Abstract
Fumarate hydratase-deficient renal cell carcinoma (FH-deficient RCC) is a rare yet highly lethal kidney cancer. To deepen our understanding of FH-deficient RCC, we conduct a comprehensive integrated genomic study. We analyze the association of FH alteration patterns with tumor heterogeneity and develop a CpG site-specific methylation signature for precise identification of FH-deficient RCC. Transcriptomic analysis unveils three distinctive molecular subtypes characterized by enrichment of immune/Angiogenic/Stromal (C1), WNT/Notch/MAPK (C2), and proliferation/stemness (C3) pathways, respectively. Tumors in C1 derive the most substantial survival benefit from a combination of immune checkpoint blockade (ICB) and anti-angiogenic therapy. Tumors in C2 display moderate response to this therapeutic approach. In contrast, tumors in C3 exhibit an unfavorable response to anti-angiogenic monotherapy and its combination with ICB. These findings contribute to a profound understanding of the aggressive nature of FH-deficient RCC, offering insights into potential precision medicine approaches for disease management.
Similar content being viewed by others
Introduction
Fumarate hydratase-deficient renal cell carcinoma (FH-deficient RCC) is a rare RCC entity, caused by biallelic inactivation of the FH gene1. Despite its rarity, FH-deficient RCC is almost universally lethal, exhibiting a pronounced tendency for invasion and metastasis even with small tumor sizes, posing a significant therapeutic challenge. The loss of FH leads to the intracellular accumulation of fumarate, a recognized oncometabolite that profoundly affects oncogenic signaling, antioxidant response, and phenotypic changes. These effects include: 1) inhibition of αKG-dependent dioxygenases, resulting in global DNA and histone hypermethylation, as well as stabilization of HIF1α2; 2) succination of proteins, which activates antioxidant pathways3; 3) induction of a pseudohypoxic state that drives metabolic rewiring and cell proliferation2,4,5,6,7,8,9; and 4) promotion of a hypermethylated phenotype, leading to epithelial-to-mesenchymal transition (EMT) and defects in homologous recombination DNA repair10,11,12; 5) induction mitochondrial DNA and RNA releasing that drive innate immunity and interferon production13,14. Nevertheless, the translation of these insights into effective targeted therapies remains challenging.
While the combination of erlotinib with bevacizumab demonstrated promising activity for FH-deficient RCC in a phase II clinical, data from real-world studies demonstrated inconsistent therapeutic efficacy15,16,17,18. Our previous investigations highlighted an immune-enriched tumor microenvironment in FH-deficient RCC, suggesting the potential effectiveness of immune checkpoint blockade (ICB)-based regimens19,20. However, subsequent studies revealed varying responses among patients treated with ICB-based therapies18,21. Previous studies, including our own, have identified frequently mutated genes in FH-deficient RCC18,19,21,22; however, their specific roles in tumor behavior and patient outcomes remain unclear. The exploration of treatment strategies for FH-deficient renal cell carcinoma still requires further research, and understanding the oncogenic genomic landscape of FH-deficient RCC is imperative for advancing therapeutic strategies.
In this work, we conduct comprehensive molecular profiling of 126 FH-deficient RCC samples, to comprehensively explore the molecular landscape and biological subtypes of FH-deficient RCC, offering insights for potential therapies. Our analyses reveal the prognostic value of FH variant patterns, establish a methylation signature for FH-deficient RCC identification, and identify three molecular clusters with distinct transcriptional signatures which hold the potential to distinguish clinical responses to anti-angiogenic monotherapy and its combination with ICB, thereby aiding the prediction of promising therapeutic strategies.
Results
Cohort description
From 2014 to 2023, a total of 126 treatment naïve FH-deficient RCC patients with genomic and transcriptomic sequencing were extracted from our multi-center database (details in methods, Fig. S1a and S1b). Formalin-fixed paraffin-embedded (FFPE) samples were obtained from all 126 patients, of whom 116 underwent nephrectomy and 10 received kidney biopsy. IHC staining of 2SC, and FH for the diagnosis of FH-deficient RCC as previously described23. For cases exhibiting FH positive immunohistochemistry (IHC) in the tumor cells, we performed further 2SC IHC staining (Fig. S2). Further analysis of FH alterations was also conducted in these cases. Baseline characteristics are summarized in Supplementary Data 1. The median age at initial diagnosis was 37 years (range: 13–75 years), and the male: female ratio was 1.8:1. In our FH-deficient RCC cohort, 81 (64.3%) patients harbored FH germline mutations, and the remaining 45 (36.7%) exhibited FH somatic mutations. Among female patients, when compared to FH somatic mutation, patients with germline mutated FH-deficient RCC were predisposed to higher frequency of uterine leiomyomas (24.7% vs. 13.3%, P = 0.041), with the other baseline characteristics showing high similarity (Supplementary Data 2). Additionally, in our cohort, the median age at diagnosis for patients with a history of uterine leiomyoma was 25 years. Out of these, 17 cases had the documented leiomyoma characteristics, which included the following: (1) 15 out of 16 cases had multiple uterine leiomyomas; (2) 13 out of 17 patients underwent surgery (4 myomectomy, 6 hysterectomy, 3 myomectomy and followed by hysterectomy); (3) average diameter of the largest leiomyoma was 5.1 cm; (4) the median age at surgery was 35 years.
After a median follow-up of 12.8 months (range 1.7–142.7 months), 109 (86.5%) patients were diagnosed with metastatic FH-deficient RCC, including 66 (52.4%) synchronous and 43 (34.1%) metachronous metastases, respectively. Unlike clear cell RCC (ccRCC)24, the most common metastatic sites for FH-deficient RCC were lymph nodes (70.6%, 77/109) and bone (48.6%, 53/109), with less than one fifth of cases (20/109, 18.3%) were diagnosed with lung metastasis, indicating unique metastatic and evolutionary patterns. Indeed, in our previous study, we found the evolutionary pattern of FH-deficient RCC20 was different from classical subtypes of RCC25,26,27,28. For patients with metastatic disease, 86/109 (78.9%) received systemic treatments. The combination of Immune Checkpoint Blockade (ICB) with tyrosine kinase inhibitor (TKI) was the most common first-line therapeutic regimen (70.9%, 61/86).
Landscape of somatic mutations and copy-number alterations in FH-deficient RCC
Whole-exome sequencing (WES) was conducted on 71 samples to detect genetic alterations, while panel sequencing was performed on another nine samples (Fig. S1b). In addition to FH alteration, the most frequently mutated genes in FH-deficient RCC were TTN (20%, 16/80), NF2 (15%, 12/80), FAT1 (9%, 7/80), KMT2D (9%, 7/80) and MUC16 (9%, 7/80), as shown in Fig. 1a and Supplementary Data 3. While TTN and MUC16 were also detected as most frequently muted genes in the current cohort (Fig. 1a), they are mostly reported as passenger genes29. Analysis across RCC types revealed the mutational spectrum of FH-deficient RCC was quite different from KIRC, KIPP, or KICH (Kidney Chromophobe Carcinoma) in TCGA cohorts (Figs. 1a and S3a–c). Furthermore, we found there were no significant differences between somatic and germline mutated FH-deficient RCC on genomic features (Fig. S4a–c).
a Heatmap shows clinicopathological features of 77 FH-deficient RCC cases (top). Oncoplot shows the global landscape of somatic alterations of 77 FH-deficient RCC cases (bottom). The top 20 genes with the highest mutation frequency were selected for display. For the FH gene, the types of germline mutation were displayed in the oncoplot as well. The diagnosis of FH-deficient RCC were confirmed by IHC staining of FH and 2SC as previously described (Zheng, L. et al. Mod. Pathol., 2023). b Kaplan-Meier curves show PFS of patients with and without NF2 somatic mutation when receiving first-line ICB+TKI or TKI monotherapy treatment. c Kaplan-Meier curves show OS of patients with and without NF2 somatic mutation when receiving first-line ICB+TKI or TKI monotherapy treatment. d The three predominant mutational signatures detect in 71 FH-deficient RCC cases that conducted WES, namely SBS6, SBS22, SBS40. e Box plots depict the TMB, truncating burden and fslNDEL burden for patients with different mutational signatures that have the highest contribution using the Kruskal-Wallis test. Patients are divided into 3 subgroups: SBS6 (n = 25), SBS22 (n = 14), SBS40 (n = 32). Box plots show median levels (middle line), 25th and 75th percentile (box), 1.5 times the interquartile range (whiskers) as well as outliners (single points). P-values are determined by the two-sided Kruskal-Wallis test. f Kaplan-Meier curves show the DFS of localized patients with different mutational signatures. P-values are determined by two-sided log-rank test (b, c, f). RCC renal cell carcinoma, ISUP International Society of Urological Pathology, PFS progression-free survival, OS overall survival, ICB immune checkpoint blockade, TKI tyrosine kinase inhibitor, NF2-wt NF2 wild type, NF2-mut NF2 mutant, WES, whole exome sequencing, TMB tumor mutational burden, fsINDEL frameshift mutation by insertion or deletion, DFS disease-free survival. Source data are provided as a Source Data file.
We evaluated the association of frequent mutated genes with clinicopathologic features and prognosis in our FH-deficient RCC cohort (Fig. S5a and Supplementary Data 4). We found that tumors with KMT2D mutation were correlated with higher ISUP nuclear grade (ISUP ≥ 3, 3/6 vs. 7/64, P = 0.003). NF2 (neurofibromatosis type 2), encoding the protein named as merlin, is a tumor suppressor gene and has demonstrated a role in the oncogenesis of multiple tumors including RCC19,21. Mutation in NF2 has been reported as the most important co-current muted gene in FH-deficient RCC18,21,30. In the current whole cohort (n = 126), a total of 22 cases (17.5%, 22/126) harboring NF2 mutation were detected (Fig. S5b). The majority of NF2 mutations in FH-deficient RCC were truncating mutations (92%,23/25), followed by missense mutations (8%, 2/25). Notably, all the truncating mutations were located within or upstream of the C-terminal residues ___domain (CTD), suggesting a probably deleterious effect on NF2 function21. Accordingly, compared to those with wild type NF2, we observed significantly lower mRNA expression of NF2 in tumors with NF2 truncating mutation (Fig. S5c). Previous studies suggested that somatic mutation of NF2 facilitated tumor metastasis31,32. In our cohort, correlation analysis demonstrated that, tumors with NF2 mutations exhibited higher ISUP/WHO grade (ISUP Grade 4: 31.2% vs. 9.2%, P = 0.058), higher Ki-67 index (Ki67 ≥ 20%: 76.9% vs. 44.6%, P = 0.033) and higher incidence of synchronous metastasis (85.0% vs. 48.5%, P = 0.009), especially bone metastasis (75.0% vs. 43.7%, P = 0.023), compared to those without NF2 mutations (Supplementary Data 4). Survival analysis showed that co-current NF2 mutation could impede the effectiveness of TKI monotherapy, while exhibiting no discernible impact on therapeutic efficacy of ICB plus TKI combination therapy for FH-deficient RCC (Fig. 1b, c). However, due to the limited sample size and the fact that the median survival for NF2 wild-type patients treated with ICB plus TKI was not reached, these differences require further validation.
