Abstract
Adenosine (Ado), a nucleoside bridging intracellular metabolism with intercellular communication, plays an essential role in regulating processes such as sleep and seizure. While the functions of extracellular Ado (“eAdo”) are well documented, our knowledge about the distribution and regulatory functions of intracellular Ado (“iAdo”) is limited by a lack of methods for detecting iAdo in vivo. Here, we develop HypnoS, a genetically encoded fluorescent sensor for iAdo characterized by its high sensitivity, specificity, spatiotemporal resolution, and rapid response (sub-seconds). HypnoS enables real-time visualization of iAdo dynamics in live cultures, acute brain slices, flies, and freely moving mice. Using HypnoS for dual-color mesoscopic imaging in mice, we show that seizure-induced iAdo waves propagated across the cortex, following calcium signals. Additionally, two-photon imaging reveals that iAdo decays more rapidly in astrocytes than in neurons during seizures. Moreover, by recording iAdo dynamics in the basal forebrain during the sleep-wake cycle, we observe that iAdo signals are present during wakefulness and rapid eye movement (REM) sleep, regulated by equilibrative nucleoside transporters (ENT1/2). Thus, HypnoS is a versatile and powerful tool for investigating the biological functions of iAdo across a range of physiological and pathological states.
Similar content being viewed by others
Introduction
Adenosine (Ado) is a multifaceted molecule playing a critical function in numerous physiological and pathological processes. It influences astrocyte activity, neuronal plasticity, motor functions, learning and memory, and sleep regulation1,2,3,4,5,6. Beyond its role as a neuromodulator, Ado serves as a metabolic sensor in the brain, facilitating intercellular communication and modulating synaptic networks and neuroinflammation5,7. Additionally, Ado is implicated in conditions such as epilepsy, stroke, and neurodegeneration2,8,9,10. Understanding how Ado is produced and released in the brain is therefore of critical importance. Recent studies suggest that the concentration of Ado in the extracellular space results from various biological processes, including the production of extracellular Ado (“eAdo”, e.g., via extracellular ATP degradation)7,11,12 and the formation of intracellular Ado (“iAdo”), which is then released through equilibrative nucleoside transporters (ENT1/2)13,14,15. ENT2-mediated adenosine signaling has been reported studies of 40 Hz flickering-induced sleep and of glymphatic regulation in the brain16,17. However, discrepancies frequently arise concerning the cellular origin of Ado, the triggers for its release, and the release mechanisms, largely due to differences in experimental methodologies18. One of the current challenges in this area of research is to more precisely identify the different sources of eAdo and elucidate the mechanisms governing Ado release in response to both physiological and pathological stimuli.
Determining the relative contributions of eAdo and iAdo has been challenging due to the lack of techniques with sufficient spatiotemporal resolution to distinguish between their effects19. Traditional methods often rely on homogenized tissue samples, which obscure spatial information and may not accurately reflect iAdo or eAdo changes within specific cell populations7,20,21. In contrast, genetically encoded fluorescent sensors offer significant advantages, allowing researchers to target specific cell types and monitor real-time changes in a minimally invasive manner within living organisms20,22,23,24. Recent studies have utilized the GRABAdo sensor to monitor eAdo dynamics during sleep-wake cycles in vivo, uncovering that glutamatergic neurons contribute substantially more to the eAdo increase than other cell types14,25. However, the role of iAdo in contributing to eAdo across different cell types remains unclear18,26,27. This underscores the need for new tools to selectively, specifically, and in real-time monitor the metabolism and release pathways of iAdo with cell-type specificity.
In response to this need, we developed HypnoS (Hypersensitive intracellular adenosine Sensor), a genetically encoded green fluorescent sensor engineered by coupling a conformationally sensitive circular-permutated EGFP (cpEGFP) to Plasmodium vivax adenosine deaminase (PvADA). HypnoS is specifically designed for real-time monitoring of iAdo dynamics in living cells and model organisms. It enables the real-time detection of iAdo in living cells, acute brain slices of mice, and intact brains of animal models including flies and mice. HypnoS allows for precise spatiotemporal resolution of iAdo dynamics with cell-type specificity, such as during chemically induced seizures in vivo. Additionally, our application of HypnoS has provided insights into the regulation of iAdo dynamics by ENT1/2 during sleep-wake cycles in mice. By facilitating the study of iAdo dynamics in real-time and across various scales, HypnoS offers a powerful tool for investigating the roles of iAdo in both physiological and pathological states.
Results
Development of the iAdo sensor HypnoS
To develop a genetically encoded sensor for iAdo, we utilized Plasmodium vivax adenosine deaminase (PvADA) as a sensing module, as a prediction based on the crystal structure of PvADA indicated that the α7 structural gate and/or the β3/α12 loop of PvADA would undergo substantial conformational changes upon Ado binding28 (Supplementary Fig. 1a). We planned to insert a circularly permuted enhanced green fluorescent protein (cpEGFP) flanked with linkers29 into the α7 structural gate or into the β3/α12 loop (Fig. 1a), anticipating that an Ado-induced conformational change might alter fluorescence.
a Schematic of the genetically-encoded iAdo sensor, HypnoS. The N-terminal peptide (RSET) with a His tag originates from the bacterial expression vector pET28a. The X-ray crystal structures of Ado-bound PvADA (PDB: 2PGF) and cpEGFP (PDB: 3WLD) are shown as cartoon with Ado and chromophore of cpEGFP shown as stick and sphere, respectively. The loop bearing the insertion site in PvADA is marked in red. b The top panel outlines the strategy for creating iAdo sensors. The bottom panel shows the responses of various sensor candidates to 100 μM Ado (relative to HypnoS’s response). c One-photon Excitation and emission spectra of Ado-free and Ado-bound HypnoS in test buffer. AU, arbitrary units. d Two-photon excitation spectra of HypnoS. Data were collected from HEK293T cells expressing HypnoS. n = 30 cells for 0 μM Ado and n = 21 cells for 100 μM Ado, respectively. AU, arbitrary units. e Fluorescence response of HypnoS and HypnoS-mut to Ado. The fluorescence response (F488/F398) of HypnoS and a control sensor (HypnoS-mut) upon excitation at 488 nm and 398 nm is plotted against increasing Ado concentrations (n = 3). Emission was measured at 513 nm. f Binding kinetics of HypnoS to Ado. Each curve represents a different Ado concentration (bottom to top: 0.5, 1, 5, 10, 50, and 100 μM). g Nucleotide selectivity of HypnoS. The responses of HypnoS to various nucleotides (n = 3) are shown. Ado (adenosine), Ade (adenine), cAMP (cyclic adenosine monophosphate), AMP (adenosine monophosphate), ADP (adenosine diphosphate), ATP (adenosine triphosphate), Ino (inosine), IMP (inosine monophosphate), Guo (guanosine), GMP (guanosine monophosphate), GDP (guanosine diphosphate), GTP (guanosine triphosphate), Thd (thymidine), Urd (uridine), Cyd (cytidine), NAD+ (nicotinamide adenine dinucleotide), NADH (reduced NAD+), NADP+ (nicotinamide adenine dinucleotide phosphate), and NADPH (reduced NADP+) were added to the sensor solution. h Adenosine deaminase enzyme activity tests (n = 3). PvADA, HypnoS, or HypnoS-mut (1 μM) were added to test buffer containing 100 μM Ado. The change in light absorbance at 265 nm (Abs265 nm) was monitored to track Ado concentration changes. Representative images (left, scale bar 20 μm), the time course (middle, F488/F405) and a summary of the peak ΔR/R0 (right) for HypnoS and HypnoS-mut fluorescence before and after Ado (100 μM, i) or 5-Iodotubercidin (5-ITu, 5 μM, j) application in HEK293T cells. The fluorescence response (HypnoS and HypnoS-mut) is presented as a pixel-by-pixel ratio between the images obtained with 488 nm and 405 nm excitation (F488/F405). n = 75 (HypnoS) and 67 (HypnoS-mut) cells from 3 cultures in i and n = 97 (HypnoS) and 82 (HypnoS-mut) cells from 3 cultures in j. Two-tailed Student’s t-test: in i P = 2.6 × 10−130; in j P = 4.1 × 10−75 between HypnoS and HypnoS-mut. All the data are shown as mean ± s.e.m. with the error bars or shaded regions indicating the s.e.m. ***P < 0.001. Source data are provided as a Source Data file.
To identify optimal cpEGFP insertion sites, we leveraged the PvADA structure28 and analyzed the Cα dihedral angle differences between Ado-bound and Ado-free states (Supplementary Fig. 1b), indicating local conformational changes30. Ten sites with significant conformational changes were selected for cpEGFP insertion (Supplementary Fig. 1c). Among these, variant 175 (cpEGFP inserted between residues 175–176) showed a 4% response. We improved this response through linker randomization, screening over 2500 variants, and ultimately identified variant FFS-AN, which exhibited a 405% response with an EC50 of 4.9 mM (Fig. 1b and Supplementary Fig. 1d, e).
Subsequently, to enhance the affinity of FFS-AN, mutations were introduced into the Ado-binding pocket (L85A, G200V, L284C)28, yielding variants AV and AVC with improved affinities (EC50 of 9.18 μM and 1.51 μM, respectively; Supplementary Fig. 1f–i). We then performed random mutagenesis of cpEGFP based on AVC and identified a variant (I23W mutation of cpEGFP) with a 929% response (Fig. 1b and Supplementary Fig. 2). We named this sensor HypnoS (Hypersensitive adenosine Sensor). We also generated HypnoS-mut, an Ado-insensitive control (Supplementary Fig. 1j, k), harboring V200G and G201K mutations in the Ado-binding pocket, that displayed intense fluorescence but no response to Ado.
