023).
Smit, A. et al. Combining automatic labelers and expert annotations for accurate radiology report labeling using BERT. In Proceedings of the 2020 Conference on Empirical Methods in Natural Language Processing (EMNLP), (eds Webber, B. et al.) 1500–1519 (2020)
Jain, S. et al. Radgraph: Extracting clinical entities and relations from radiology reports. In Thirty-fifth Conference on Neural Information Processing Systems Datasets and Benchmarks Track (2021).
Irvin, J. et al. Chexpert: A large chest radiograph dataset with uncertainty labels and expert comparison. In Proc of the AAAI Conference on Artificial Intelligence. 33, 590–597 (2019). https://doi.org/10.1609/aaai.v33i01.3301590.
Reis, E. P. et al. Brax, brazilian labeled chest x-ray dataset. Sci. Data 9, 487 (2022).
Feng, S. et al. CANDID-PTX. Radiology: Artificial Intelligence. https://auckland.figshare.com/articles/dataset/CANDID-PTX/14173982 (2021).
Nguyen, H. et al. Vinbigdata chest x-ray abnormalities detection. Kaggle Competition. https://www.kaggle.com/c/vinbigdatachest-xray-abnormalities-detection (2020).
Healthcare, J. Object-cxr - automatic detection of foreign objects on chest x-rays. https://web.archive.org/web/20201127235812/.
Johnson, A. E. et al. Mimic-cxr, a de-identified publicly available database of chest radiographs with free-text reports. Sci. data 6, 317 (2019).
Bustos, A., Pertusa, A., Salinas, J.-M. & De La Iglesia-Vaya, M. Padchest: A large chest x-ray image dataset with multi-label annotated reports. Med. image Anal. 66, 101797 (2020).
Liu, H., Li, C., Li, Y. & Lee, Y. J. Improved baselines with visual instruction tuning. In 2024 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), 26286–26296 (2024).
Hu, E. J. et al. LoRA: Low-rank adaptation of large language models. In Proc International Conference on Learning Representations. https://openreview.net/forum?id=nZeVKeeFYf9 (2022).
Zheng, L. et al. Judging llm-as-a-judge with mt-bench and chatbot arena. In Proc 37th International Conference on Neural Information Processing Systems, NIPS ’23 (Curran Associates Inc., Red Hook, NY, USA, 2024). https://doi.org/10.5555/3666122.3668142.
Chen, Z. et al. Chexagent: Towards a foundation model for chest x-ray interpretation. Preprint at https://arxiv.org/abs/2401.12208 (2024).
Hyland, S. L. et al. Maira-1: a specialised large multimodal model for radiology report generation. Preprint at https://arxiv.org/abs/2311.13668 (2023).
Zambrano Chaves, J. M. et al. Rales: a benchmark for radiology language evaluations. In Advances in Neural Information Processing Systems, 36 (eds Oh, A. et al.) 74429–74454 (Curran Associates, Inc., 2023).
Demner-Fushman, D., Antani, S., Simpson, M. & Thoma, G. R. Design and development of a multimodal biomedical information retrieval system. J. Comput. Sci. Eng. 6, 168–177 (2012).
Liu, Y. et al. G-eval: NLG evaluation using gpt-4 with better human alignment. In Proceedings of the 2023 Conference on Empirical Methods in Natural Language Processing (eds Bouamor, H., Pino, J. & Bali, K.) 2511–2522 (Association for Computational Linguistics, Singapore, 2023).
Wang, J. et al. Is ChatGPT a good NLG evaluator? a preliminary study. In Proceedings of the 4th New Frontiers in Summarization Workshop, (eds Dong, Y. et al.) 1–11 (Association for Computational Linguistics, Singapore, 2023).
Gilardi, F., Alizadeh, M. & Kubli, M. Chatgpt outperforms crowd workers for text- annotation tasks. Proceedings of the National Academy of Sciences 120, e2305016120 (2023).
Yu, F. et al. Radiology report expert evaluation (rexval) dataset. PhysioNet (2023).
Mohsan, M. M. et al. Vision transformer and language model based radiology report generation. IEEE Access 11, 1814–1824 (2022).
Wang, J., Bhalerao, A. & He, Y. Cross-modal prototype driven network for radiology report generation. In Proc European Conference on Computer Vision. (eds Avidan, S. et al.) 563–579 (Springer, 2022).
Pan, R. et al. S3-net: A self-supervised dual-stream network for radiology report generation. IEEE J. Biomed. Health Inform. (2023).
Hou, X. et al. Mkcl: Medical knowledge with contrastive learning model for radiology report generation. J. Biomed. Inform. 146, 104496 (2023).
Huang, J. et al. Generative artificial intelligence for chest radiograph interpretation in the emergency department. JAMA Netw. open 6, e2336100 (2023).
Aksoy, N., Ravikumar, N. & Frangi, A. F. Radiology report generation using transformers conditioned with non-imaging data. In Medical Imaging 2023: Imaging Informatics for Healthcare, Research, and Applications, (eds Park, B. & Yoshida, H.) 12469, 146–154 (SPIE, 2023).
Chen, Z., Song, Y., Chang, T.-H. & Wan, X. Generating radiology reports via memory-driven transformer. In Proceedings of the 2020 Conference on Empirical Methods in Natural Language Processing (EMNLP), (eds Webber, B. et al.) 1439–1449 (Association for Computational Linguistics, Online, 2020).