We identified three prominent mutational signatures (M1, M2 and M3) in FH-deficient RCC (Fig. 1d). Signature M1 and M3 correspond COSMIC (the Catalogue of Somatic Mutations in Cancer) Signature SBS6 (DNA mismatch repair) and SBS40 (unknown), respectively. Signature M2 was highly similar to COSMIC Signature SBS22 (cosine similarity = 0.897), which is associated with exposure to aristolochic acid, a Chinese herbal ingredient linked to renal injury and RCC carcinogenesis33,34,35. By contrast, previous study has reported that SBS5 (clock-like) and SBS124 (unknown) were more predominant in ccRCC and papillary RCC (pRCC), and SBS17 (unknown) was predominant in chRCC36, indicating a different mutation profile of FH-deficient RCC comparing to other subtypes of RCC. In contrast to patients with germline FH mutations, those with somatic FH alterations showed a higher contribution from SBS22 (39.7% vs. 7.1%, P = 0.005), corroborating that aristolochic acid exposure might be a carcinogenic factor for somatic FH-deficient RCC in Chinese cohort. Moreover, we found that patients with signature M2 had higher TMB and truncating burden compared to those with signature M1 and M3 (Fig. 1e). Survival analysis further indicated that patients with M2 signature had higher risk of recurrence than those with other signatures (8.3 months vs. 12.2 months, P = 0.039, Fig. 1f).
FACETS and GISTIC analysis were used to investigate somatic copy number alterations (SCNA) in FH-deficient RCC19,21. FH-deficient RCC exhibits distinct genomic alterations compared to other RCC subtypes. We observed a higher SCNA burden (median 0.244, range 0–0.96) in FH-deficient RCC compared to other RCC subtypes in TCGA cohorts (Fig. S6a). In addition, frequent arm-level gains in 2q, 17q, 16p, and 16q, and losses in 18q, 18p, and 22q were observed in FH-deficient RCC (Fig. S6b, c). At the focal level, it showed significant amplifications and deletions at 51 loci, including notable gains in 17q21.2 and 17q12, and losses in 9p12 and 22q11.1 (Fig. S6d). These unique genomic characteristics suggest that FH-deficient RCC may have distinct biological behaviors from other RCC subtypes.
Comprehensive analysis of FH variants in FH-deficient RCC
Recent studies suggest that different alterations of FH result in varying degrees and mechanisms of reduced catalytic activity, which may link to tumor behavior of FH-deficient kidney cancer37,38. Thus, we performed a comprehensive evaluation of the patterns of FH alterations in FH-deficient kidney cancer in our cohort. A total of 159 FH variants were detected among 126 cases in the study, including 72 (45.3%) somatic FH alterations and 87 (54.7%) germline FH alterations (Fig. 2a and Supplementary Data 5). Notably, we observed the FH alteration spectrum was different between FH somatic and germline-mutated patients. In contrast to somatic FH variants, missense mutation contributed most to germline FH alterations (25.0% (18/72) vs. 62.1% (54/87), P < 0.001), whereas nonsense mutations and large deletions were more common in somatic FH variants compared to germline FH alterations (nonsense mutations 13.9% vs. 2.3%, P = 0.006, large deletions 25.0% vs. 2.3%, P < 0.001, Fig. 2b and Supplementary Data 5). A total of 66 cases had evidence of two hits in the FH gene, among which, 8 (12.1%) cases had more than one FH mutations, 37 (56.1%) cases had concurrent FH mutation(s) and loss of heterozygosity (LOH), 7 (10.6%) cases had concurrent FH mutation and hypermethylation of the promoter of FH gene, 1 (1.5%) case had concurrent FH mutation and 1q loss, 11 (16.7%) cases had concurrent large deletion and LOH, 1 (1.5%) case had concurrent FH mutation, large deletion and LOH, 1 (1.5%) case had concurrent FH mutation, large deletion and hypermethylation of the promoter of FH gene (Supplementary Data 5). For those with FH missense mutations, we detected several hot spots mutations, including p.N154H, p.N188I, p.H196R, p.G397R, and p.S419L. Among them, p.N188I was located in catalytic active site, p.N154H in proximity to the active site (within 10 Å from the active site), and p.H196R, p.G397R, and p.S419L were located on the inter-/intra-subunit interface of FH protein (Fig. S7a). Moreover, patients with FH p.S419L showed a higher incidence of lymph node metastasis (85.7% vs. 43.2%, P = 0.047) and shorter disease-free survival (DFS) (9 months vs. 20.4 months, P = 0.012, Fig. S7b) compared to those without FH p.S419L.
a Lollipop plot shows types and locations of FH germline and somatic alterations. *Large deletions were not shown. Sites with no less than 3 mutations are annotated. b Pie charts show the distribution of alteration types of somatic (top) and germline (bottom) FH variants. c Pie chart shows the proportion of the three FH alteration subtypes (non-truncating mutation, truncating mutation, large deletion). d Box plot depicting the FH mRNA expression level in patients with different FH alteration subtypes. Patients are divided into 3 subgroups: Non_Truncating_Mutation (n = 18), Truncating_Mutation (n = 28), Large_Deletion (n = 10). Box plots show median levels (middle line), 25th and 75th percentile (box), 1.5 times the interquartile range (whiskers) as well as outliners (single points). P-values were determined by the two-sided Kruskal-Wallis test. e Bar chart depicting the proportions of FH protein expression by IHC in patients with different FH alteration subtypes. Patients are divided into 3 subgroups: Large deletion (n = 19), Truncating (n = 55), Non-truncating (n = 52). f Bar chart depicting the proportions of T stage <3 and ≥3 in patients with different FH alteration types. Patients are divided into 3 subgroups: Large deletion (n = 19), Truncating (n = 55), Non-truncating (n = 47). g Kaplan-Meier curves show PFS of patients with different FH alteration types who received first-line ICB + TKI treatment. h Kaplan-Meier curves show OS for patients with different FH alteration types who received first-line ICB + TKI treatment. P-values were determined by the two-sided Pearson’s Chi-squared test with continuity correction (e, f). P-values were determined by two-sided log-rank test (g, h). IHC immunohistochemistry, PFS progression-free survival, OS overall survival, ICB immune checkpoint blockade, TKI tyrosine kinase inhibitor. Source data are provided as a Source Data file.
We further reviewed previously reported FH variants in FH-deficient RCC and compared differences between the Chinese (n = 256) and Western cohorts (n = 268, Fig. S7c, d, Supplementary Data 6). We observed that the proportion of missense mutation in the Chinese cohort was lower than that in the Western cohort (50.8% vs. 68.7%, P < 0.001, Fig. S7e and Supplementary Data 7). In contrast, the Chinese cohort had higher proportions of frameshift mutation (18.8% vs. 10.4%, P = 0.007) and large deletion (10.5% vs. 4.9%, P = 0.014). Notably, the variants p.G397R and p.R233H were hotspots in both cohorts, while several variants were found unique to each population (Supplementary Data 7). Specifically, p.N188I, p.S419L and p.K80* were unique to the Chinese cohort, whereas p.N373S and p.L132* were unique to the Western cohort.
In total, 55 (43.7%) patients exhibited FH truncating mutations, 52 (41.3%) patients showed FH non-truncating mutations and the remaining 19 (15.1%) patients harbored FH large deletions (Fig. 2c). Lower mRNA expression level of FH was observed in tumors harboring FH truncating mutations and large deletions compared with tumors harboring FH non-truncating mutations (Fig. 2d). Nonsense-mediated decay is expected to degrade mRNAs from truncating mutations, and we indeed noticed a small subset of cases (13.5%, 17/126) showed retained FH protein expression by IHC in tumor cells, and almost all of cases with weak FH protein expression by IHC harbored FH non-truncating mutation (88.2%, 15/17, Figs. 2e and S7f). Remarkably, compared to those with FH non-truncating mutations and FH large deletion, tumors with FH truncating mutations were more aggressive, characterized by larger tumor size (median tumor size: 6.1 cm vs. 5.0 cm vs.8.8 cm, P = 0.036) and higher T stage (T stage ≥ 3: 36.7% vs. 47.4% vs. 63.3%, P = 0.021, Fig. 2f and Supplementary Data 8). Additionally, we identified an association between FH mutational patterns with clinical outcomes from ICB + TKI combination therapy (Fig. 2g, h). Specifically, patients with germline non-truncating mutations demonstrated longer PFS compared to those with germline truncating mutations (median PFS: 19.0 vs. 11.4 mo, P = 0.018, Fig. S7g, h). Whereas, in patients treated with TKI monotherapy, no statistical significances for PFS and OS were found among patients with different mutational patterns (Fig. S7i, j).
Transcriptional landscape in FH-deficient RCC
RNA-seq was performed on 56 tumors and 37 paired normal adjacent tissues (NATs). A total of 1467 up-regulated and 1214 down-regulated differentially expressed genes (DEGs) were identified in FH-deficient RCC compared to NATs (Fig. S8a and Supplementary Data 9). IHC staining confirmed that top DEGs including CD44 and KRT19 (CK19) were highly expressed in FH-deficient RCC (Fig. S8b, S8c), with positive rates of 72% (18/25) and (85%) 21/25, respectively (Fig. S8b, c). Gene set enrichment analysis (GSEA) demonstrated the enrichment of immune-related pathways involving allograft rejection, IFNγ response and IL6-JAK-STAT3 signals in FH-deficient RCC (Fig. S8d). Correlation analysis further indicated that CD8 T cells, activated CD4 memory T cells, M1 and M2 macrophages signatures were associated with these immune pathways (Fig. S8e). In lines with these findings, FH-deficient RCC showed a significantly more abundant enrichment with CD8+ naive T-cells than KIRC, KIRP and KICH (Fig. S9a, b).
Comparative analysis across RCC subtypes revealed that cell cycle regulation, epithelial-mesenchymal transition (EMT), and DNA repair pathways were also enriched in FH-deficient RCC relative to other RCC subtypes (Fig. S9c). Coalition analysis of gene expression with SCNAs identified that cell cycle, mTOR, TGF-β, EMT, NRF2-ARE, EGF/EGFR, and ERBB signaling were significantly enriched in FH-deficient RCC (Fig. S10a and Supplementary Data 10). Visualization Pipeline for RNA-seq (VIPER) analysis further confirmed the upregulation of several master regulators involved in cell cycle (MDM2), mTOR (RHEB), TGF-beta (TGFA and SMAD3), and ErbB (EGFR) pathways in FH-deficient RCC (Supplementary Data 10). These findings indicated that these aberrant signaling may underpin oncogeneses and lethal behavior of FH-deficient RCC.
Snice the activation of ERBB2 pathway underpins tumor aggressive features and serves as a druggable target for anti-tumor therapy39. We subsequently validated HER2 expression in FH-deficient RCC and other RCC subtypes using IHC staining. We observed HER2 expression in 17.9% (5/28) of FH-deficient RCC but rare expressed in other RCC subtypes (Fig. S10b, c and Supplementary Data 11). These findings demonstrated that a subset of FH-deficient RCC exhibited activation of ERBB2 signaling, implying the potential of HER2 targeting.
We proceeded to validate the efficacy of anti-HER2 by using patient-derived organoids of FH-deficient RCC (Fig. S10d). In comparison to ccRCC organoids (KI-284, KI-273, both with HER2 negative expression), organoids of FH-deficient RCC (FHRCC128 with strong HER2 expression, FHRCC46 and FHRCC118 with weak HER2 expression) exhibited a lower IC50 when treated with lapatinib (a pan-ERBB inhibitor). Notably, the organoid derived from FHRCC128, characterized by strong HER2 positivity, exhibited the lowest IC50 value (0.09 uM), more than 7.5-fold lower than the other two FH-deficient RCC organoids (Fig. S10d). These findings suggested the potential sensitivity of FH-deficient RCC with ERBB2 positivity to agents specifically targeting HER2.