In vitro characterization of HypnoS and monitoring of iAdo dynamics in mammalian cells
To characterize the properties of HypnoS in vitro, we purified the protein and conducted a comprehensive analysis. HypnoS exhibited excitation peaks at 398 nm and 500 nm, with emission at 513 nm (Fig. 1c), similar to other cpEGFP-based sensors31. Upon Ado binding, HypnoS displays a ratiometric response with increased fluorescence at 488 nm and decreased fluorescence at 398 nm (Supplementary Fig. 1l, m). Absorbance spectra revealed two peaks at 398 nm and 500 nm in both Ado-bound and Ado-free states (Supplementary Fig. 1n,o). Ado binding increases HypnoS’s extinction coefficient (EC) by 1.5-fold (9408 vs. 6316 mM⁻1 cm−1 at 500 nm) and increases its quantum yield (QY) by 1.8-fold (0.44 vs. 0.25) (Supplementary Table 1). Under two-photon excitation, HypnoS in HEK293T cells and neurons displays peaks at ~800 nm and ~930 nm, with fluorescence increases at 930 nm and decreases at 800 nm upon Ado binding (Fig. 1d and Supplementary Fig. 3e). HypnoS showed intense, uniform fluorescence, with no detectable photobleaching during long-term (60 min), continuous two-photon imaging (Supplementary Fig. 3f).
HypnoS fluorescence increased with pH from 7.0 to 8.0 when excited at 488 nm (Supplementary Fig. 1p–s), while excitation at 398 nm allowed pH-independent Ado measurements32 (Supplementary Fig. 1s,t). Temperature changes between 22 °C and 37 °C did not significantly affect HypnoS’s fluorescence response to Ado (Supplementary Fig. 1u). HypnoS exhibited an affinity for Ado with an EC50 of 10.9 μM (Fig. 1e), suitable for detecting physiological Ado fluctuations (estimated as 1.8 ± 1.4 μM)33. Stopped-flow analysis revealed fast response kinetics, with apparent association (kon) and dissociation (koff) rate constants of 0.31 μM−1s−1 and 1.84 s−1, respectively, resulting in a ton of 0.22 s under 10 μM Ado and toff of 0.38 s, enabling sub-second reporting of Ado dynamics (Fig. 1f). The sensor demonstrated high specificity for Ado, with no detectable response to other tested nucleotides like inosine (Ino), AMP, ADP, and ATP (Fig. 1g). Moreover, neither HypnoS nor HypnoS-mut exhibited ADA enzymatic activity34 (Fig. 1h).
Next, we examined the cellular localization and brightness of HypnoS in mammalian cells. We observed that HypnoS displayed strong cytoplasmic fluorescence in HEK293T cells and showed an 800% increase in the F488/F405 ratio upon the application of 100 μM Ado. In contrast, HypnoS-mut showed no response (Fig. 1i). To assess HypnoS’s ability to monitor endogenous iAdo dynamics, cells expressing HypnoS were treated with the adenosine kinase (AdK) inhibitor 5-Iodotubercidin (5-ITu, Supplementary Fig. 3a). The application of 5 μM 5-ITu increased fluorescence of HypnoS by 30%. Still, not HypnoS-mut (Fig. 1j). In addition, 5-ITu did not induced detectable response of purified HyponoS sensor (Supplementary Fig. 3b, c), excluding the possibility of 5-ITu and HypnoS interactions. To measure the sensor affinity in cell cultures and to assess HypnoS sensor performance in quantifying iAdo concentration in living cells, we adopted a previously reported approach35,36 in which cells were permeabilized with digitonin to allow equilibration of iAdo with a standardized extracellular concentration prior to monitoring fluorescence by flow cytometry. Ado was added at a range of concentrations from 0.5 μM to 1 mM in permeabilized HEK293T cells expressing HypnoS. Titration curves (generated with flow cytometry data) indicated that the fluorescence ratio (F488/F405) of HypnoS increased following Ado addition in a dose-dependent manner, with apparent EC50 values of ~16.4 μΜ (Supplementary Fig. 3d), comparable the EC50 measured using purified HypnoS. Subsequently, the value of the fluorescence ratio (F488/F405) for HypnoS in nonpermeabilized HEK293T cells was interpolated into the titration curve to reveal a cytosolic iAdo concentration of 1.03 ± 0.45 μΜ (Supplementary Fig. 3d). These results suggest that HypnoS is a sensitive, specific, and rapid sensor for iAdo in mammalian cells.
HypnoS reveals distinct iAdo dynamics in neurons and astrocytes
After validating the HypnoS sensor for detecting iAdo dynamics in HEK293T cells, we explored its performance in rat primary neuron-glia cocultures. Using adeno-associated viruses (AAVs) under the control of the human synapsin (hSyn) promoter or glial fibrillary acidic protein (GfaABC1D) promoter, we selectively expressed HypnoS in neurons and astrocytes, respectively (Fig. 2a and Supplementary Fig. 4a). Both cell types exhibited similar fluorescence responses (Supplementary Fig. 4a–d) and kinetics (τon 22.0 s vs 50.8 s; τoff 38.8 s vs 146.3 s, Supplementary Fig. 3g–j) to 100 μM Ado, confirming that HypnoS’s sub-second response is sufficient for capturing kinetic differences across various cell types. Crucially, field stimuli-evoked calcium signaling remained unaltered between HypnoS-expressing and non-transfected neurons (Supplementary Fig. 4e, f), confirming that HypnoS expression does not interfere with calcium signaling. Additionally, qPCR analysis shows that HypnoS sensors did not alter the mRNA levels of Ent1, Ent2, Ada, or Adk (Supplementary Fig. 4g, h), suggesting that HypnoS does not interfere with normal adenosine metabolism. These results demonstrate that HypnoS can be reliably expressed and functional in both neurons and astrocytes.
a Schematic diagram illustrates the experimental protocol. DIV refers to days in vitro. b Representative pseudocolor images of HypnoS- or PercevalHR-expressing neurons before and after treatment with 10 mM 2-DG. c Left panel shows example traces of the fluorescence response for both sensors. Right panel summarizes the peak ΔR/R0 change. The HypnoS and PercevalHR fluorescence response is presented as a pixel-by-pixel ratio between the 488 nm excitation image and the 405 nm excitation image (F488/F405). ΔR/R0 denotes the change in the fluorescence ratio excited at 488 nm and 405 nm. n = 112 region of interest (ROIs) from 3 cultures. Two-tailed Student’s t-test: for iAdo, P = 3.1 × 10−83 between baseline and 2-DG, P = 5.0 × 10−146 between baseline and Ado; for ATP/ADP, P = 4.4 × 10−106 between baseline and 2-DG. Scale bar: 100 μm. d Representative pseudocolor images of HypnoS- or PercevalHR-expressing neurons before and after electrical field stimulation (10 Hz). e Left panel shows example traces of the fluorescence response for both sensors. Right panel summarizes the peak ΔF/F0 and ΔR/R0 change. For HypnoS, ΔF/F0 denotes the change in fluorescence intensity excited at 405 nm. For PercevalHR, ΔR/R0 denotes the change in the fluorescence ratio excited at 488 nm and 405 nm. Data are from n = 110 ROIs from 3 cultures. Two-tailed Student’s t-test: for iAdo, P = 1.1 × 10−68 between baseline and 600 pulses; for ATP/ADP, P = 3.4 × 10−104 between baseline and 600 pulses. Scale bar: 100 μm. f, g Same to b, c except HypnoS was expressed in astrocytes. Data are from n = 90 ROIs from 3 cultures. Two-tailed Student’s t-test: for iAdo, P = 1.1 × 10−22 between baseline and 2-DG, P = 2.9 × 10−80 between baseline and Ado; for ATP/ADP, P = 3.4 × 10−30 between baseline and 2-DG. Scale bar: 100 μm. h Representative pseudocolor images of cultured hippocampal astrocytes co-expressing HypnoS (upper panel) and jRGECO1a (calcium indicator, lower panel) in response to 100 μM ATP or 30 nM Thrombin. i Left panel shows example traces of the fluorescence response for HypnoS and jRGECO1a. Right panel summarizes the peak ΔF/F0 change for HypnoS. Data are from n = 102 cells from 3 cultures. Scale bar: 100 μm. All the data are shown as mean ± s.e.m. with the error bars or shaded regions indicating the s.e.m. ***P < 0.001. Source data are provided as a Source Data file.
To evaluate the ability of HypnoS to monitor endogenous iAdo dynamics in response to cellular stress and activity in neurons and astrocytes, we applied neuron-glia cocultures with two distinct perturbations: metabolic inhibition and electrical stimulation. By blocking glycolysis with 10 mM 2-deoxyglucose (2-DG)37,38, resulted in a significant increase in iAdo levels in neurons, as reported by fluorescence (F488/F405 ratio) of HypnoS; no such increases were detected in control cells expressing HypnoS-mut (Fig. 2b, c and Supplementary Fig. 4i, j). This increase corresponded with a decrease in the intracellular ATP: ADP ratio, as measured by PercevalHR (an ATP: ADP ratio sensor)37 (Fig. 2b, c).