Alayrac, J.-B. et al. Flamingo: a visual language model for few-shot learning. Adv. Neural Inf. Process. Syst. 35, 23716–23736 (2022).
Fleming, S. L. et al. Medalign: A clinician-generated dataset for instruction following with electronic medical records. In Proc AAAI Conference on Artificial Intelligence, 38, 22021–22030 (2024). https://doi.org/10.1609/aaai.v38i20.30205.
Wollek, A. et al. Attention-based saliency maps improve interpretability of pneumothorax classification. Radiol Artif. Intell. 5, e220187 (2023).
Saporta, A. et al. Benchmarking saliency methods for chest x-ray interpretation. Nat. Mach. Intell. 4, 867–878 (2022).
Zhang, J. et al. Revisiting the trustworthiness of saliency methods in radiology AI. Radiol. Artif. Intell. 6, e220221 (2024).
Ramesh, V., Chi, N. A. & Rajpurkar, P. Improving radiology report generation systems by removing hallucinated references to non-existent priors. In Machine Learning for Health, (eds Parziale, A. et al.) 456–473 (PMLR, 2022).
Zhang, S. et al. A multimodal biomedical foundation model trained from fifteen million image–text pairs. NEJM AI 2, AIoa2400640 (2025).
Oquab, M. et al. Dinov2: Learning robust visual features without supervision. https://arxiv.org/abs/2304.07193 (2024).
Chiang, W.-L. et al. Vicuna: An open-source chatbot impressing GPT-4 with 90%* chatgpt quality. https://lmsys.org/blog/2023-03-30-vicuna/ (2023).
Liu, H., Li, C., Wu, Q. & Lee, Y. J. Visual instruction tuning. In Proc 37th Conference on Neural Information Processing Systems. https://openreview.net/forum?id=w0H2xGHlkw (2023).
Dettmers, T., Pagnoni, A., Holtzman, A. & Zettlemoyer, L. Qlora: efficient finetuning of quantized llms. In Proceedings of the 37th International Conference on Neural Information Processing Systems, NIPS ’23 (Curran Associates Inc., Red Hook, NY, USA, 2023).
Lu, Y. et al. An empirical study of scaling instruct-tuned large multimodal models. https://arxiv.org/abs/2309.09958 (2023).
Chambon, P. et al. Chexpert plus: Augmenting a large chest x-ray dataset with text radiology reports, patient demographics and additional image formats. https://arxiv.org/abs/2405.19538 (2024).
Tu, T. et al. Towards generalist biomedical ai. NEJM AI 1, AIoa2300138 (2024).
OpenAI. GPT-4V(ision) system card. OpenAI. https://cdn.openai.com/papers/GPTV_System_Card.pdf (2023).
Wang, P. et al. Qwen2-vl: Enhancing vision-language model’s perception of the world at any resolution. Preprint at https://arxiv.org/abs/2409.12191 (2024).
Dubey, A. et al. The llama 3 herd of models. Preprint at https://arxiv.org/abs/2407.21783 (2024).
Abdin, M. et al. Phi-3 technical report: A highly capable language model locally on your phone. Preprint at https://arxiv.org/abs/2404.14219 (2024).
Zhang, Y., Merck, D., Tsai, E., Manning, C. D. & Langlotz, C. Optimizing the factual correctness of a summary: A study of summarizing radiology reports. In Proc 58th Annual Meeting of the Association for Computational Linguistics, (eds Jurafsky, D. et al.) 5108–5120 (Association for Computational Linguistics, 2020).
Delbrouck, J.-B. et al. Improving the factual correctness of radiology report generation with semantic rewards. In Findings of the Association for Computational Linguistics: EMNLP 2022, 4348–4360 (Association for Computational Linguistics, Abu Dhabi, United Arab Emirates, 2022). https://aclanthology.org/2022.findings-emnlp.319.
Chaudhry, H. et al. Forty-one million radpeer reviews later: what we have learned and are still learning. J. Am. Coll. Radiol. 17, 779–785 (2020).
Bannur, S. et al. Learning to exploit temporal structure for biomedical vision-language pro- cessing. In 2023 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), 15016–15027 (2023).
Yang, X. et al. A large language model for electronic health records. NPJ Digit. Med. 5, 194 (2022).
Acknowledgements
The authors acknowledge the contributions of the patients and institutions that provided the datasets used in this study. This research received no specific grant from any funding agency.
Author information
Authors and Affiliations
Contributions
J.M.Z.C., S.C.H., Y. Xu, H.X., N.U., S.Z. are core contributors to this work, performing equal contributions in conception, experimental planning, execution, presentation of results and manuscript drafting. F.W., Y. Xie, M.K., Z.Y., H.A., J.G., H.H., contributed to data acquisition and curation and carried out experiments, J.Y., C.L., J.G., Y.G., C.W., M.W., T.N., M.C., M.L., A.C., S.Y., C.P., provided technical expertise and contributed to technical discussions, M.L. and C.P. provided clinical insights, and S.W., H.P. supervised the work, providing experimental and manuscript drafting oversight. All authors participated in manuscript drafting and approved the final draft.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Yixiao Ge and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zambrano Chaves, J.M., Huang, SC., Xu, Y. et al. A clinically accessible small multimodal radiology model and evaluation metric for chest X-ray findings. Nat Commun 16, 3108 (2025). https://doi.org/10.1038/s41467-025-58344-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-58344-x