Methylation patterns in FH-deficient RCC and identification of FH-deficient RCC methylation signature
EPIC array was performed on 56 tumors and paired 32 NATs (Fig. S1b). In line with previous findings, we observed a sharp global hypermethylation pattern in FH-deficient RCCs compared with NATs (Fig. S11a). To explore the impact of CpGs methylation changes on the transcriptome, we performed integrative methylation and transcriptomic analysis and identified significantly methylated 64,407 genes (Fig. S11b and Supplementary Data 12). Of note, several up-regulated oncogenes were hypomethylated in tumors compared to NATs (Supplementary Data 12), including EGFR, RAC1, CTNNB1, CDK4, MYC, XPO1, CUL1, and NFE2L2. GSEA analysis showed that the upregulated and hypomethylated genes were enriched in proliferation (MYC targets, E2F targets and G2M checkpoint), glycolysis, and hypoxia signaling, whereas downregulated and hypermethylated genes were predominantly linked to lipid metabolic pathways (xenobiotic and fatty acid metabolism, Fig. S11c), indicating that the aberrant methylation might participate in the pathogenesis of FH-deficient RCC.
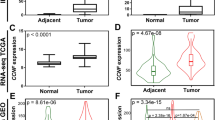
Upon merging data from our FH-deficient RCC and TCGA KIRP cohorts, we identified that the majority of FH-deficient RCC cases (82.1%, 46/56) were in the CpG island methylator phenotype (CIMP) cluster, while the remaining ten cases (17.9%, 10/56) were in the non-CIMP cluster (Fig. 3a). In contrast to non-CIMP tumors, tumors with CIMP phenotype had higher TMB burden (median: 1.24 vs. 0.67, P = 0.036), truncating burden, (Fig. 3b) and frequency of loss in 22q (14/46 vs. 0/9, P = 0.055) (Supplementary Data 13). Transcriptomic analysis indicated that upregulated genes involved in cell cycle and EMT pathways were more abundant in CIMP tumors compared to non-CIMP ones, suggesting the aggressive biology of tumor with CIMP phenotype (Fig. S11d). Consistent with these findings, CIMP tumors were associated with higher propensity to recur (91.2% vs. 50%, P = 0.005) and synchronous metastasis (51.4% vs. 20%, P = 0.002) compared to those with non-CIMP phenotype (Fig. 3c, d and Supplementary Data 13). Moreover, there was some evidence that the DNA methylation signature was notably higher in CIMP tumors compared to their non-CIMP counterparts (Fig. S11e). In addition, we noticed that CIMP tumors had higher PD-L1 positive rate than non-CIMP tumors (36/46 vs. 3/9, P = 0.004, Fig. 3e).
a One-dimensional hierarchical clustering of the 2000 most variant DNA methylation probes revealing three clusters of tumor samples of FH-deficient RCC (n = 56) in our cohort and KIRP (n = 276) in the TCGA database. In contrast to cluster 1 and 2, cluster 3 shows widespread DNA hypermethylation patterns characteristic of CIMP-associated tumors. The FH-deficient RCC tumors in our cohort are classified into two subsets: one, characterized by a global DNA hypermethylation phenotype, referred to as “CIMP”; the other, showing relatively low genome-wide DNA methylation, referred to as “non-CIMP”. The term “CIMP-RCC” refers to pRCC tumors from KIRP database, with a global DNA hypermethylation phenotype, which were used as controls in the clustering analysis. Each row represents a probe; each column represents a sample. b Bar chart depicts the prevalence truncating burden in CIMP (n = 46) and non-CIMP (n = 10) groups. c Bar chart depicts the prevalence recurrence status in CIMP (n = 22) and non-CIMP (n = 8) groups. Both CIMP and non-CIMP are from FH-deficient RCC tumors. d Bar chart depicts the prevalence metastatic status in CIMP (n = 46) and non-CIMP (n = 10) groups. Both CIMP and non-CIMP are from FH-deficient RCC tumors. e Bar chart depicts the prevalence PD-L1 expression by immunohistochemistry in CIMP (n = 45) and non-CIMP (n = 9) groups. P-values were determined by the two-sided Pearson’s Chi-squared test with continuity correction (b, c–e). Both CIMP and non-CIMP are from FH-deficient RCC tumors. f One-dimensional hierarchical clustering of the 42 CpG sites reveal three clusters of samples from TCGA KIRP cohort (tumors n = 276, normal tissues n = 45) using the 42 CpG sites in our FH-deficient RCC methylation signature. Five tumor samples of KIRP carry FH mutations and are known to be FH-deficient RCC, and are defined as “KIRP_FH” (n = 5). Five tumors samples of KIRP has not been previously recognized as FH-deficient RCC but are clustered together with KIRP_FH, and are defined as “KIRP_suspicious_FH” (n = 5). The rest tumor samples are defined as “KIRP” (n = 266). g Principal component analysis of KIRP (n = 266), KIRP_FH (n = 5), KIRP_suspicious_FH (n = 5) and FH-deficient RCC (n = 56) in our cohort performed on 42 CpG sites derived from our FH-deficient RCC methylation signature. h Box plot depicts the FH mRNA expression level in KIRP (tumors, n = 266; normal tissues, n = 45), KIRP_FH (n = 5), KIRP_suspicious_FH (n = 5) in the TCGA database. Box plots show median levels (middle line), 25th and 75th percentile (box), 1.5 times the interquartile range (whiskers) as well as outliners (single points). P-values were determined by the two-sided Kruskal-Wallis test. i Representative H&E staining demonstrates morphological patterns of the 5 “KIRP suspicious FH” samples. Magnification ×200. Scale bar = 100 μm. RCC: renal cell carcinoma, CIMP CpG island methylator phenotype, KIRP kidney renal papillary cell carcinoma, KIRP_FH KIRP with FH mutations, KlRP_suspicious_FH KIRP suspicious to FH-deficient RCC using the 42-CpG methylation signature. Source data are provided as a Source Data file.
Due to the distinct methylation phenotype, we subsequently tempted to construct an FH-deficient RCC-specific methylation signature through combination analysis of our FH-deficient RCC and RCCs in TCGA cohorts (n = 665, details in methods). All cases were divided into development cohort and validation cohort, respectively (Supplementary Data 14). Eventually, a 42 CpG sites-based methylation signature were developed, including 26 hypermethylated and 16 hypomethylated CpGs (Fig. S12a and Supplementary Data 15). Utilizing this signature, we could distinguish FH-deficient RCC from other RCC subtypes regardless of CIMP or non-CIMP phenotype (Fig. S12b). The methylation signature could further distinguish FH-deficient RCC (including known HLRCC in TCGA cohort, namely “KIRP_FH”) from other RCC subtypes in the validation cohort (Fig. S12c). We also validated four CpGs sites (CG08548498, CG17209188, CG22321237 and CG00592510) in an independent RCC cohort by using bisulfite pyrosequencing (Fig. S12d).
Interestingly, utilizing our FH-deficient RCC methylation signature, five cases within the KIRP cohort were categorized to FH-deficient RCC, designated as “KIRP_suspicious_FH” (Fig. 3f, g). The FH mRNA expression in “KIRP_suspicious_FH” cases were significantly lower than KIRP, but comparable to that of known HLRCC in TCGA cohort (Fig. 3h). Of note, all five tumors exhibited typical morphological patterns of FH-deficient RCC, including abundant and eosinophilic cytoplasm, thickened nuclear membraneeosinophilic large nucleolieosinophilic macronuclei and perinuclear halo (Fig. 3i). Furthermore, we identified that two FH-deficient RCC cases with copy number loss of the FH gene (Fig. 3i), emphasizing the necessity for additional FH IHC staining in the diagnosis of FH-deficient RCC.
Since the EPIC 850 K data available for RCC were limited to the TCGA dataset, which led us focus on CpG probes shared between the 450 K and 850 K arrays. To address this limitation, we validated the 42-CpGs signature in the GEO dataset (GSE253101) that includes 850 K profiles across different RCC subtypes, confirming its effectiveness in distinguish FH-deficient RCC (Fig. S13a, b). Additionally, we identified 40 CpGs sites (TOP40 based on P value between tumor vs. normal) exclusive to the 850 K array in our FH-deficient RCC samples (Supplementary Data 15). The 850 K and combined 450 K and 850 K signatures performed well in discriminating FH-deficient RCC from other RCC subtypes (Fig. S13c–f).
Identification and characterization of three molecular subtypes of FH-deficient RCC
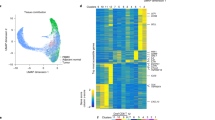
To gain insight into the biologic characteristics of FH-deficient RCC, we employed an unsupervised consensus partitioning approach40 to identify and refine transcription-based subgroups of patients with FH-deficient RCC. Consequently, three distinct clusters were identified using 110 features from Gene set variation analysis (GSVA) (details in methods, Fig. S14a). To elucidate the transcriptional characteristics underpinning these subtypes, we integrated DEG analysis (Supplementary Data 16), quantitative set analyses for gene expression (QuSAGE) and weighted gene co-expression network analysis (WGCNA) by comparing each subtype against others (Fig. S14b–d). By combining the results of these analyses, we summarized and refined seven representative gene signatures related to T cell-inflamed, angiogenesis, stroma, MAPK/ERK, NOTCH, Wnt/β-catenin, and proliferation (Fig. 4a, b).
a Heatmap representing ssGSEA scores of MSigDb hallmark gene set for each cluster. b Heatmap of expression level of genes comprised in transcriptional signatures for each cluster. Samples were grouped by cluster. c Box plots depicte the Z-scores of T effector, angiogenesis, stroma, MAPK/ERK, NOTCH, Wnt/β-catenin, proliferation signatures for different clusters (C1-C3). C1, n = 27; C2, n = 23; C3, n = 6. Box plots show median levels (middle line), 25th and 75th percentile (box), 1.5 times the interquartile range (whiskers) as well as outliners (single points). P-values were determined by the two-sided Mann-Whitney test. d Bar chart depicting the proportions of tumors with average CD8 IHC expression ≤50 cells/mm2 and >50 cells/mm2 in different molecular subtypes of FH-deficient RCC. C1 (n = 20), C2 (n = 15), C3 (n = 4). P-value was determined by the two-sided Fisher’s exact test. e Bar chart depicting the proportions of patients with negative (TPS < 1%) or positive (TPS ≥ 1%) PD-L1 expression by IHC in different molecular subtypes of FH-deficient RCC. C1 (n = 27), C2 (n = 20), C3 (n = 6). P-value was determined by the two-sided Pearson’s Chi-squared test with continuity correction. f Box plots depicting the number of CD8+CXCL13+, CD8+ TIGIT+, CD8+PD1+, CD4+TIGIT+, CD4+PD1+ cells/mm2 in different molecular subtypes of FH-deficient RCC. CD8+/PD-1+, CD4+/PD-1+, CD8+/TIGIT+ and CD4+/TIGIT+ markers to depict exhausted CD8+ and CD4+ T cells; while CD8+/CXCL13+ to represent anti-tumor CD8+ T cells. Box plots show median levels (middle line), 25th and 75th percentile (box), 1.5 times the interquartile range (whiskers) as well as outliners (single points). P-values were determined by the two-sided Kruskal-Wallis test. g, h Box plots depicting the ssGSEA scores of CCP score (g) and cancer stemness score (h) in different molecular subtypes of FH-deficient RCC. C1, n = 27; C2, n = 23; C3, n = 6. Box plots show median levels (middle line), 25th and 75th percentile (box), 1.5 times the interquartile range (whiskers), and specific score of each sample (single dots). P-values were determined by the two-sided Kruskal-Wallis test. RCC renal cell carcinoma, ssGSEA single-sample gene set enrichment analysis, IHC immunohistochemistry, TPS tumor proportion score, CCP cell cycle progression. Source data are provided as a Source Data file.