Next, we investigated the relationship between neuronal activity and iAdo by stimulating cultures with electrical pulses or application of high extracellular K+ (75 mM). Imaging HypnoS at 405 nm (a pH-insensitive channel) revealed that electrical stimuli and application of high extracellular K+ (75 mM) each evoked significant, long-lasting iAdo elevations in somata and in neuronal processes (Fig. 2d, e and Supplementary Fig. 4k, l). Increasing the number of stimuli (from 30 to 1200) at 10 Hz progressively elevated iAdo levels as reported by HypnoS fluorescence (F405) (Fig. 2d, e). A concurrent decrease in the ATP:ADP ratio (Fig. 2d, e) was observed as a control. These findings suggest a direct link between neuronal energy metabolism and iAdo dynamics18,26,39,40. In contrast, astrocytes exhibited minimal changes in iAdo levels and ATP:ADP ratio in response to metabolic stress and neuronal stimulation (Fig. 2f, g and Supplementary Fig. 5). Additionally, while astrocytes showed robust calcium responses to various stimuli (e.g., ATP or thrombin)41,42, HypnoS detected no corresponding changes in iAdo levels (Fig. 2h, i and Supplementary Fig. 5). These results underscore the distinct iAdo metabolic processes in neurons versus astrocytes and highlight HypnoS’s utility for cell type-specific iAdo detection in cocultures.
Detecting iAdo dynamics ex vivo and in Drosophila in vivo with HypnoS
To assess HypnoS’s sensitivity for detecting iAdo dynamics in brain tissues, we expressed the sensor in hippocampal CA1 neurons of mice using AAVs. Two-photon imaging of acute brain slices 2–4 weeks post-transduction (Fig. 3a, b) revealed a dose-dependent increase in intracellular fluorescence with single-cell resolution in response to Ado, while no significant changes were observed in response to inosine (Fig. 3c, d). Similar to cultured neurons, a gradual elevation of iAdo levels was observed in neurons in acute brain slices in response to 2-DG-induced metabolic inhibition and to electrical stimulation (Fig. 3e–h). And a combination of these stimuli resulted in a synergistic iAdo increase (Fig. 3g, h), consistent with previous findings that neuronal activity drives ATP synthesis43. In contrast, astrocytes exhibited minimal changes in iAdo levels in response to electrical stimulation (Fig. 3i, j), yet did respond to the application of 100 μM Ado (Supplementary Fig. 6a, b), findings similar to our observations with cultured astrocytes. These results indicate that distinct iAdo metabolic processes are occurring in neurons versus astrocytes and highlight HypnoS’s utility for cell-type-specific iAdo measurement in brain tissue.
a Schematic illustration depicting the experimental design for (c–h). b Representative images showing HypnoS expression in the hippocampus CA1. Scale bar, 1 mm. Representative images (c), representative traces (d, top), and group summary (d, bottom) of the change in HypnoS fluorescence to indicated concentration of Ado or 100 μM inosine (Ino) in sensor-expressing brain slices. Scale bar, 100 μm (c, top) and 20 μm (c, middle and bottom). n = 20, 30, 30, 30, and 30 neurons from 3 slices of 2 mice for 1 μM, 10 μM, 100 μM, 1 mM Ado, and 100 μM Ino. Two-tailed Student’s t-test: P = 3.4 × 10−13 between Ino and Ado. Representative images (e), representative traces (f, top), and group summary (f, bottom) of the change in HypnoS fluorescence to 25 mM 2-DG. Scale bar, 100 μm (e, top) and 20 μm (e, middle and bottom). n = 60 neurons from 6 slices of 4 mice. Two-tailed Student’s t-test: P = 1.2 × 10−18 between ACSF and 2-DG. Representative images (g), representative traces (h, left), and group summary (h, right) of fluorescence response of HypnoS to electrical stimulation either in artificial cerebrospinal fluid (ACSF) or in 25 mM 2-DG. White dashed line, electrode placement. Scale bar, 100 μm. n = 5 slices from 5 mice. Paired two-tailed Student’s t-test: P = 0.0086 between ACSF and 2-DG. Representative images (i), representative traces (j, left), and group summary (j, right) of fluorescence response of astrocytic HypnoS to electrical stimuli. White dashed line, electrode placement. Scale bar, 100 μm. n = 38 cells of 5 slices from 5 mice. Two-tailed Student’s t-test: P = 0.22 between Basal and 20 pulses; P = 0.61 between Basal and 600 pulses. Data are shown as mean ± s.e.m. in (d, f, h, j) with the error bars or shaded regions indicating the s.e.m. **P < 0.01; ***P < 0.001; ns not significant. Source data are provided as a Source Data file.
Given that neuronal activity and energy metabolism are closely linked in various species, including Drosophila44, we examined this relationship in vivo. Transgenic flies expressing HypnoS in mushroom body (MB) Kenyon cells (KCs) were generated (Fig. 4a, b). Two-photon imaging of the MB medial lobe showed that HypnoS specifically responded to Ado, without interference from inosine (Fig. 4c, d). In control experiments, we did not observe a response in KCs expressing HypnoS-mut (Supplementary Fig. 6c, d). Inhibiting AdK increased iAdo levels, confirming HypnoS’s ability to report endogenous Ado changes in Drosophila (Fig. 4e and Supplementary Fig. 6e). To monitor iAdo dynamics during neuronal activation, we expressed the optogenetic tool CsChrimson45 in KCs (Fig. 4f, g and Supplementary Fig. 6f, g). Upon 635 nm laser excitation, a time-locked fluorescence increase was observed in the MB medial lobe. Increasing the stimulus frequency (0.2, 0.5, 1, 2, 4 Hz) elicited progressively stronger responses (Fig. 4h, i), supporting the coupling between neuronal activity and iAdo dynamics in KCs; no such increases were detected in KCs expressing HypnoS-mut (Fig. 4i and Supplementary Fig. 6h, i). These findings demonstrate HypnoS’s effectiveness in monitoring neuronal activity-induced iAdo increases with high specificity and sensitivity, both ex vivo in acute mouse hippocampal slice and in vivo in Drosophila.
a Schematic illustration depicting the experimental setup for imaging in Drosophila. b Representative fluorescence images of HypnoS expressed in Kenyon cells (KCs), with an expanded view of the olfactory MB medial lobe. Scale bars, 100 µm. Representative images (top), traces (bottom), and group summary of ΔF/F0 (bottom) in response to 100 µM Ado (c), 100 µM Ino (d), and 5 µM 5-ITu (e) perfusion. Scale bar, 20 µm. n = 3 flies. Two-tailed Student’s t-test: (c) P = 0.0040; (d) P = 0.0728; (e) P = 0.00020 between HypnoS and HypnoS-mut. f Schematic illustration depicting the strategy for imaging optogenetic stimuli induced iAdo elevations. g Representative fluorescence images of HypnoS and CsChrimson-mCherry expressed in KCs. Scale bars, 100 μm (left) and 20 μm (right). Representative response images (h), representative traces (i, top), and group summary (i, bottom) of HypnoS fluorescence in response to the optogenetic stimulation. Scale bar, 20 µm. n = 8 flies. Two-tailed Student’s t-test: P = 5.6 × 10−7 between HypnoS and HypnoS-mut. Data are shown as mean ± s.e.m. in (c–e, i) with the error bars or shaded regions indicating the s.e.m. **P < 0.01; ***P < 0.001; ns not significant. Source data are provided as a Source Data file.
Spatiotemporal dynamics of iAdo during seizures
Previous studies suggest that iAdo plays a protective role against seizures in various brain regions, including the hippocampus and cortex, with elevated levels in neurons (a 2–6-fold elevation in hippocampus46) and clearance primarily regulated by astrocytes10,47,48,49. To elucidate the spatiotemporal dynamics of iAdo during seizures and its potential role in seizure propagation and termination, we employed HypnoS in combination with calcium imaging and electrophysiological recordings using a kainic acid (KA)-induced seizure model in mice50.
We co-expressed HypnoS with the red-shifted calcium sensor mjRGECO1a14,51 by injecting AAVs into neonatal (postnatal day 0 (P0)–P1) mice52. Imaging the mouse dorsal cortex while performing electroencephalography (EEG) and electromyography (EMG) recordings, we identified seizures induced by KA injection (Fig. 5a). Similar to previous reports53,54,55, we observed an increase in calcium signals during the KA-induced seizure, followed by a spreading calcium wave of larger magnitude that closely tracked the flattening the epileptic EEG signal. Concurrently, we detected a spreading iAdo wave, reflected in the HypnoS signals, that followed the calcium wave’s propagation (Fig. 5b–d and Supplementary Movie 1). The waves reported by HypnoS and mjRGECO1a originated from approximately the same ___location, propagated at similar speeds (at ~75 μm/s and ~85 μm/s, respectively), and moved from the lateral cortex towards the medial region (Fig. 5e, f). Furthermore, iAdo waves followed calcium waves with a delay of ~20 s and exhibited slower decay kinetics (~100 s) (Fig. 5g, h), suggesting that neuronal activity drives iAdo accumulation during seizures in vivo. As a control, HypnoS-mut showed little fluorescence change during the KA-induced seizure (Fig. 5d).
a Schematic illustration depicting the experimental design of in vivo mesoscopic imaging for (b–d). b Representative cortical expression images of neuronal expression of iAdo sensor HypnoS and membrane-targeted calcium sensor jRGECO1a (mjRGECO1a) after kainic acid (KA) injection. Two ROIs are labeled. c The response of the HypnoS and mjRGECO1a in two ROIs. The solid and dashed lines in traces correspond to ROI 1 and ROI 2 shown in b, respectively. The red shading in the EEG, EMG, and velocity trace indicates the epileptic seizure. d HypnoS and HypnoS-mut peak responses in mice during the epileptic seizure. 8 seizure events from n = 4 mice for HypnoS and 10 seizure events from 3 mice for HypnoS-mut. Two-tailed Student’s t-test: for maximum response, P = 0.0057 between HypnoS and HypnoS-mut. e Representative images of the wave propagation detected by the indicated sensor. The white circle indicates the origin of waves; small white arrows indicate the wave-propagating velocity vector; green arrows indicate example propagating trajectories. L lateral, M medial, A anterior, P posterior. f Probability distributions of wave-propagating speeds and directions calculated using indicated sensors. g Probability distributions of the rise, peak, and decay time for hSyn-HypnoS (green) and mjRGECO1a (red). The rise time is defined as the time required to achieve 50% of the maximum response, starting from the point when mjRGECO1a reaches 50% of its maximum response. Similarly, the decay time is defined as the time required to return to 50% of the maximum response in the descent phase. The peak time is defined as the time required to reach maximum responses. h Representative pseudocolor images (top) and probability distributions (bottom) of time intervals between HypnoS and mjRGECO1a in the rising phase, at peak response, and in the decaying phase. n = 3 mice for hSyn-HypnoS and mjRGECO1a. All the data are shown as mean ± s.e.m. with the error bars or shaded regions indicating the s.e.m. **P < 0.01. Source data are provided as a Source Data file.
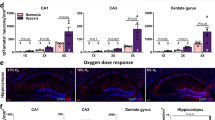
Next, we investigated differences in iAdo dynamics between neurons and astrocytes by expressing HypnoS under neuron-specific (hSyn) or astrocyte-specific (GfaABC1D) promoters (Fig. 6a). Two-photon imaging of HypnoS in the M1/M2 motor cortex revealed that both neuronal and astrocytic HypnoS showed significant fluorescence increases upon KA injection (Fig. 6b–g and Supplementary Movie 2), while HypnoS-mut displayed no detectable responses (Supplementary Fig. 7). Notably, Ado elevation in neurons exhibited a longer decay time compared to astrocytes (Fig. 6h–j), implying distinct iAdo metabolic pathways in neurons and astrocytes in vivo2,47. These results demonstrate HypnoS’s capability to report iAdo dynamics in mice in vivo with high sensitivity, molecule, and cell-type specificity during seizure activity.
a Left, schematic illustration depicting the experimental design of in vivo two-photon imaging in the mouse motor cortex for b–j. Right, images of neuron expressing HypnoS from −90 μm to −170 μm. b Representative images of expression and responses of HypnoS after KA or saline injection. 6 ROIs are labeled, and their traces are shown in d. f Group summary of the peak fluorescence changes of HypnoS in response to KA or saline injection. 120 ROIs are equally selected from 3 mice. Two-tailed Student’s t-test: for maximum response, P = 6.5 × 10−55, between saline and KA injections. c, e, g, Similar to b, d, f except for astrocyte expressing HypnoS. 60 ROIs are equally selected from 4 mice. Two-tailed Student’s t-test: for maximum response, P = 2.1 × 10−38, between saline and KA injections. h Representative images show neuronal (upper panels) and astrocytic (bottom panels) HypnoS expression and responses during epileptic seizure at a single cell level. i Representative responding traces of astrocytic and neuronal expressed HypnoS presented in h. j Group summary of decay time (τoff) of HypnoS expressed in astrocytes and neurons. For astrocytes, n = 60 ROIs from 3 mice; for neurons, n = 113 ROIs from 3 mice. Two-tailed Student’s t-test: for decay time, P = 2.4 × 10−37, between astrocytes and neurons. Scale bars, 25 µm (b, c) and 20 µm (h). All the data are shown as mean ± s.e.m. with the error bars or shaded regions indicating the s.e.m. ***P < 0.001. Source data are provided as a Source Data file.
Monitoring iAdo dynamics in freely behaving mice during sleep-wake cycles
To evaluate HypnoS in fiber photometry within deep brain areas of freely moving mice56,57, we expressed HypnoS or HypnoS-mut in the CA1 of the hippocampus and recorded fluorescence signals through an implanted optical fiber in a KA-induced seizure model (Fig. 7a and Supplementary Fig. 8a, b). Following KA injection, mice expressing HypnoS showed an increase in fluorescence, that was absent in mice expressing HypnoS-mut (Fig. 7b, d and Supplementary Fig. 9a). During successive seizures, the iAdo amplitude during the second seizure is lower than during the first seizure (Fig. 7c), findings consistent with the eAdo patterns observed in a microdialysis-based study58, where kainic acid-induced successive eAdo elevation was significantly attenuated. These findings all support coupling between iAdo and eAdo during seizure13,14,16. Notably, the HypnoS signal increase coincided with seizure termination as measured by EEG (Fig. 7e), supporting Ado’s role in seizure termination59. The signal kinetics revealed a rise time (τon) of ~25 s and a decay time (τoff) of ~80 s (Fig. 7f), indicating HypnoS’s compatibility with real-time iAdo dynamics detection in freely behaving mice.
a Schematic diagram depicting the strategy for in vivo fiber photometry recording during the seizures. b Example traces of simultaneous EEG (scale, 0.5 mV), EMG (scale, 0.5 mV), HypnoS (top, green) and HypnoS-mut (bottom, gray) signals (scale, 50% ΔR/R0) measured during the indicated seizure stages. c Peak responses of HypnoS in twin seizures. n = 6 mice for each treatment. Paired two-tailed Student’s t-test: P = 0.037 between the first and the second seizure. d Peak responses of HypnoS and HypnoS-mut. n = 6 mice for each treatment. Paired two-tailed Student’s t-test: P = 0.00020 between saline and KA for HypnoS. e HypnoS signals aligned to seizure termination for all mice. (Left panel) Heat map plot of 6 individual trials; (right panel) averaged signal; red line, seizure termination. n = 6 trials from 6 mice. f Summary of rise and decay time constants measured for the fluorescence change of HypnoS to seizures. n = 13 seizure events in 6 mice. Two-tailed Student’s t-test: P = 5.3 × 10−8 between τon and τoff. Data are shown as mean ± s.e.m. in e, f with the error bars or shaded regions indicating the s.e.m. * P < 0.05; ***P < 0.001. Source data are provided as a Source Data file.
To study iAdo dynamics during physiological sleep-wake cycles, we virally expressed HypnoS or HypnoS-mut AAVs in the basal forebrain (BF), a region regulating sleep homeostasis via Ado signaling60,61,62,63. Using fiber photometry (fluorescence channel) alongside EEG and EMG recordings (Fig. 8a and Supplementary Fig. 8c, d), we observed significantly higher iAdo levels during wakefulness and rapid eye movement (REM) sleep, compared to non-rapid eye movement (NREM) sleep (Fig. 8b, c, e; wake vs NREM, P = 0.0005; REM vs NREM, P = 0.0006). No such differences were detected in mice expressing HypnoS-mut (Fig. 8b, d, e and Supplementary Fig. 9b). Benefiting from the high temporal resolution of HypnoS, we captured rapid and significant iAdo changes during sleep-wake transitions (Fig. 8e, f), with average transition times (t50) of 22.8 s (wake to NREM), 11.5 s (NREM to wake), 38.3 s (NREM to REM,) and 19.3 s (REM to wake), suggesting a neural activity-dependent increase in iAdo.
a Schematic diagram depicting the strategy for in vivo fiber photometry recording during the sleep-wake cycles. b EEG power spectrogram, EMG (scale, 1 mV), HypnoS (top, green) and HypnoS-mut (bottom, gray) signals (scale, 5 z-score) during the sleep-wake cycles. In this and subsequent figures, the wake state is shaded in blue, REM sleep is shaded in pink, and NREM sleep is unshaded. Summary of the average HypnoS (c; n = 7 sessions from 5 mice) and HypnoS-mut (d; n = 7 sessions from 5 mice) fluorescence responses measured in the indicated sleep-wake states. Each symbol represents data from one recording. Paired two-tailed Student’s t-test: for HypnoS, P = 0.00046 between wake and NREM and P = 0.00057 between REM and NREM; for HypnoS-mut, P = 0.066 between wake and NREM and P = 0.16 between REM and NREM. e Time course and average signals measured for the HypnoS (green) or HypnoS-mut (gray) during the indicated transitions between sleep-wake states. The vertical dashed lines at time 0 represent the transition time. Unpaired two-tailed multiple t-test is performed between HypnoS and HypnoS-mut trace with shaded regions indicating P ˂ 0.05. The exact P values are provided in the Source Data file. Paired two-tailed Student’s t-test is performed between different brain states: REM→wake P = 0.075; NREM→wake P = 0.00041; wake→NREM P = 0.00055; NREM→REM P = 0.0017. n = 7 sessions from 5 mice for each panel. f Summary of t50 measured for the change in HypnoS fluorescence. n = 7 sessions from 5 mice. Data are shown as mean ± s.e.m. in e, f with the error bars or shaded regions indicating the s.e.m. ***P < 0.001; ns not significant. Source data are provided as a Source Data file.