Nearly one half of tumors were categorized in cluster 1 (C1: n = 27, 48.2%), characterized by high immunogenicity and angiogenic activity, exhibiting strong enrichment in T cell-inflamed (e.g., CD8A, IFNG, GZMB and CXCL13) and angiogenesis-related genes (e.g., PECAM1 and CXCL8, Fig. 4b). These tumors showed the highest expression of both adaptive and innate immune cell signatures, including CD8+, CD4+, regulatory T cells, B cells, macrophages, and dendritic cells, and the highest angiogenesis scores (Figs. 4c, S15a–b).
Consistently, IHC revealed increased CD8+ T cells infiltration (Fig. 4d), increased PD-L1 expression (Fig. 4e), strongest nuclear HIF1A staining, and increased vessel density in these tumors (Fig. S16c). Differentiated from the other two clusters, tumors in cluster 1 exhibited high expression of the exhausted-associated genes (CD274, TIGIT, HAVCR2, BTLA and PDCD1LG2) and exhausted score (Fig. S16d, e). Multiple immunofluorescences further confirmed these tumors had increased infiltration of exhausted (PD-1+ and TIGIT+) and tumor-reactive (CXCL13+) CD8+ and CD4+ T cells in the tumor stroma (Figs. 4f, g and S17a). Additionally, tumors in cluster 1 showed higher stroma-specific genes expression, exemplified by activation of collagens and activated stroma-associated genes (e.g., FN1, COL8A1, and MMP1). We thus labeled this cluster as T effector/Angiogenic/Stromal.
About two fifths of tumors (C2: n = 23, 41%) were classified into cluster 2, characterized by moderate expression of both angiogenesis and immune genes and low expression of both stroma and cell-cycle genes (Figs. 4b, c and S16a). These tumors showed increased expression of genes associated MAPK/ERK (eg. MAPK3, MAPK7 and MAPK12), Notch (eg. NOTCH1 and NOTCH3), and Wnt/β-catenin (eg. WNT5B and DDK4), which associated with aggressive features of clear cell RCC (Fig. 4b, c). We labeled this cluster as the WNT/MAPK/Notch cluster.
A small portion of tumor were classified as cluster 3 (C3: n = 6, 10.7%), characterized by low expression of angiogenesis, stroma, and immune genes, but enrichment of genes involved in proliferation transcriptional pathways (E2F targets and G2M checkpoint, MYC targets and p53). Compare with tumors in the other two clusters, tumors in cluster 3 had higher cell cycle progression (CCP) score and stemness score (Fig. 4b, c, g, h). We therefore labeled this cluster as Proliferation/Stemness.
FH-deficient RCC molecular subtypes associate with molecular features and clinical outcomes to PD-1 inhibitor and TKI
We subsequently explored the association of clinical and genetic features with different molecular subtypes (Figs. 5a and S17a and Supplementary Data 17). Of note, compared with the other clusters, tumors in the Proliferation/Stemness (C3) cluster were highly aggressive, characterized by a larger tumor size (median 7.2 cm vs. 10.2 cm, P = 0.092) and a higher frequency of lymph node metastasis (24/50 vs. 6/6, P = 0.091). Morphologically, tumors presented with low-grade eosinophilic patterns were reported to be associated with favorable prognosis41,42,43. Interestingly, all tumors with low-grade eosinophilic patterns (n = 7) were identified in the T effector/Angiogenic/Stromal (C1) and WNT/MAPK/Notch (C2) clusters. In contrast to the other clusters, tumor in the Proliferation/Stemness (C3) cluster showed a higher frequency of copy number gain in 17q (21/50 vs. 6/6, P = 0.034), 16p (18/50 vs. 6/6, P = 0.015), and 16q (16/50 vs. 5/6, P = 0.006), and FH truncating mutations (23/50 vs. 5/6, P = 0.096).
a Heatmap shows the genomic, transcriptomic and clinicopathological features for each molecular clusters (C1, n = 27; C2, n = 23; C3, n = 6). b Bar chart depicts the proportions of patients who achieved ORR when receiving first-line ICB + TKI combination therapy or TKI monotherapy in each molecular cluster. C1: ICB + TKI (n = 18), TKI (n = 3); C2: ICB + TKI (n = 8), TKI (n = 5); C3: ICB + TKI (n = 2), TKI (n = 3). c Kaplan-Meier curves show PFS of patients in different clusters when receiving first-line ICB + TKI (C1, n = 18; C2, n = 8; C3, n = 2). d Kaplan-Meier curves show PFS of patients in different clusters when receiving first-line TKI monotherapy (C1, n = 3; C2, n = 5; C3, n = 3). P-values were determined by two-sided log-rank test (c, d). RCC renal cell carcinoma, ORR objective response rate, ICB immune checkpoint blockade, TKI tyrosine kinase inhibitor, PFS progression-free survival. Source data are provided as a Source Data file.
We proceeded to evaluate clinical benefits of ICB plus TKI and TKI monotherapy within each cluster. Regarding the efficacy of combination therapy, patients in C1 exhibited an improved objective response rate (ORR) and prolonged PFS, whereas those in C3 demonstrated the poorest ORR and shortest PFS. The clinical benefits of patients in C2 were between those observed in C1 and C3 (C1 vs. C2 vs. C3: ORR: 66.7% vs. 50% vs. 0%, P = 0.053; median PFS: 23.8 vs. 16.2 vs. 6.3 months, P = 0.022, Fig. 5b, c). In contrast, patients with FH-deficient RCC could not achieve acceptable survival benefits from TKI monotherapy, regardless of molecular classification (Fig. 5d).
Drawing from the above analyses, we have summarized the clinical, pathological, genomic, and transcriptomic features of the three subtypes, along with potential therapeutic targets, as illustrated in Fig. 6.
Integrative analysis of 56 FH-deficient RCC resulted in 3 different subtypes with distinct biological and clinical characteristics. Radar charts in the RNA profile panel represent mean Z-scores for each gene signature in the respective subtypes. RCC, renal cell carcinoma, PFS progression-free survival, TPS tumor proportion score of PD-L1 by IHC, IMDC International Metastatic RCC Database Consortium Risk Model.
Discussion
In this study, we presented a comprehensive integrative analysis of 126 untreated primary FH-deficient RCCs, elucidating the unique molecular characteristics of this highly aggressive RCC entity. Our findings provided an in-depth understanding of key biological features underlying disease progression, revealed the comprehensive nature of FH alteration patterns, developed an FH-deficient RCC-specific DNA methylation signature with prospects for differential diagnosis. More importantly, we defined three distinct molecular subtypes based on differential transcriptional features, which might not only facilitate predicting tumor response to current therapeutic strategies, but also pinpoint promising potential targets for personalized therapy (Fig. 6).
Somatic or germline inactivating mutations in FH, an enzyme of the citric acid cycle, represent the initial steps in oncogenic transformation of FH-deficient RCC44. Previous studies indicated a strong association between tumorigenesis and the restricted catalytic activity caused by FH variants37. However, the impact of catalytic activity on tumor aggressiveness remained unclear37,45. Human FH formed as a homotetramer with four subunits46. It is reasonable to posit that FH alterations could disrupt protein folding, stability or change protein dynamics, thereby interfering with tetramerization and leading to enzymatic dysfunction. Nonsense-mediated decay is expected to degrade mRNAs from truncating mutations. Missense mutations, occurring within binding sites or hinge regions of proteins, could impede the quaternary structure of FH. We found an association of FH mutational patterns and structural integrity with clinical outcomes. Notably, FH truncating mutations and mutations near hinge regions (such as p.S419L), were predominantly associated with more aggressive tumor behavior and poorer prognosis (Figs. 2f and S7b). Therefore, these findings highlighted the significance and necessity of determining FH alteration patterns and structural integrity at the time of initial diagnosis.
Owing to its structural similarity to α-ketoglutarate (αKG), the aberrantly accumulated fumarate in FH-deficient RCC can inhibit αKG-dependent dioxygenases, such as histone lysine demethylases (KDMs) and DNA demethylation enzymes (TETs)47. The global DNA hypermethylation resulting from the inhibition of KDMs and TETs has been recognized as a hallmark of FH-deficient RCC19,48. In our study, based on the hypermethylation phenotype, we successfully developed a DNA methylation signature comprising 42 CpG sites for accurate identification of FH-deficient RCC (Fig. S12). Recent investigations on differentially methylated regions highlighted the potential of cell-free DNA (cfDNA) methylation sequencing for early-stage RCC detection49. Ricketts et al. previously identified a methylation signature for FH-deficient RCC patients with germline FH mutation (HLRCC) using the 450 K methylation array, incorporating only hypermethylated CpGs50. In contrast, by utilized the 850 K methylation array and incorporated both hypermethylated and hypomethylated CpGs, we established a methylation signature that distinguish both germline or somatic FH mutated FH-deficient RCC from other RCC subtypes. Further exploration of early detection in larger cohorts particularly through liquid biopsy, using the current DNA methylation signature is warranted. In addition, we observed an association of the CIMP phenotype with tumor aggressiveness and metastatic capacity, whereas those with a non-CIMP phenotype exhibited relatively indolent clinical behaviors (Fig. 3c, d). Epigenetic dysregulation, which gives rise to inappropriate transcriptional activity and chromosomal instability, can influence tumor progression and lethal phenotypic switches2,51. Notably, we identified significant transcriptional differences between CIMP and non-CIMP tumors, particularly in processes related to EMT and cell-cycle processes (Fig. S11d). Further exploration of the mechanism underlying epigenetic regulation in FH-deficient RCC could pave the way for more effective strategies targeting CIMP and non-CIMP tumors.
Given the absence of a standard of care, bevacizumab plus erlotinib and ICB plus TKI could be considered as therapeutic options for the treatment of advanced or metastatic FH-deficient RCC. However, both in the clinical practice and trials, evident heterogeneity in tumor response to these therapies was observed. In the present study, unsupervised transcriptomic analysis identified three molecular clusters, which were capable of distinguishing prognosis and response to current effective therapies. Previous researches have demonstrated that the accumulated fumarate could inhibit prolyl hydroxylase, activating HIF1A and angiogenesis signal in FH-deficient RCC52,53,54. Indeed, in the present study, not all cases demonstrated the enrichment of angiogenesis signal. Transcriptomic classification revealed nearly a half of FH-deficient RCCs (tumors in C1) exhibited high degree of angiogenesis (Figs. 4a, c and S16a). At the same time, tumors in C1 were also characterized by abundant T effector and stroma-related genes (Figs. 4a–c and S16a). These transcriptomic features in C1 implied the potential therapeutic efficacy with anti-angiogenetic and immunotherapy. Patients with FH-deficient RCC were hard to gain survival benefits from monotherapy, probably due to the increased stroma signature together with interaction between angiogenesis and immune signatures. In summary, the distinct transcriptional features and clinical evidence with C1 tumors provided a biological rationale for the use of TKI combined with ICB in this setting.