ENT1/2 regulated iAdo levels during sleep-wake cycles
Equilibrative transporter proteins (ENT1/2) are critical in regulating iAdo levels and their coupling with eAdo26. Neuronal iAdo dynamics during different sleep-wake states largely mirror neuronal activity related eAdo dynamics recorded by a separated sensor GRABAdo25,64, suggesting a tight coupling between iAdo and eAdo. However, how iAdo is increased and contributes to eAdo remains unclear. To investigate ENT1/2’s impacts on Ado transport, we measured iAdo levels with HypnoS (Fig. 9a, b) both in cultures and in vivo. We observed significant increases in iAdo levels in response to exogenous Ado; and this increase was significantly attenuated in neurons lacking ENT1, ENT2, or both ENT1 and ENT2 (Fig. 9c). We also found that iAdo levels increased significantly in ENT1/2 knockout neurons compared to WT neurons under both the glycolysis inhibition and neuronal activation conditions (Fig. 9d, e). In contrast, no increase in eAdo was detected in a previous study in ENT1/2 KO neurons14 or upon our use of the ENT1/2 inhibitor S-(4-nitrobenzyl)-6-thioinosine (NBTI, 5 μM) and dipyridamole (DIPY, 10 μM) (Supplementary Fig. 10). These findings support that iAdo contributes to eAdo trafficking via both ENT1 and ENT2 in cultured neurons.
a Schematic illustration depicting the experimental design for c–e. b Schematic drawing depicting the transmembrane transport of Ado via ENT1/2. Representative pseudocolor images (c, left), traces (c, middle) and group summary (c, right) of HypnoS ΔF/F0 in response to 100 μM Ado in cultured hippocampal neurons from wild-type (WT) mice, ENT1 knockout (ENT1 KO) mice, ENT2 knockout (ENT2 KO) mice and ENT1/2 double knockout (ENT2 KO & sgENT1 KO, dKO) mice. ΔF/F0 denotes the change in fluorescence ratio excited at 488 nm and 405 nm. n = 150, 130, 148, and 150 cells from 3 cultures for WT, ENT1 KO, ENT2 KO, and ENT1/2 KO, respectively. Two-tailed Student’s t-test: P = 0.0000026 between ENT1 KO and ENT2 KO; P = 2.2 × 10−140 between WT and ENT1/2 dKO. Scale bar, 100 μm. Representative pseudocolor images (d, left), traces (d, middle) and group summary (d, right) of HypnoS ΔF/F0 in response to 10 mM 2-DG in cultured hippocampal neurons from WT mice, ENT1 KO mice, ENT2 KO mice and ENT1/2 dKO mice. ΔF/F0 denotes the change in fluorescence response excited at 405 nm. n = 150, 110, 148, and 150 cells from 3 cultures for WT, ENT1 KO, ENT2 KO, and ENT1/2 dKO, respectively. Two-tailed Student’s t-test: P = 2.1 × 10−61 between ENT1 KO and ENT2 KO; P = 2.8 × 10−103 between WT and ENT1/2 dKO. Representative pseudocolor images (e, left), traces (e, middle) and group summary (e, right, 600 pulses) of HypnoS ΔF/F0 in response to 10 Hz, indicated number of pulses in cultured hippocampal neurons from WT mice, ENT1 KO mice, ENT2 KO mice and ENT1/2 dKO mice. ΔF/F0 denotes the change in fluorescence response excited at 405 nm. n = 150, 117, 148, and 144 cells from 3 cultures for WT, ENT1 KO, ENT2 KO, and ENT1/2 KO, respectively. Two-tailed Student’s t-test: P = 4.0 × 10−11 between ENT1 KO and ENT2 KO; P = 3.3 × 10−41 between WT and ENT1/2 dKO. All scale bars, 100 µm. Data are shown as mean ± s.e.m. in c–e with the error bars or shaded regions indicating the s.e.m. ***P < 0.001. Source data are provided as a Source Data file.
To examine the role of ENT1/2 in regulating iAdo dynamics in vivo, we selectively ablated ENT1/2 in the BF by injecting an AAV virus expressing SaCas9-sgENT1 in ENT2 knockout mice (ENT1/2 dKO) (Supplementary Fig. 8e, f)14. We compared iAdo dynamics recorded by HypnoS with that in the BF of wild-type mice (WT) during sleep-wake cycles, observing that ENT1/2 dKO significantly increased iAdo during wakefulness and REM sleep in neurons (Fig. 10a–d; Wake: P = 0.0023; REM: P = 0.0020; NREM: P = 0.12), while significantly decreased iAdo during wakefulness and REM sleep in astrocytes (Fig. 10e, h; Wake: P = 0.000040; REM: P = 0.00052; NREM: P = 0.0078). Together, these results demonstrate HypnoS’s ability to monitor ENT1/2-dependent Ado transport with cell-type specificity in freely behaving mice.
a Schematic diagram depicting the strategy for fiber photometry recording of HypnoS fluorescence in neurons in the BF of wild-type (WT) or ENT1/2 double knockout (dKO) mice during the sleep-wake cycle. b EEG power spectrogram, EMG (scale, 2 mV), and HypnoS recordings (scale, 20 z-score) in WT (bottom) or ENT1/2 dKO (top) mice. In this and subsequent figures, the wake state is shaded in blue, REM sleep is shaded in pink, and NREM sleep is unshaded. c Time course for the HypnoS sensor measured in neurons in WT (green; n = 10 sessions from 8 mice) or ENT1/2 dKO (bule; n = 7 sessions from 4 mice) mice during the indicated transitions between sleep-wake states. The vertical dashed lines at time 0 represent the transition time. d Summary of the average HypnoS fluorescence responses measured in neurons in WT (green; n = 10 sessions from 8 mice) and ENT1/2 dKO (blue; n = 7 sessions from 4 mice) mice in the indicated sleep-wake states. Each symbol represents data from one recording. Two-tailed Student’s t-test: for wake, P = 0.0023; for NREM, P = 0.1204; for REM, P = 0.0020 between WT and ENT1/2 dKO mice. e–h Similar to a–d except for astrocyte expressing HypnoS. g Time course for the HypnoS sensor measured in astrocytes in WT (green; n = 8 sessions from 3 mice) or ENT1/2 dKO (blue; n = 8 sessions from 4 mice) mice during the indicated transitions between sleep-wake states. h Summary of the average HypnoS fluorescence responses measured in astrocytes in WT (green; n = 7 sessions from 3 mice) and ENT1/2 dKO (blue; n = 8 sessions from 4 mice) mice in the indicated sleep-wake states. Each symbol represents data from one recording. Two-tailed Student’s t-test: for wake, P = 0.000040; for REM, P = 0.00052; for NREM, P = 0.0078 between WT and ENT1/2 dKO mice. Data are shown as mean ± s.e.m. in (c, d, g, h) with the error bars or shaded regions indicating the s.e.m. **P < 0.01; ***P < 0.001; ns not significant. Source data are provided as a Source Data file.
Discussion
We present the development, characterization, and in vivo application of HypnoS, a genetically encoded sensor for intracellular adenosine (iAdo). HypnoS exhibits an impressive response (>900%), high affinity for Ado (EC50 ~ 11 μM), rapid kinetics (sub-second), high specificity, and spatial resolution, all while minimally impacting cellular physiology. This makes it highly suitable for monitoring pathologically and physiologically relevant iAdo dynamics in vivo. We demonstrated HypnoS’s capability to detect iAdo dynamics in response to diverse stimuli and under various behavioral conditions across multiple animal models. To our knowledge, this study represents the first demonstration of iAdo activity in cortical neurons and astrocytes at single-cell resolution in living animals. Additionally, it highlights the dynamic fluctuations of iAdo levels during seizures and sleep-wake cycles in the mammalian brain. Due to its high sensitivity and direct measurement capabilities, HypnoS is poised to be an invaluable tool for screening pharmaceuticals that target iAdo metabolic pathways through high-throughput screening assays.
Our in vivo two-photon imaging experiments revealed cell-type specific regulation of iAdo dynamics in a seizure model (Fig. 6h–j). This regulation correlates with higher adenosine kinase (AdK) expression in astrocytes compared to neurons, which facilitates iAdo clearance10,47,48. Simultaneous imaging of neuronal HypnoS and mjRGCO1a allowed us to visualize iAdo and calcium dynamics throughout the entire dorsal cortex, revealing a robust positive correlation between the two. Electrical and optogenetic stimulation experiments, conducted in vitro, ex vivo, and in vivo, demonstrated that action potentials are sufficient to elevate iAdo levels. This suggests that increases in iAdo induced by neuronal activity may be mediated by calcium-dependent synthesis or degradation. Notably, neuronal iAdo waves persist for considerably longer (>100 s) than calcium waves, allowing for the integration and storage of information through iAdo increases in neurons rather than astrocytes.
A well-recognized role of Ado is its involvement in sleep homeostasis. Traditionally, iAdo has been conceptualized as a “retaliatory metabolite” that is released during wakefulness to promote sleep drive. However, the dynamics of iAdo during sleep-wake cycles have yet to be documented. Utilizing the sensor, we recorded iAdo changes throughout the sleep-wake cycle in the basal forebrain, revealing elevated iAdo levels during both wakefulness and rapid eye movement (REM) sleep in neurons, and show that these increases are regulated by equilibrative nucleoside transporters ENT1/2 (Fig. 10). These results highlight the crucial role of ENT1/2 in maintaining the balance between iAdo and eAdo, particularly in sleep regulation. Future studies are necessary to elucidate the precise mechanisms by which ENT1/2 is involved in sleep-wake regulation.
Although HypnoS is already suitable for sensitively monitoring iAdo levels in vivo in both physiological and pathological conditions, it is conceivable that further improvements would be enabling for further, specialized applications. For example, we could generate HypnoS variants with improved properties through structure-guided protein engineering: perhaps a HypnoS variant with higher affinity would enable more sensitive detection of subtle physiological changes of iAdo at submicromolar concentration, as is thought to occur in erythrocytes, adipose tissue, and liver33,65,66,67. In addition, iAdo is a ubiquitous metabolite in living cells, functioning as both a neuromodulator—shaping communication between neurons—and a regulator of intracellular metabolism2,68. Expanding the spectral range of iAdo sensors to encompass red, far-red, and near-infrared options should enable monitoring of multiple signals in living systems, likely offering deeper insights into iAdo’s role in both physiological and pathological conditions, which would be similar to achievements with calcium sensors69,70,71.
Methods
This research complies with all relevant ethical regulations approved by Peking University and the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences.
Animals
All procedures for animal surgery and experimentation were performed in accordance with and approved by the laboratory animal care and use committees of Peking University and the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences. Both male and female postnatal day 0 (P0) Sprague-Dawley rats were used to prepare neuron-glia cocultures; wild-type C57BL/6N mice (Beijing Vital River Laboratory) and ENT2 knockout mice14 were used in this study. All animals were housed at 18–23 °C in 40–60% humidity under a normal 12-h light–dark cycle with food and water available ad libitum.
The transgenic Drosophila lines UAS-HypnoS and UAS-HypnoS-mut were generated using Phi-C31-directed integration into attp40 or VK00005 at the Core Facility of Drosophila Resource and Technology, Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences. The following Drosophila lines were also used in this study: 30y-Gal472 (a gift from Yi Rao, Peking University, Beijing); and UAS-CsChrimson-mCherry (a gift from Chuan Zhou, Institute of Zoology, Chinese Academy of Sciences, Beijing). The flies were raised on standard cornmeal-yeast medium at 25 °C and 70% relative humidity under a 12-h/12-h light/dark cycle. Adult female flies within 2 weeks after eclosion were used for fluorescence imaging.
Chemicals and reagents
Adenosine and other nucleotides were purchased from Sigma and prepared just before use. 2-DG was purchased from MCE. 5-ITu was purchased from TargetMol Chemicals Inc. Thrombin was purchased from Sigma. Kainic acid was purchased from Cayman. The mouse anti-HA-tag antibody (Cat. No. M20003) was purchased from Abmart Shanghai Co., Ltd. iFluor® 647 goat anti-mouse IgG (H + L) (Cat. No. 16837) was purchased from AAT Bioquest. All the restriction enzymes used in plasmid cloning were purchased from NEB. Primers were synthesized by GENEWIZ. The PvADA and PercevalHR gene fragments were synthesized by GENEWIZ. All the PCR reactions were performed using KOD-Plus-Neo from TOYOBO. All plasmids were confirmed by commercial sequencing (GENEWIZ).
Plasmid construction
Plasmodium vivax Adenosine deaminase (PvADA) cDNA was synthesized and cloned into the pET-28a vector using the T7 promoter and a N-terminal 6x His tag. The gene fragment encoding cpEGFP from GRABDA1h29 with GG/TGAA linkers was PCR amplified and inserted into various positions of PvADA gene using the BsaI Golden Gate Assembly method73. For expressing HypnoS in mammalian cells, a gene fragment encoding HypnoS was inserted between the NheI and EcoRI restriction sites of the pcDNA3.1 vector by T4 ligation. HypnoS-mut was cloned into pET-28a and pcDNA3.1 vectors with the same restriction enzymes sites as HypnoS constructs for expression in E. coli and mammalian cells, respectively. The gene fragment encoding PercevalHR was synthesized according to the published sequence37 and cloned into the pAAV vectors to make pAAV-EFS-DIO-PercevalHR and pAAV-GfaABC1D-PervevalHR. cDNAs of HypnoS and HypnoS-mut were subcloned into pAAV vectors to make pAAV-hSyn-HypnoS, pAAV-hSyn-HypnoS-mut, pAAV-GfaABC1D-HypnoS, and pAAV-GfaABC1D-HypnoS-mut. All the resulting plasmids products were confirmed by commercial sequencing (GENEWIZ).
Protein expression and purification
Protein expression of various PvADA constructs was carried out according to a previously published method28. In brief, pET-28a vectors carrying ORF were transformed into E. coli strain BL21(DE3). Transformed cells were grown in TB media containing 50 μg/mL kanamycin to an OD600 of 2–3. IPTG was then added to a final concentration of 1 mM to induce protein expression. Cells were grown at 18 °C for an additional 36–48 h before harvested by centrifugation at 4 °C for 10 min. The harvested cells were resuspended in binding buffer (50 mM HEPES/NaOH, pH 7.3, 300 mM NaCl, 5% glycerol, 10 mM imidazole), lysed by sonication, and clarified by centrifugation. The supernatant was loaded onto a HisTrap HP column (GE Healthcare), washed with 10 column volumes binding buffer and eluted with a linear gradient of imidazole (10–500 mM) of 10 column volumes. The protein preparations were subsequently pooled and desalted in test buffer (50 mM HEPES/NaOH, pH 7.3, 100 mM NaCl, 1 mM DTT). All purification steps were performed at 4 °C or on ice. Protein purity was verified as ≥95% by SDS-PAGE. Protein quantification was performed using Nanodrop A280 test in three times and diluted to a concentration of 1 μM in test buffer for subsequently use immediately.
Linker library construction and screening
A library of PvADA-cpEGFP constructs with different N-terminal and C-terminal linkers was generated according to a published method74, with modifications. Briefly, the gene fragment for cpEGFP was PCR amplified with degenerate primers containing 1–3 NNC codons for the linker sequence and inserted into PvADA gene using the BsaI Golden Gate Assembly method. DNA libraries were assembled into pET-28a vector, and products were transformed to E. coli strain DH5α cells and grown in solid medium plate at 37 °C for 12 h and colonies were washed and collected. Then, plasmids were extracted using a miniprep kit according to the manufacturer’s instructions. The extracted plasmids libraries were transformed to E. coli strain BL21(DE3) cells and grown in solid medium plate at 37 °C for 12 h, followed by cultured at 18 °C for 1 day. Colonies showed high fluorescence intensity were picked and cultured in deep well plates in TB media containing 50 μg/mL kanamycin at 37 °C for 12 h. IPTG was then added to a final concentration of 1 mM to induce protein expression. Colonies were then transferred to 18 °C, 1000 rpm shaking for 48 h. Cultures were then pelleted by centrifugation and finally were resuspended in 200 μL test buffer (50 mM HEPES/NaOH, pH 7.3, 100 mM NaCl, 1 mM DTT), then repeated freezing and thawing 3 times. Cell debris was pelleted by centrifugation, and the supernatant (protein containing solution) was transferred to a clean 96-well plate. 30 μL protein containing solution was added into optically clear 384-well plates and mixed with 100 μM Ado or the same volumes of test buffer as control wells. Plates were read on Synergy H1 microplate reader (BioTek). Emission signals at 513 nm with excited by 398 nm and 488 nm respectively of each well was then recorded after 5 min of shaking.
In vitro characterization of HypnoS
Fluorescence spectra were measured on Cary Eclipse Fluorescence Spectrophotometer (Agilent Technologies). Purified HypnoS was diluted in test buffer to a concentration of 1 μM and added to fluorescent cuvettes. Excitation spectra was scanned from 380 nm to 512 nm with emission wavelength of 513 nm, and emission spectra was scanned from 501 nm to 600 nm with excitation wavelength of 500 nm at room temperature both before and 1 min after the addition of Ado or other ligands dissolved in test buffer. UV-Vis absorbance spectra were measure on Cary Series UV-Vis Spectrophotometer (Agilent Technologies). Purified HypnoS was diluted in test buffer to a concentration of 1 μM and added to cuvettes. Stock solution of Ado was added to sensor proteins and diluted more than 50 times into indicated concentrations to maintain solution volumes unchanged. Spectra was scanned from 550 nm to 300 nm at room temperature. Plate reader assays were performed on Synergy H1 microplate reader (BioTek). Purified sensor protein was diluted in test buffer at pH 7.3 (or at various pH values, for pH sensitivity assays) to a concentration of 1 μM. 100 μL of diluted sensor was added to each well of a black 96-well plate, to which 100 μL of 2 times concentration of nucleotides dissolved in the same buffer was added in three duplicates. Emission signals at 513 nm with excited by 398 nm and 488 nm respectively of each well was then recorded after 5 min of shaking unless otherwise noted.