Unlike tumors in C1, those in C2 were characterized by moderate T cell inflamed tumor microenvironment, but with high expression of WNT/Notch/MAPK signature (Figs. 4a–c and S16a). Survival analysis also revealed modest benefit from TKI plus ICB among patients in C2 (Fig. 5c). Compelling evidence has indicated negative immunomodulatory effects of WNT and Notch signaling pathways in cancers55,56. Active WNT signaling can increase intratumoral Treg cells, drive CD4+ T cells towards the protumorigenic Th17 subtype, and induce tolerogenic dendritic cells57,58,59. Activation of Notch signaling can promote polarization of M2 tumor-associated macrophage and recruit infiltration of tumor-associated neutrophils60,61,62. Therefore, these data highlighted the potential therapeutic benefit of targeting WNT and/or Notch in combination with immunotherapies in this subset of FH-deficient RCC.
In contrast to tumors in C1 and C2, those in C3 seemed to be associated with decreased expression of T effector and angiogenesis signatures (Figs. 4a–c and S16a). In agreement with our clinical findings, these tumors showed worse clinical outcomes and poorest response to either TKI plus ICB or TKI monotherapy. Tumors in C3 showed an enrichment of cell cycle signature. Thus, targeting cell cycle checkpoints, such as CDK4/6 inhibitors63, may be options for this type of tumor. Further studies are needed to evaluate the potential efficacy of CDK4/6 inhibitors or combination therapies in treating this aggressive subtype. In addition, we observed a subset of tumors with FH-deficient RCC presented positive HER2 expression and were sensitive to ERBB inhibitor (Lapatinib) using organoids models (Fig. S10). Evidence in other cancers demonstrated that only those with relatively strong HER2 IHC staining achieve definite treatment response64. Therefore, HER2-targeted therapies could be an alternative option for patients with strong HER2 expression and warrant further investigation.
This study has several limitations. Due to the rarity of FH-deficient RCC, it is challenging to assemble an adequate number of cases with various genetic/molecular subtypes for further analysis, thereby limiting the overall sample size for comprehensive investigation. In the survival analysis, we acknowledge that the majority of patients received ICB + TKI as the primary systemic therapy. The imbalance in treatment modalities may have influenced survival outcomes, particularly in the TKI monotherapy group, potentially impacting the robustness of those results. Additionally, due to availability of drugs in China, TKI monotherapy was predominantly used prior to 2019. Following the survival benefit demonstrated in our previous study19 and the reduced prices of patented drugs, ICB + TKI therapy was gradually became the primary treatment strategy from 2019 onwards. What’s more, this study systematically analyzed the transcriptional features of FH-deficient RCC. The roles of FH loss triggers fumarate accumulation, which activates oncogenic pathways such as pseudohypoxic, EMT, DNA repair, and antioxidant pathways, as reported in previous studies. Our findings validated these pathways in clinical tissue specimens, emphasizing their significance in carcinogenesis and development of FH-deficient RCC while highlighting their potential as therapeutic targets warranting further investigation. Nonetheless, our study represents the largest cohort to date investigating molecular landscape of FH-deficient RCC. Moreover, the relatively short follow-up time and the small number of total death events limited the statistic power of survival analysis. Future studies with larger sizes are needed to further elucidate the molecular mechanisms of FH-deficient RCC.
In summary, the current study has uncovered valuable insights into the molecular underpinnings of FH-deficient RCC, emphasized the importance of mutational pattern FH and dysregulation of DNA methylation on tumor behavior. This study also provided a molecular basis for understanding the diverse prognosis and resistance mechanisms associated with TKI and ICB, as well as their combinations in patients with advanced FH-deficient RCC. Our findings may pave the way for promising opportunity in developing more effective and personalized therapies for this lethal disease.
Methods
Patient identification
The study was conducted in accordance with the Declaration of Helsinki and was approved by the Ethics Committee of West China Hospital of Sichuan University. All patients or family members provided written consent for genetic and clinical analysis, and publication of relevant clinical information before enrollment. In our multi-center database established at West China Hospital, cases who were suspicious for FH-deficient RCC were further reviewed by two experienced genitourinary pathologists (Ni Chen and Mengni Zhang). In total, 126 cases of FH-deficient RCC from 2014 to 2023 were identified, originating from 48 medical centers spread across 21 provincial administrative regions in China. Among these 126 cases, some cases have been previously reported22, both of which and the current one are companion studies. In detail, 87 cases were shared between these two studies. This group of cases made up the majority of the subjects in this study, but the two studies investigated distinct research objectives.
The diagnosis of FH-deficient RCC was based on IHC evidence of negative FH and/or positive 2SC, with subsequent DNA sequencing confirmation of FH alterations23. FH staining was assessed in all RCC cases using whole sections. Negative FH staining in tumor cells was considered truly negative in the presence of an internal positive control. For cases with FH positivity in the tumor cells, we performed further 2SC staining. For 2SC, samples with both cytoplasmic positivity and nucleocytoplasmic positivity were interpreted as positive.
All cases were confirmed by Next-generation, Sanger sequencing or Multiplex Ligation-dependent Probe Amplification (MLPA) for FH alteration. Untreated primary tumor tissues and adjacent normal samples were collected. To avoid potential variability from metastatic lesions at different sites, we exclusively included samples from untreated primary tumors. Prior to genomic and transcriptional sequencing, tumor sections (H&E staining) were reviewed by pathologist Xiaoxue Yin to ensure that tumor sections contained > 70% tumor cells and <10% necrosis. Whole-exome sequencing (WES) was performed on 71 Formalin-fixed paraffin-embedded (FFPE) tumor tissues and 69 matched adjacent normal (n = 34)/blood (n = 35) samples. Targeted sequencing was performed on 9 tumor tissues and matched blood samples due to insufficient tumor DNA for WES (Fig. S1b). EPIC array was performed on 56 tumors and 32 adjacent normal tissues. RNA-seq of FFPE tissues was performed on 56 tumors and 37 adjacent normal tissues.
Clinicopathological characteristics and outcomes
We retrospectively gathered clinicopathological information encompassed FH and NF2 variant information (mutation site, type, etc.), age, gender, family history, metastatic sites, TNM stage, histological features, WHO/ISUP grade, International Metastatic RCC Database Consortium (IMDC) risk score, time of metastasis, metastatic sites, and approaches to systemic treatment. Synchronous metastasis referred to the detection of distant metastasis concurrently with the primary diagnosis of renal cell carcinoma. Metachronous metastasis, on the other hand, was characterized by its occurrence three months or more after the surgical intervention. Tumor response was assessed using Response Evaluation Criteria in Solid Tumors (RECIST) 1.1, objective response rate (ORR) was defined as complete response (CR) rate + partial response (PR) rate. Disease-free survival (DFS) was defined from diagnosis to local or regional recurrence or distance metastasis or death. Progression-free survival (PFS) was defined from the start of systemic therapy to disease progression or death. Overall survival (OS) was defined from the start of systemic therapy to death from any cause. The last follow-up was on July, 2023.
Identification of FH biallelic inactivation
Biallelic inactivation of FH was characterized by deep deletion, two or more deleterious mutations within the same tumor sample, one mutation plus 1q loss, one mutation plus large deletion or hypermethylation in the promoter of FH gene, one mutation or large deletion plus loss of heterozygosity (LOH), assessed through the FACETS algorithm65.
DNA extraction
All tumor sections were reviewed by two pathologists to ensure tumor sections with at least 70% tumor nuclei. Representative sections of FFPE tumor (8 μm) and matched normal tissues/blood samples were collected. High-quality genomic DNA was extracted by using the GeneRead DNA FFPE Kit (180134, QIAGEN, Hilden, GER) according to the manufacturer’s instructions. Germline DNA was extracted from white blood cells using the Blood Genomic DNA (gDNA) Mini Kit (CW2087, Cwbiotech, Beijing, China).
Whole-exome sequencing
Exome capture was performed using xGen Exome Research Panel v1.0 (IDT), and this was followed by paired-end sequencing using Illumina Hiseq Xten platform (Illumine Inc, CA, USA). Mean sequencing depth was 278v (range from 36× to 576×). For panel sequencing, DNA was extracted and estimated by a targeted sequencing strategy capturing all exons of 642 tumor-related genes19.
Multiplex ligation-dependent probe amplification (MLPA)
MLPA analysis was conducted using SALSA MLPA Probemix P198 FHn kit (MRC-Holland, The Netherlands) to access gene dosage of FH, following the manufacturer’s instructions. 50 ng of gDNA sample was denatured at 98 °C for 5 minutes and then were subjected to hybridization with the probe mix at 60 °C for 16 hours. Subsequently, the probes underwent ligation and amplification through polymerase chain reaction (PCR). The amplification products were further denatured at 86 °C for 3 min and analyzed by using SeqStudio Genetic Analyzer (Applied Thermo Fisher Scientific, USA). The MLPA fragments analysis was performed by utilizing Coffalyzer software packages (MRC-Holland, The Netherlands), and dosage quotients (DQ) were calculated for each target probe by comparison with normal controls. The standard deviation of each individual probe across all the reference samples (n = 5) should not exceed 0.10, and the DQ of each individual reference probe in the patient samples should fall within the range of 0.80 to 1.20, with ratio values < 0.65 indicating genomic deletion and values > 1.30 indicating genomic duplication.
Read alignment, BAM file generation and post-alignment optimization
Clean reads were aligned to the reference human genome hg19 (Genome Reference Consortium GRCh37) using BWA 0.7.17 (Burrows-Wheeler Aligner) MEM algorithm with default parameters. BAM was coordinate sorted and PCR duplicates were removed with Sambamba version 0.8.0. After the initial alignment of WES data, we followed GATK v3.8 Best Practice to process all BAMs from the same patient together for a post-alignment optimization process called “co-cleaning” which includes GATK IndelRealigner and BaseQualityScoreRecalibration (BQSR). Local realignment of all reads at loci close to indels was performed using IndelRealigner to improve mapping quality, and systematic errors made by the sequencer when it estimated the quality score of each base call were detected and fixed by BQSR19,20.
Somatic mutations analysis
The GATK MuTect2 (v4.2.0.0) pipeline was run for paired tumor-normal somatic mutation calling with gnomAD database and a panel of normals made from all normal samples to filter common germline mutations and recurrent technical artifacts. The resulting VCFs were filtered by Mutect2 FilterMutectCalls module, and FilterByOrientationBias module was used to filter out false-positive calls from OxoG and FFPE. Same as our previous studies19,20, the resulting somatic SNVs and indels were further filtered according to the flowing criteria: read depth ≥10 in both tumor and normal samples, mapping quality ≥40 and base quality ≥20, variants allele frequency (VAF) ≥ 5% and supporting reads ≥5 in tumor, VAF in tumor was ≥5 times than that of the matched normal VAF. Variants were annotated with Oncotator v1.9.9.0. To further avoid miscalling germline variants at least 19 read depth in the normal sample in dbSNP sites. For FH and NF2, truncating mutation was defined as frameshift mutation, splice site and nonsense mutation.
Somatic mutation signature profiling
The R package MutationalPatterns (v3.0.1)66 was used to extract the somatic motifs of these samples. Non-negative Matrix Factorization (NMF) was used to estimate the optimal number of mutation signatures extracted from WES samples. Cosine similarity was calculated to measure the similarity between our identified signatures and the catalogue of somatic mutations in cancer (COSMIC) signatures v3.2 [cancer.sanger.ac.uk/cosmic/signatures].
Germline mutation analysis
Germline SNVs and indels were called by GATK HaplotypeCaller. The vcfs were annotated by InterVar v2.0.267 to classify variants based on five-tiered categorization system: pathogenic, likely pathogenic, uncertain significance, likely benign, and benign. Variants were selected if the InterVar or ClinVar annotation matched “Likely_pathogenic” or “Pathogenic”. The possible pathogenic variants in normal samples with read depth ≥10, genotype quality ≥60, supporting allele reads ≥ 2 and VAF ≥ 0.1 were finally kept.
Somatic copy number alterations (SCNA) analysis
FACETS (v0.5.14)64,65 was used to estimate tumor cellularity and ploidy from paired tumor and normal WES data, and calculated allele-specific somatic copy number alterations. Copy number (CN) gains were defined as alterations showing total CN > 2 and CN losses were defined as alterations showing total CN < 2. Arm-level events were identified when autosomal arms demonstrated CN gains or losses spanning at least 10% of the arm35. To identify significantly focal SCNAs, we used the GISTIC2 (v2.0.23)68 with the following modified parameters “-smallmem 1 -broad 1 -brlen 0.7 -cap 1.5 -conf 0.99 -ta 0.2 -td 0.25—armpeel 1 -genegistic 1 -savegene 1 -gcm extreme -js 4 -maxseg 2000 -qvt 0.25 -rx 0”. CN gains were defined as alterations showing 0.1 < log2 CN ratio <0.7 and CN losses were defined as alterations showing −1.1 < log2 CN ratio < −0.135.
Sanger sequencing
The presence of somatic mutations of FH was validated by Sanger sequencing. For this analysis, primers to amplify a 220-bp fragment covering each exon (1–10) of FH gene were designed. PCR amplification was conducted under standardized thermal cycling conditions: a 95 °C for 5 minutes, 35 cycles of 95 °C for 20 seconds, 60 °C for 30 seconds, 72 °C for 30 seconds, and a 72 °C for 7 minutes. Reactions employed 10 ng of genomic DNA template with Premix Taq polymerase (RR901A, Takara) in a Veriti96 thermal cycler (Life Technologies). PCR fragments were purified using the ChargeSwitch PCR Clean-Up Kit (CS12000, Invitrogen), and the sequencing reactions were performed on an ABI 3730XL automatic sequencer (Life Technologies) according to the manufacturer’s instructions. All analyses were performed in duplicate. Sequences of the forward and reverse strands were analyzed using Chromas (Technelysium)19.
Evaluation of FH mutations on structure of FH protein
To evaluate how these mutations disrupt the structure of FH protein, we conducted molecular dynamics simulations using Gromacs version 2018.169. FoldX-predicted ∆∆G values was calculated with the PositionScan command within FoldX4, which was run sequentially on each residue in the protein structure. Rosetta-predicted ∆∆G values were determined using the cartesian_ddg method70.
FH mutation types
Patients were stratified into three groups according to the types of FH mutation. Those with FH truncating mutations (nonsense mutations, frameshift mutations, splice site mutations) were classified into the “Truncating_Mutation” group. Patients with FH large deletions, but without truncating mutations were categorized into the “Large_Deletion” group. Those with non-truncating mutations (missense mutations, in-frame mutations) without truncating mutations or large deletions, were stratified into the “Non_Truncating_Mutation” group.
Infinium methylationEPIC BeadChip assay
Genomic DNA (gDNA) was treated with bisulfite using the EpiTect Fast Bisulfite Conversion Kits (59802, Qiagen). All samples were processed in the same batch. Genome-wide DNA methylation profiles of samples were generated using Infinium MethylationEPIC BeadChip assay (EPIC array, Illumina). The assay determines DNA methylation levels at >850,000 CpG sites and provides coverage of CpG islands, RefSeq genes, ENCODE open chromatin, ENCODE transcription factor-binding sites, and FANTOM5 enhancers. The assay was performed according to the manufacturer’s instructions and scanned on an Illumina HiScan.
EPIC array data processing and DMP identification
Raw EPIC array data were preprocessed using the ChAMP R/Bioconductor package with default settings. Different methylation positions (DMPs) analysis was performed on beta (β) value. We used a linear model (limma) with the empirical Bayes approach with normal control samples as the reference group. Genomic annotation of CpG sites were annotated using HumanMethylationEPICm probe annotations through ChAMP. The percentage of DMPs in each annotation region were calculated with R software and visualized with ggplot2. A probe was considered significantly differentially methylated if the methylation difference (β-values) between the tumor and normal control samples were at least 15% with an FDR-adjusted (Benjamini-Hochberg) P value < 0.01. The methylation of candidate probes for immune genes among different groups were compared using Kruskal-Wallis Test and Dunn’s test. The differential methylated probes between paired samples were also compared using paired t test or Wilcoxon signed ranked test. P value < 0.05 was considered as significant difference.
Methylation profiling analysis
DeepTools271 was used for methylation profiling analysis. In detail, the region between the transcription start site (TSS) and transposable elements (TES) of whole genome genes or specific genes (for example, hallmark gene sets) were normalized into a relative equal length and extending 3,000 bp of upstream and downstream. Regions were divided into 50 windows and average methylation was calculated in each window, and then visualized as profile line plot.
Bisulfite pyrosequencing
Bisulfite modification of gDNA was performed by standard methods. PCR primers were designed with the PyroMark Assay Design V.2.0 software (QIAGEN) (Supplementary Data 18). PCR products were pyrosequenced with the PyromarkTM Q24 system (QIAGEN), according to the manufacturer’s protocol19.
Consensus cluster analysis for methylation
As our previous studies19,20, consensus cluster analysis was run for FH-deficient RCC cohort alone and combined with TCGA-KIRC/KIRP/KICH cohort, respectively. Methylation β-values matrix was adjusted firstly by removing features with small variance and impute missing value with K-Nearest Neighbor (KNN). Three feature selection methods (SD, MAD and CV) and five cluster methods (hclust, kmeans, skmeans, pam and mclust) were chose to infer possible stable consensus subgroup from 2 to 6 clusters with different number of top features. The best stable partitions from all methods were chose based on check the membership matrix.
Screening of CpG sites for establishment of methylation signature
The construction of FH-deficient RCC-specific methylation signature was based on combination analysis of our FH-deficient RCC and RCCs in TCGA cohorts (KIRC, KIRP, and KICH). By comparing DMPs across these cohorts and extracting the set that is present in all FH-deficient RCC cohort but absent in the TCGA cohort, a Venn diagram was employed to illustrate this unique CpG site list for FH-deficient RCC. The gene set included in the signature was ultimately determined based on statistical significance (P < 0.01) and various delta beta values (−0.15, −0.45, 0.45, and 0.3). All cases were divided into development cohort and validation cohort, respectively (Supplementary Data 14). For the analysis of 850 K CpG sites and GEO validation, we selected the top 40 CpG sites based on the P-values of DMPs in our cohort, focusing exclusively on sites present in the 850 K array and not in the 450 K array. The original 42-CpG signature, the 40-CpG signature, and the combined 82-CpG signature were then validated using the GEO dataset (GSE253101), which includes 850 K profiles across various RCC subtypes, confirming its effectiveness in distinguish FH-deficient RCC (Fig. S13a, b). Additionally, we identified 40 methylation sites (TOP40 based on P value between tumor vs. normal) exclusive to the 850 K array in our FH-deficient RCC samples (Supplementary Data 15). The 450 K, 850 K and combined signatures performed well in discriminating FH-deficient RCC from other RCC subtypes (Fig. S13c–f). These findings have been added in the revised Results and Methods section.
Over represented analysis (ORA)
Gene Ontology, KEGG and MSigDB Hallmark (50 gene sets) analysis were performed with ORA using R clusterprofiler packages72 on the annotated genes from hypo or hyper different methylated CPGs (DMCs). Right-tailed Fisher’s exact test was used to calculate P values, with a P < 0.05 considered statistically significant.
Total RNA isolation, library preparation and sequencing
RNAs were extracted and purified from FFPE tissues by the RNeasy FFPE Kit (73504, Qiagen, Germany), according to the manufacturer’s instructions, and quantified with Qubit RNA HS Assay Kit (Thermo Fisher Scientific). RNA quality and integrity were characterized using the Bioanalyzer and High Sensitivity RNA ScreenTape (Agilent Technologies). Total RNA was isolated from each sample using the Qiagen RNeasy FFPE Kit (73504, Qiagen, Hilden, Germany), following the protocol from the manufacturer. Purity and quantity of total RNA were measured by Nanodrop. Integrity of RNA was evaluated using the RNA Nano6000 Assay Kit on the Bioanalyzer 2100 system (Agilent Technologies, CA, USA). 1 μg RNA of per sample was used as input for the RNA sample preparations. Strand-specific RNA sequencing libraries were generated using the Whole RNA-seq Lib Prep kit for Illumina (RK20303, ABclonal, Shanghai, China). Library quality was evaluated on the Agilent Bioanalyzer 2100 system (Agilent, USA). Final libraries were sequenced at the Novogene Bioinformatics Institute (Beijing, China) on an Illumina Hiseq X10 platform by 150 bp paired-end reads.
Gene expression quantification
Raw RNA-Seq reads were trimmed the adapter sequences and filtered low-quality bases using FASTP (v0.20.1)73, followed by mapping to human genome reference hg19 with STAR (v2.7.9a)74. During alignment, STAR was supplied with transcript models GENCODE v19 from https://data.broadinstitute.org/Trinity/CTAT_RESOURCE_LIB/__genome_libs_StarFv1.3. The quality control metrics were obtained using FastQC (v0.11.9) (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/), and alignment quality metrics of bam files were measured using RSeQC (v4.0.0)75. RNA abundance was calculated using RSEM (v1.2.28)76 and the RSEM results were converted with Bioconductor package tximport (v4.1)77.
Analysis of differentially expressed genes (DEG) and enrichment of signaling pathways
DEGs were determined using the R package “limma” with cutoff P-value < 0.05. Up-regulated genes and down-regulated genes were used to perform ontology and pathway enrichment analysis based on Gene Ontology and KEGG databases using R package “ClusterProfiler”. Coalition analysis of gene expression with SCNAs was visualized by using the online tool of Enrichment Analysis Visualizer (https://maayanlab.cloud/Enrichr/).
Gene set enrichment analysis (GSEA) and single-sample GSEA
GSEA was conducted using the GSEA software version 4.2.178. KEGG, GO, WikiPathway and Hallmark gene sets from MSigDB database79 were utilized. The ssGSEA method is an extension of the GSEA method80, which works at single sample level rather than a sample population. Normalized RNA-Seq data was used as input without further processing (i.e., no standardization or log transformation).
Immune gene signature analysis
Single-sample gene set enrichment analysis (ssGSEA) was used for quantifying immune infiltration and activity in tumors using markers reported by Wang et al.80 and Ricketts et al.81. RSEM upper quartile-normalized, log2-transformed, and mean-centered RNA-seq data were matched to predefined immune gene signature clusters via Entrez IDs. Each gene signature was calculated as the average value of all genes included in the signature.
Visualization pipeline for RNA sequencing analysis (VIPER) analysis
Protein activity was inferred for each sample by running the VIPER algorithm with KIRC ARACNe network on the normalized gene expression matrix.
Coalition analysis of gene expression with SCNAs
We implemented the DIGGIT algorithm to identify functional SCNAs by coalition analysis of gene expression with SCNAs, as previously reported by Paull et al.82.