The association rate constant (kon) and dissociation rate constant (koff) between HypnoS and Ado were determined using an SX20 Stopped-Flow accessory (Applied Photophysics Ltd). Briefly, 2 μM of protein solution was mixed 1:1 with Ado of different concentrations (1, 2, 10, 20, 100, and 200 μM), and the fluorescence excited at 488 nm and 405 nm were measured with a 520/30 nm filter. The data were fitted using the following single-exponential function75: R(t) = R0 + Aobs × exp(−kobs × t), where R(t) is the value of fluorescence ratio (F488/F405) increase at time t, R0 is the final value of fluorescence ratio increase, Aobs is the amplitude of the exponentially decreasing part and kobs is the observed first-order rate constant. The kon and koff were fitted using the following equation: kobs = kon × [Ado] + koff, where [Ado] is the concentrations of Ado used. The association and dissociation half-time ton and toff were calculated as ln2/(kon × [Ado]) and ln2/koff, respectively.
To get the excitation wavelength-dependent brightness under two-photon excitation, HEK293T cells expressing HypnoS were excited with wavelengths from 700 to 1100 nm with a 10 nm step size on a Leica -TCS SP8 DIVE two-photon microscope equipped with a MaiTai DeepZ laser and a 25 × 0.95 NA water immersion objective. Images were captured using LAS X software. The fluorescence were collected and the intensities were then ploted to different excitation wavelengths.
Cell culture, DNA transfection, and virus infection
HEK293T cells were acquired from ATCC and maintained in DMEM medium (Gibco) supplemented with 10% FBS, 1% penicillin and streptomycin (Gibco) in an incubator at 37 °C with 5% CO2. Plasmid transfections of cultured cells were performed using Hieff TransTM Liposomal Transfection Reagent (Yeasen) according to the manufacturer’s instructions.
Primary neuron-glia cocultures were prepared and cultured as described previously25. In brief, rat primary neuron-glia cocultures were prepared from 0-day-old (P0) rat pups (male and female, randomly selected) purchased from Beijing Vital River Laboratory (Beijing, China). Hippocampal cells were dissociated from the dissected brains in 0.25% Trypsin-EDTA (Gibco) and plated on 12-mm glass coverslips coated with poly-D-lysine (Sigma-Aldrich) in neurobasal medium (Gibco) containing 2% B-27 supplement (Gibco), 1% GlutaMAX (Gibco), and 1% penicillin-streptomycin (Gibco). Based on glial cell density, after ~4 days in culture (DIV4) cytosine β-D-arabinofuranoside (Sigma) was added to the hippocampal cultures in a 50% growth media exchange, with a final concentration of 2 μM. Primary neuron-glia cocultures were cultured at 37°C in 5% CO2.
Confocal imaging of cultured cells
Before imaging, the culture medium was replaced with Tyrode’s solution contained (in mM): 150 NaCl, 4 KCl, 2 MgCl2, 2 CaCl2,10 HEPES, and 10 glucose (pH 7.3–7.4). HEK293T cells expressing HypnoS and HypnoS-mut were plated on 35 mm glass-bottomed dishes. Imaging was performed using a Zeiss LSM880 laser scanning confocal microscope equipped with a PFS, motorized XY stage, and a stage incubator set at 37 °C. A Plan Apochromat 20×/0.45 NA objective was utilized. HypnoS and HypnoS-mut were imaged by continuous excitation at 405 nm and 488 nm, respectively, with emission ranges from 502 nm to 544 nm. Images were acquired in a 512 × 512 format with 8-bit depth and a gain range of 650 to 800. The raw data in 8-bit CZI format were exported to ImageJ (https://fiji.sc) for further analysis. Primary neuron-glia cocultures grown on 12-mm coverslips were imaged using a Ti-E A1 confocal microscope (Nikon) equipped with a 10×/0.45 NA objective, a 20×/0.75 NA objective, a 40×/1.35 NA oil-immersion objective, a 405-nm laser, a 488-nm laser, and a 561-nm laser; fluorescence excited at 405 nm and 488 nm (HypnoS, HypnoS-mut and PercevalHR) were recorded using a 525/50-nm emission filter, and red fluorescence (jRGECO1a) were recorded using a 595/50-nm emission filter.
The following compounds were applied by either bath application or a custom-made perfusion system: Ado, ATP, Ino, 5-ITu, 2-DG, and thrombin. Between experiments, the recording chamber was cleaned thoroughly using Tyrode’s solution and 75% ethanol. For field stimulation of cultured neurons, parallel platinum electrodes positioned 1-cm apart were controlled using a Grass S88 stimulator (Grass Instruments), and 1-ms pulses were applied at 80 V. Except where indicated otherwise, all experiments were performed at room temperature.
Flow cytometry
Total RNA was isolated from each sample by the acid phenol-guanidinium-chloroform method using the TRIeasy Total RNA Extraction reagent (Yeasen, China) according to the manufacturer’s instructions. The cDNA was synthesized from total RNA (2000 ng) using Hifair AdvanceFast One-step RT-gRNA Digestion SuperMix (Yeasen, China) according to the manufacturer’s instruction. The cDNA was mixed with RNase-free water, HiEff UNICON Universal Blue qPCR SYBR Green Master Mix (Yeasen, China), and forward and reverse primers at a final concentration of 0.5 μM each. The primer used for RT-qPCR are listed76: ADA (5′- GAA GGC AAA GGA GGG CGT GGT CTA -3′ and 5′- GAT GTC CAC AGC CTC ACG CAC AAC -3′); ADK (5′- GTG CTA TTT GGA ATG GGG AAT -3′ and 5′- CAA CCA CTG AGC CAC TTT CAT -3′); ENT1 (5′- CTT GGA ATT CAG GGT CAG AA -3′ and 5′- ATC AGG TCA CAC GAC ACC AA -3′); ENT2 (5′- CAT GGA AAC TGA GGG GAA GA -3′ and 5′- GTT CCA AAG GCC TCA CAG AG -3′); Beta-actin (5′- TGA GAG GGA AAT CGT GCG TGA C-3′ and 5′- GAA GGA AGG CTG GAA AAG AG-3′). The relative mRNA amount of the target gene was normalized to that of a reference gene (Beta-actin).
RNA isolation and quantitative real-time PCR
Data acquisition was performed using a Cytoflow LX flow cytometer, utilizing the 488 channel (excitation: 488 nm, emission: 525/40 nm) and the 405 channel (excitation: 405 nm, emission: 525/40 nm) for the biosensor. To ensure data quality, cells were gated to exclude debris, and a standard doublet-exclusion method was applied. A total of 10,000 cells per sample were evaluated. Data analysis and visualization were conducted using CytExpert 2.5 software. Ratiometric fluorescence values (R405/488) were calculated for each individual cell using the derived function obtained from CytExpert 2.5.
Preparation and fluorescence imaging of acute brain slices
Wild type adult C57BL/6 N mice (6–12 weeks old) were anesthetized via intraperitoneal injection of Avertin (500 mg/kg body weight) and secured in a stereotaxic frame. Using a Nanoliter 2000 microinjector (WPI), AAV vectors were delivered into the hippocampal CA1 region at stereotaxic coordinates AP −1.8 mm from Bregma; ML ±1.2 mm; DV −1.25 mm from the dura.
Two to four weeks post injection, mice were re anesthetized with Avertin (500 mg/kg, i.p.) and transcardially perfused with ice cold, oxygenated slicing buffer composed of (in mM): 110 choline chloride, 2.5 KCl, 0.5 CaCl2, 7 MgCl2, 1.3 NaH2PO4, 25 NaHCO3, 20 glucose, 1.3 Na ascorbate, and 0.6 Na pyruvate. Brains were rapidly excised and submerged in the same ice cold, oxygenated buffer, then sectioned into 200 µm slices using a Leica VT1200 vibratome. Slices were incubated for at least 40 min at 34 °C in oxygenated artificial cerebrospinal fluid (ACSF) containing (in mM): 125 NaCl, 2.5 KCl, 2 CaCl2, 1.3 MgCl2, 1.3 NaH2PO4, 1.3 Na ascorbate, 0.6 Na pyruvate, 20 glucose, and 25 NaHCO3.
For imaging, slices were transferred to a perfusion chamber on an Olympus FV1000MPE two photon microscope equipped with a 20× water immersion objective and a mode locked Mai Tai Ti:Sapphire laser (Spectra Physics). HypnoS fluorescence was elicited by excitation at 920 nm, and emission was collected through a 495–540 nm band pass filter. A bipolar stimulation electrode (WE30031.0A3, MicroProbes for Life Science) was positioned in the CA1 stratum radiatum under fluorescent guidance for electrical stimulation. Frame acquisition (320 × 320 pixels) and stimulus timing were synchronized via a custom Arduino control script, with images captured at 1.3 frames per second.
Two-photon imaging in Drosophila
Female Drosophila within 2 weeks of eclosion, were used for imaging experiments. The fly was mounted on a customized chamber by tape, such that the antennae and abdomen were exposed to the air. The cuticle between the compound eyes, as well as the air sacs and fat bodies, were removed to expose the brain, which was then bathed in saline (AHLS) containing (in mM) 108 NaCl, 5 KCl, 5 HEPES, 5 trehalose, 5 sucrose, 26 NaHCO3, 1 NaH2PO4, 2 CaCl2 and 2 MgCl2. The same Olympus two-photon microscope used for brain slice imaging was also used here. A 920-nm laser was used for excitation. A 495–540-nm filter was used for HypnoS imaging, and a 575–630-nm filter was used for mCherry imaging. For Ado and Ino application, a patch of the blood–brain barrier of the fly was carefully removed with tweezers before imaging, and saline containing Ado or Ino was delivered to the brain to a 100 μM final concentration. For 5-ITu application, a patch of the blood–brain barrier of the fly was carefully removed with tweezers before imaging, and saline containing 5-ITu was delivered to the brain to a 5 μM final concentration. For optogenetic stimulation, the 635-nm laser was used to deliver pulses (10-ms pulses at 0.2, 0.4, 1, 2, and 4 Hz) through optical fibers placed near the fly’s brain. Flies were fed with cornmeal containing 10 μM β-carotene (Sigma-Aldrich) 1 d before optogenetics experiments. The sampling rates of imaging were set at 2 Hz during Ado, Ino, and 5-ITu application. The sampling rates of imaging were set at 4 Hz during optogenetic stimulation. Arduino was used to synchronize stimulation delivery and imaging with a custom program.