Unsupervised consensus clustering
Consensus cluster analysis was run for cases with RNA-seq data. Gsva score matrix was first to be adjusted by removing features with small variance and impute missing value with KNN. Four feature selection methods (SD, MAD ACT and CV) and five cluster methods (hclust,kmeans,skmeans,pam and mclust) were choosed to infer possible stable consensus subgroup at different k (k = 2,3,4) with different number of top features (n = 1000, 2000, 3000, 4000, 5000, 10000). The best stable partions from all methods was choosed based on check the membership matrix. After choosed partions, signature analysis with conducted. We look for features show A distinct difference in one subgroup compared to others (by using F test), they can be called signature gene, kmeans was used to partions all found signature gene to infer possible pattern of gene. Consesus matirx, which measures how consistent two samples are in the same groups, was visulized as heatmap with sample membership, partitons and clinical information as row labels. In addition to consensus matrix heatmap, one-dimension hierarchical clustering of the 10000 most variant probes choosed from SD methods, with retention of the same sample order as in consensus matrix were conducted to show the gene expression value siganl in different partions.
Quantitative set analysis for gene expression (QuSAGE)
This package is an implementation the Quantitative Set Analysis for Gene Expression (QuSAGE) method described by Yaari G. et al.83. This is a novel Gene Set Enrichment-type test, which is designed to provide a faster, more accurate, and easier to understand test for gene expression studies. QuSAGE accounts for inter-gene correlations using the Variance Inflation Factor technique proposed by Wu et al.84. In addition, rather than simply evaluating the deviation from a null hypothesis with a single number (a P value), qusage quantifies gene set activity with a complete probability density function (PDF).
Weighted gene co-expression network analysis (WGCNA) analysis
Datasets were constructed using following method. A signed weighted correlation network was constructed by first creating a matrix of pairwise correlations between all pairs of genes chosen by variance. The resulting Pearson correlation matrix was transformed into a matrix of connection strengths (e.g., an adjacency matrix) using a power of *. Then the topological overlap was calculated to measure network interconnectedness. Average linkage hierarchical clustering was used to group genes on the basis of the topological overlap dissimilarity measure (1-topological overlap) of their network connection strengths. Using a dynamic tree-cutting algorithm and merging threshold function at 0.25, we identified * modules in the dataset. The freely available statistical analysis software (WGCNAR package) and R tutorials for constructing a weighted gene coexpression network have been described previously85. We summarized the expression profile of each module by the corresponding module eigengene (i.e., the first principal component obtained by singular value decomposition). We then defined the module membership for each gene with respect to each module as the Pearson cor- relation between the expression level of the gene and the module eigengene, also known as module eigengene-based connectivity (kME). This measure was naturally scaled to lie in the interval [−1, 1]. Genes with the greatest module membership values are referred to as intramodular hub genes. We used Cytoscape to visualize the top 40 gene connections (based on topological overlap). Intramodular hub genes (i.e., genes with the highest kME values) usually are centrally located inside the module. To identify modules associated with external traits, we first calculated the module eigengenes of each module, then correlated these with the external traits using Pearson’s correlation coefficients, modules with P-values < 0.05 were identified as trait-related modules.
IHC and multiple immunofluorescence
IHC was performed using an automatic staining platform, Ventana NexES (Roche)19,20. Multiplex immunofluorescence staining was obtained using PANO 7-plex Kit (0004100100, Panovue)19,20. Commercially available primary anti-FH (1: 800, sc-100743, Santa Cruz, TX, USA), ki-67 (clone MIB-1, 1:100, MXB biotechnologies, Fujian, China), CD8 (clone C8/144B, ready to use, Dako, Copenhagen, DEN), HIF1A (clone NB100-449, 1:5000, Novus Biologicals, Colorado, USA), CD31 (clone JC-70A, ready to use, MXB biotechnologies), PD-L1 (clone 22C3, ready to use, Dako, Copenhagen, DEN), HER2 (clone 4B5, ready to use, Roche, Switzerland), (clone 42, ready to use, Zhongshan Golden Bridge, Beijing, China) and KRT19 (CK19) (clone UMAB2, 1:100, Zhongshan Golden Bridge, Beijing, China), and 2SC (1:2000, crb2005017e, Discovery Antibodies) were used in this study. Multiplex immunofluorescence staining was performed using primary anti-CD8 (clone SP16, Zhongshan Golden Bridge, Beijing, China), CD4: (clone ST0488, 1:500, Huabio, Hangzhou, China), PD-1: (clone UMAB199, Zhongshan Golden Bridge, Beijing, China), TIGIT: (clone E5Y1W, CST, Massachusetts, USA), CXCL13 (clone EPR23400-92, abcam, Cambridge, UK). PD-L1 expression was assessed by tumor proportion score, which was defined as the percentage of tumor cells with membranous PD-L1 staining. PD-L1 expression > 1% was defined as positivity. For the quantification of checkpoint molecules, we used CPS and TPS for the assessment of PD-L1 expression, and measured the numbers of TIGIT+ cells from random five 0.045 mm2 fields of lesions. Ki-67 index was calculated based on the percentage of Ki-67 positive nuclei of tumor cells.
Statistics
All analyses were conducted using R software (v3.6.0) and SPSS (v16.0). All comparisons for continuous variables were performed using the two-sided Mann-Whitney test for two groups or Kruskal-Wallis test for more than two groups. For categorical variables, two-sided Pearson’s Chi-squared test with continuity correction or Fisher’s exact test was used. Survival analyses were conducted using Kaplan–Meier method and the difference was tested using log-rank. Cox proportional hazards regression (forward likelihood ratio model) was used to determine the independent predictor of PFS. All clinicopathological parameters and biomarkers at P < 0.05 were then further tested on multivariate Cox regression analysis. A P value less than 0.05 was considered statistically significant. Key resources, including software, algorithms, and databases used in the project are summarized in Supplementary Table 1.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The WES and gene expression data are publicly deposited and are available in the National Genomics Data Center (NGDC) Genome Sequence Archive (GSA) at HRA006641 and the Gene Expression Omnibus (GEO) at GSE273036. The DNA methylation data is also publicly deposited and is available in the GEO at GSE291073. The remaining data are available within the Supplementary Information. Source data are provided with this paper.
Code availability
The custom code/software used in this study has been uploaded to GitHub: https://github.com/Xingming1990/code and has been assigned a DOI for permanent reference: https://doi.org/10.5281/zenodo.1497013486.
References
Gardie, B. et al. Novel FH mutations in families with hereditary leiomyomatosis and renal cell cancer (HLRCC) and patients with isolated type 2 papillary renal cell carcinoma. J. Med. Genet. 48, 226–234 (2011).
Yong, C., Stewart, G. D. & Frezza, C. Oncometabolites in renal cancer. Nat. Rev. Nephrol. 16, 156–172 (2020).
Adam, J. et al. Renal cyst formation in Fh1-deficient mice is independent of the Hif/Phd pathway: roles for fumarate in KEAP1 succination and Nrf2 signaling. Cancer Cell 20, 524–537 (2011).
Ashrafian, H. et al. Expression profiling in progressive stages of fumarate-hydratase deficiency: the contribution of metabolic changes to tumorigenesis. Cancer Res. 70, 9153–9165 (2010).
Shanmugasundaram, K. et al. The oncometabolite fumarate promotes pseudohypoxia through noncanonical activation of NF-kappaB signaling. J. Biol. Chem. 289, 24691–24699 (2014).
Bratslavsky, G., Sudarshan, S., Neckers, L. & Linehan, W. M. Pseudohypoxic pathways in renal cell carcinoma. Clin. Cancer Res. 13, 4667–4671 (2007).
Schmidt, C., Sciacovelli, M. & Frezza, C. Fumarate hydratase in cancer: a multifaceted tumour suppressor. Semin. Cell Dev. Biol. 98, 15–25 (2020).
Pollard, P. J. et al. Targeted inactivation of fh1 causes proliferative renal cyst development and activation of the hypoxia pathway. Cancer Cell 11, 311–319 (2007).
Ratcliffe, P. J. Fumarate hydratase deficiency and cancer: activation of hypoxia signaling? Cancer Cell 11, 303–305 (2007).
Sciacovelli, M. et al. Fumarate is an epigenetic modifier that elicits epithelial-to-mesenchymal transition. Nature 537, 544–547 (2016).
Sulkowski, P. L. et al. Krebs-cycle-deficient hereditary cancer syndromes are defined by defects in homologous-recombination DNA repair. Nat. Genet. 50, 1086–1092 (2018).
Sulkowski, P. L. et al. Oncometabolites suppress DNA repair by disrupting local chromatin signalling. Nature 582, 586–591 (2020).
Zecchini, V. et al. Fumarate induces vesicular release of mtDNA to drive innate immunity. Nature 615, 499–506 (2023).
Hooftman, A. et al. Macrophage fumarate hydratase restrains mtRNA-mediated interferon production. Nature 615, 490–498 (2023).
National Comprehensive Cancer Network. Bone Cancer (Version 1.2024). https://www.nccn.org/professionals/physician_gls/pdf/kidney.pdf (2023).
Srinivasan, R. et al. Results from a phase II study of bevacizumab and erlotinib in subjects with advanced hereditary leiomyomatosis and renal cell cancer (HLRCC) or sporadic papillary renal cell cancer. J. Clin. Oncol. 38, 5004–5004 (2020).
Carril-Ajuria, L. et al. Response to systemic therapy in fumarate hydratase-deficient renal cell carcinoma. Eur. J. Cancer 151, 106–114 (2021).
Xu, Y. et al. Genomic profiling and response to immune checkpoint inhibition plus tyrosine kinase inhibition in fh-deficient renal cell carcinoma. Eur. Urol. 83, 163–172 (2023).
Sun, G. et al. Integrated molecular characterization of fumarate hydratase-deficient renal cell carcinoma. Clin. Cancer Res. 27, 1734–1743 (2021).
Liang, J. et al. Genomic and transcriptomic features between primary and paired metastatic fumarate hydratase-deficient renal cell carcinoma. Genome Med. 15, 31 (2023).
Gleeson, J. P. et al. Comprehensive molecular characterization and response to therapy in fumarate hydratase-deficient renal cell carcinoma. Clin. Cancer Res. 27, 2910–2919 (2021).
Chen, J. et al. Memory/active T-cell activation is associated with immunotherapeutic response in fumarate hydratase-deficient renal cell carcinoma. Clin. Cancer Res. 30, 2571–2581 (2024).
Zheng, L. et al. AKR1B10 is a new sensitive and specific marker for fumarate hydratase-deficient renal cell carcinoma. Mod. Pathol. 36, 100303 (2023).
Dudani, S. et al. Evaluation of clear cell, papillary, and chromophobe renal cell carcinoma metastasis sites and association with survival. JAMA Netw. Open 4, e2021869 (2021).
Turajlic, S. et al. Tracking cancer evolution reveals constrained routes to metastases: TRACERx renal. Cell 173, 581–594.e512 (2018).
Turajlic, S. et al. Deterministic evolutionary trajectories influence primary tumor growth: TRACERx renal. Cell 173, 595–610.e511 (2018).
Mitchell, T. J. et al. Timing the landmark events in the evolution of clear cell renal cell cancer: TRACERx renal. Cell 173, 611–623.e617 (2018).
Zhu, B. et al. The genomic and epigenomic evolutionary history of papillary renal cell carcinomas. Nat. Commun. 11, 3096 (2020).
Shyr, C. et al. FLAGS, frequently mutated genes in public exomes. BMC Med. Genomics 7, 64 (2014).
Satter, K. B., Xi, L., Rickets, C., Linehan, W. M. & Merino, M. J. Abstract 5048: NF2 mutations are enriched in FH deficient renal cell cancer. Cancer Res. 84, 5048 (2024).
Luo, Z. L. et al. A splicing variant of Merlin promotes metastasis in hepatocellular carcinoma. Nat. Commun. 6, 8457 (2015).
Quan, M. et al. Merlin/NF2 suppresses pancreatic tumor growth and metastasis by attenuating the FOXM1-mediated wnt/beta-catenin signaling. Cancer Res. 75, 4778–4789 (2015).
Qu, Y. et al. A proteogenomic analysis of clear cell renal cell carcinoma in a Chinese population. Nat. Commun. 13, 2052 (2022).
Wang, X. M. et al. Integrative genomic study of Chinese clear cell renal cell carcinoma reveals features associated with thrombus. Nat. Commun. 11, 739 (2020).
Sun, G. et al. Integrated exome and RNA sequencing of TFE3-translocation renal cell carcinoma. Nat. Commun. 12, 5262 (2021).
Fukagawa, A. et al. Genomic and epigenomic integrative subtypes of renal cell carcinoma in a Japanese cohort. Nat. Commun. 14, 8383 (2023).
Wilde, B. R. et al. FH variant pathogenicity promotes purine salvage pathway dependence in kidney cancer. Cancer Discov. 13, 2072–2089 (2023).
Bezwada, D. & Brugarolas, J. Reporting on FH-deficient renal cell carcinoma using circulating succinylated metabolites. J. Clin. Invest. 133, e170195 (2023).
Oh, D. Y. & Bang, Y. J. HER2-targeted therapies - a role beyond breast cancer. Nat. Rev. Clin. Oncol. 17, 33–48 (2020).
Gu, Z., Schlesner, M. & Hubschmann, D. cola: an R/Bioconductor package for consensus partitioning through a general framework. Nucleic Acids Res. 49, e15 (2021).
Lau, H. D. et al. A clinicopathologic and molecular analysis of fumarate hydratase-deficient renal cell carcinoma in 32 patients. Am. J. Surg. Pathol. 44, 98–110 (2020).
Smith, S. C. et al. A distinctive, low-grade oncocytic fumarate hydratase-deficient renal cell carcinoma, morphologically reminiscent of succinate dehydrogenase-deficient renal cell carcinoma. Histopathology 71, 42–52 (2017).
Li, Y. et al. Re-evaluation of 33 ‘unclassified’ eosinophilic renal cell carcinomas in young patients. Histopathology 72, 588–600 (2018).
Pasini, B. & Stratakis, C. A. SDH mutations in tumorigenesis and inherited endocrine tumours: lesson from the phaeochromocytoma-paraganglioma syndromes. J. Intern. Med. 266, 19–42 (2009).
Zheng, L. et al. Circulating succinate-modifying metabolites accurately classify and reflect the status of fumarate hydratase-deficient renal cell carcinoma. J. Clin. Invest. 133, e165028 (2023).
Picaud, S. et al. Structural basis of fumarate hydratase deficiency. J. Inherit. Metab. Dis. 34, 671–676 (2011).
Xiao, M. et al. Inhibition of alpha-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev. 26, 1326–1338 (2012).
Cancer Genome Atlas Research, N. et al. Comprehensive molecular characterization of papillary renal-cell carcinoma. N. Engl. J. Med. 374, 135–145 (2016).
Nuzzo, P. V. et al. Detection of renal cell carcinoma using plasma and urine cell-free DNA methylomes. Nat. Med. 26, 1041–1043 (2020).
Ricketts, C. J. et al. Kidney tumors associated with germline mutations of FH and SDHB show a CpG island methylator phenotype (CIMP). PLoS ONE 17, e0278108 (2022).
Cedar, H. & Bergman, Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Genet. 10, 295–304 (2009).
Isaacs, J. S. et al. HIF overexpression correlates with biallelic loss of fumarate hydratase in renal cancer: novel role of fumarate in regulation of HIF stability. Cancer Cell 8, 143–153 (2005).
Pollard, P. J. et al. Accumulation of Krebs cycle intermediates and over-expression of HIF1alpha in tumours which result from germline FH and SDH mutations. Hum. Mol. Genet. 14, 2231–2239 (2005).
MacKenzie, E. D. et al. Cell-permeating alpha-ketoglutarate derivatives alleviate pseudohypoxia in succinate dehydrogenase-deficient cells. Mol. Cell. Biol. 27, 3282–3289 (2007).
Parsons, M. J., Tammela, T. & Dow, L. E. WNT as a driver and dependency in cancer. Cancer Discov. 11, 2413–2429 (2021).
Li, X. et al. The Notch signaling pathway: a potential target for cancer immunotherapy. J. Hematol. Oncol. 16, 45 (2023).
Ding, Y., Shen, S., Lino, A. C., Curotto de Lafaille, M. A. & Lafaille, J. J. Beta-catenin stabilization extends regulatory T cell survival and induces anergy in nonregulatory T cells. Nat. Med. 14, 162–169 (2008).
Fu, C. et al. beta-Catenin in dendritic cells exerts opposite functions in cross-priming and maintenance of CD8+ T cells through regulation of IL-10. Proc. Natl. Acad. Sci. USA 112, 2823–2828 (2015).
Hong, Y. et al. beta-catenin promotes regulatory T-cell responses in tumors by inducing vitamin A metabolism in dendritic cells. Cancer Res. 75, 656–665 (2015).
Tao, S., Chen, Q., Lin, C. & Dong, H. Linc00514 promotes breast cancer metastasis and M2 polarization of tumor-associated macrophages via Jagged1-mediated notch signaling pathway. J. Exp. Clin. Cancer Res. 39, 191 (2020).
Meng, J. et al. Tumor-derived Jagged1 promotes cancer progression through immune evasion. Cell Rep. 38, 110492 (2022).
Yang, M. et al. Tumour-associated neutrophils orchestrate intratumoural IL-8-driven immune evasion through Jagged2 activation in ovarian cancer. Br. J. Cancer 123, 1404–1416 (2020).
Ingham, M. & Schwartz, G. K. Cell-cycle therapeutics come of age. J. Clin. Oncol. 35, 2949–2959 (2017).
Yoon, J. & Oh, D. Y. HER2-targeted therapies beyond breast cancer - an update. Nat. Rev. Clin. Oncol. 21, 675–700 (2024).
Shen, R. & Seshan, V. E. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. 44, e131 (2016).
Blokzijl, F., Janssen, R., van Boxtel, R. & Cuppen, E. MutationalPatterns: comprehensive genome-wide analysis of mutational processes. Genome Med. 10, 33 (2018).
Li, Q. & Wang, K. InterVar: clinical interpretation of genetic variants by the 2015 ACMG-AMP guidelines. Am. J. Hum. Genet. 100, 267–280 (2017).
Mermel, C. H. et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 12, R41 (2011).
Abraham, M. J. et al. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1, 19–25 (2015).
Kellogg, E. H., Leaver-Fay, A. & Baker, D. Role of conformational sampling in computing mutation-induced changes in protein structure and stability. Proteins 79, 830–838 (2011).
Ramírez, F. et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Wang, L., Wang, S. & Li, W. RSeQC: quality control of RNA-seq experiments. Bioinformatics 28, 2184–2185 (2012).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011).
Soneson, C., Love, M. I. & Robinson, M. D. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Research 4, 1521 (2015).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550 (2005).
Liberzon, A. et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740 (2011).
Wang, T. et al. An empirical approach leveraging tumorgrafts to dissect the tumor microenvironment in renal cell carcinoma identifies missing link to prognostic inflammatory factors. Cancer Discov. 8, 1142–1155 (2018).
Ricketts, C. J. et al. The Cancer Genome Atlas comprehensive molecular characterization of renal cell carcinoma. Cell Rep. 43, 113063 (2024).
Paull, E. O. et al. A modular master regulator landscape controls cancer transcriptional identity. Cell 184, 334–351.e320 (2021).
Yaari, G., Bolen, C. R., Thakar, J. & Kleinstein, S. H. Quantitative set analysis for gene expression: a method to quantify gene set differential expression including gene-gene correlations. Nucleic Acids Res. 41, e170 (2013).
Wu, D. & Smyth, G. K. Camera: a competitive gene set test accounting for inter-gene correlation. Nucleic Acids Res. 40, e133 (2012).
Peter, L. & Steve, H. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008).
Zhang, X. et al. Code for “Comprehensive molecular profiling of FH-deficient renal cell carcinoma identifies molecular subtypes and novel therapeutic targets”. GitHub Repository. https://zenodo.org/records/14970134 (2025).
Acknowledgements
The authors thank all patients involved in this study, as well as their families/caregivers. This work was supported by the Natural Science Foundation of China (NSFC 82472673, 82403766, 82103097, 82202901, 82203110, 82273047, 82172785, 81902577, 81974398, 81872107 and 81872108), China Postdoctoral Science Foundation (2021M692286, 2020M673239 and 2021M692281), Research Foundation for the Postdoctoral Program of Sichuan University (2021SCU12014 and 2022SCU12042), 1.3.5 project for disciplines of excellence, West China Hospital, Sichuan University (ZYJC21020), Science and Technology Support Program of Sichuan Province (2021YFS0119, 24SYSX0223), the Natural Science Foundation of Sichuan Province (2023NSFSC1856, 2023NSFSC1858, 2023NSFSC1857, 23NSFSC2454), Post-Doctor Research Project, West China Hospital, Sichuan University (20HXBH026, 2021HXBH036 and 2021HXBH028), CAMS Innovatinn Fund for Medical Sciences (2022-I2M-C&T-B-098). We thank GloriousMed for their help in data analysis and uploading.
Author information
Authors and Affiliations
Contributions
Study concept and design: X.M.Z., J.J.Z., X.X.Y., J.Y.L., L.M.Z., P.F.S., N.C., H.Z., G.X.S.; Methodology, acquisition, analysis, or interpretation of the data: X.M.Z., J.J.Z., X.X.Y., J.Y.L., L.M.Z., P.T., Y.F.L., N.W.X., S.Z., J.R.C., J.G.Z., X.H., X.Y.P., L.N., M.N. Z., Y.T.C., Y.W.Z., H.Y.L., J.D.D., Z.P.W, H.L.L., Y.C.N., X.N.S., Y.Q.W., K.G., X.D.L., Z.B.C., Z.Y.H, Y.D.W., L.J.X, M.S.L, Q.H.Z, R.H., Q.W., X.L., J.Y.L., J.Y., B.H.L., Z.H.L., P.F.S., N.C., H.Z., G.X.S.; Financial support: X.M.Z., X.X.Y., J.Y.L., P.F.S., N.C., H.Z., G.X.S.; Drafting of the manuscript: X.M.Z., J.J.Z., X.X.Y., J.Y.L., L.M.Z.; Critical revision of the manuscript for important intellectual content: X.M.Z., J.J.Z., X.X.Y., J.Y.L., L.M.Z., S.Z., J.R.C., N.J.R., H.M., B.T., P.F.S., N.C., H.Z., G.X.S.; Statistical analysis: X.M.Z., J.J.Z., X.X.Y., J.Y.L., L.M.Z.; Study supervision: Q.W., X.L., J.Y.L., J.Y., B.H.L., Z.H.L., P.F.S., N.C., H.Z., G.X.S.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Christian Frezza, who co-reviewed with Désirée Schatton Iva Guberovic, A. Ari Hakimi and Chris Ricketts for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, X., Zhao, J., Yin, X. et al. Comprehensive molecular profiling of FH-deficient renal cell carcinoma identifies molecular subtypes and potential therapeutic targets. Nat Commun 16, 4398 (2025). https://doi.org/10.1038/s41467-025-59513-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-59513-8