Two-photon imaging in mice
AAV2/9-hSyn-HypnoS, AAV2/9-hSyn-HypnoS-mut, AAV5-GfaABC1D-HypnoS, and AAV5-GfaABC1D-HypnoS-mut viruses were packaged by Vigene Biosciences (Jinan, China). Wild-type female C57BL/6 N mice, aged 6 to 8 weeks, were anesthetized with an injection of 2.5% Avertin and maintained under anesthesia with 1.5% isoflurane. The skin and skull covering the motor cortex were retracted, and 300 nL of the respective AAVs were injected into the motor cortex (AP. 1.0 mm from Bregma; ML ±1.5 mm; DV −0.5 mm from the dura). A 3 mm diameter round coverslip was used to replace the removed skull, and a metal recording chamber was securely attached. Three weeks post-virus injection, awake mice were habituated for ~15 min in the treadmill-adapted imaging apparatus to minimize stress effects from head restraint. The motor cortex, at a depth of 100–200 μm below the pial surface, was imaged using Prairie View 5.5.64.100 software on a Bruker Ultima Investigator two-photon microscope equipped with the Spectra-Physics Insight X3 laser and a 16×/0.8 NA water immersion objective. A 920 nm laser line was used for exciting HypnoS or HypnoS-mut, and a 490–560 nm filter was employed for fluorescence collection. The sampling rate was set to 1 Hz.
For image analysis, motion artifacts were first corrected using the EZcalcium77 motion correction algorithm. Subsequently, the fluorescence intensities of regions of interest (ROIs) encompassing neuronal/astrocytic somata were extracted using ImageJ software. The background-subtracted fluorescence intensity was then used to calculate the relative fluorescence change (ΔF/F0).
Mesoscopic in vivo imaging
To achieve expression of the genetically encoded sensor throughout the cortex, the AAVs were introduced through the transverse sinus following established protocols52. Neonatal C57BL/6 N pups (postnatal days 0–1) were gently removed from their dam and placed on a heated pad. Hypothermic anesthesia was induced by laying the pup on an ice-cooled metal platform for 2–3 min, after which two small incisions were made directly above each transverse sinus. Using a pulled glass micropipette, a 1:1 mixture of AAV2/9-hSyn-HypnoS and AAV2/9-hSyn-GAP43-jRGECO1a (for dual-color imaging; total volume 6 µL) or AAV2/9-hSyn-HypnoS-mut (for single-color experiments; total volume 4 µL, 2 µL per hemisphere) was slowly infused at 1.2 µL min−1. The pipette remained in situ for at least 30 s post-injection to minimize backflow. Incisions were then closed with Vetbond tissue adhesive (3 M Animal Care Products), and pups were allowed to recover on a warmed surface before being gently rubbed with nesting material and returned to their home cage.
Approximately 8 weeks after neonatal AAV administration, mice underwent aseptic surgery for implantation of a chronic cranial window and EEG/EMG electrodes. Anesthesia was induced by intraperitoneal injection of 2,2,2-tribromoethanol (500 mg/kg, Sigma-Aldrich) and maintained with 1% isoflurane via nose cone. Animals were secured in a stereotaxic frame, and 2% lidocaine hydrochloride was infiltrated subcutaneously beneath the scalp. Erythromycin ophthalmic ointment protected the corneas throughout the procedure. After reflection of the scalp and removal of most of the skull above the dorsal cortex and the overlying muscle, a craniotomy was made over the dorsal cortex. The exposed dura was covered with a custom-cut, flat coverslip (D-shape, ~8 mm × 8 mm) to establish a stable optical window. EEG and EMG electrodes were then implanted as described above and affixed with dental cement. Post-operatively, mice were allowed at least 7 days for recovery prior to imaging sessions.
Mesoscopic fluorescence recordings were acquired with a bespoke dual-color macroscope comprising a ×2/0.5 NA objective (Olympus MVPLAPO2XC), two ×1/0.25 NA tube lenses (Olympus MVPLAPO1X), and paired sCMOS detectors (Andor Zyla 4.2 Plus; 2048 × 2048 pixels, 16 bit)78,79. Excitation at 405 nm, 488 nm, and 561 nm was provided by a multi-line, fiber-coupled laser system (Chang-chun New Industries Optoelectronics RGB-405/488/561/642 nm, 220 mW). Emission fluorescence was separated by a 567 nm cut-on long-pass dichroic (Thorlabs DMLP567L) and directed through either a 525/36 nm or 609/34 nm band-pass emission filter (Chroma) onto the respective cameras.
Laser firing and frame acquisition were orchestrated by an Arduino Uno running custom firmware, with image capture managed via Micro-Manager 2.0 software (NIH). For single-channel (green) recordings, the 488 nm line was interleaved with brief 405 nm pulses; for dual-color experiments, 488 nm and 561 nm beams were co-activated and similarly interleaved with 405 nm. Images were acquired using Micro-Manager 2.0, and data were binned 4× on-sensor to yield 512 × 512 pixel frames, acquired at 1 Hz with 40 ms exposure per channel.
During imaging sessions, mice were head-fixed but permitted to ambulate on a custom treadmill80. In seizure paradigms, an infrared video camera was positioned above the animal to capture behavioral correlates. Following ~60 min of baseline recording, kainic acid (10 mg/kg, i.p.) was administered to elicit seizures. All data streams-including mesoscopic imaging, EEG/EMG signals, optical stimulation triggers, and infrared video-were time-aligned via a Power1401 data acquisition interface (Cambridge Electronic Design).
Fiber-photometry recording of intracellular Ado dynamics during the sleep-wake cycle
Adult wild-type C57BL/6N mice or ENT2 KO mice were anesthetized with isoflurane and placed on a stereotaxic frame for AAV injection (300 nL per site). In Fig. 8, AAV2/9-hSyn-HypnoS or AAV2/9-hSyn-HypnoS-mut was injected into the BF using the following coordinates: AP 0 mm from Bregma; ML ±1.35 mm; DV −5.1 mm from the dura. In Fig. 10a–d, AAV2/9-hSyn-HypnoS was injected into the BF of both wild-type and ENT2 KO mice, and AAV2/9-CMV-SaCas9-sgENT1 was injected into the BF of ENT2 KO mice. An optical fiber cannula (200 µm, 0.37 NA, Inper) was implanted 0.05 mm above the virus injection site in BF to record sensor signals.
To record the animal’s sleep–wake state, we attached and fixed custom-made EEG and EMG electrodes to the skull through a microconnector. EEG electrodes were implanted into craniotomy holes situated over the frontal cortex and visual cortex, and EMG wires were placed bilaterally in the neck musculature. The microconnector was attached to the skull using glue and a thick layer of dental cement. After surgery, the mice were allowed to recover for at least 2 weeks.
The fiber-photometry system (Inper) was used to record the fluorescence signals in freely moving mice during the sleep–wake cycle. In Figs. 8 and 10a 10-Hz 470/10-nm filtered light (20–30 µW) was used to excite HypnoS. A 10-Hz 410/10-nm filtered light (20–30 µW) served as a control channel and a 520/20-nm bandpass filter was used to collect the fluorescence signals.
Photometry data were analyzed using a custom MATLAB program. The fluorescence intensity of HypnoS excited by 410 nm and 470 nm was recorded as F410 and F470, respectively. The ∆R/R0 was calculated under the above conditions using the formula: ∆R/R0 = ∆(F470/F410)/(F470/F410). To calculate the ∆R/R0 during sleep–wake cycles, baseline values were measured during a period of NREM sleep in which no apparent fluctuations were observed. To better quantify the fluorescence changes across multiple animals, the ∆F/F0 was further normalized using the s.d. of the baseline signals to get the z-score, with the formula: z-score = ∆R/R0/s.d.baseline.
Fiber-photometry recording of intracellular Ado dynamics during seizure
Adult wild-type C57BL/6 N mice were anesthetized with isoflurane and placed on a stereotaxic frame for AAV injection (300 nL per site). In Fig. 7, AAV2/9-hSyn-HypnoS or AAV2/9-hSyn-HypnoS-mut was injected into the hippocampal CA1 region using the following coordinates: AP −1.8 mm from Bregma; ML ±1.2 mm; DV −1.25 mm from the dura. An optical fiber cannula (200 µm, 0.37 NA, Inper) was implanted 0.05 mm above the virus injection site in BF to record sensor signals.
To record the animal’s seizure state, we attached and fixed custom-made EEG and EMG electrodes to the skull through a microconnector. EEG electrodes were implanted into craniotomy holes situated over the frontal cortex and visual cortex, and EMG wires were placed bilaterally in the neck musculature. The microconnector was attached to the skull using glue and a thick layer of dental cement. After surgery, the mice were allowed to recover for at least 2 weeks.
The same fiber-photometry system (Inper/RWD) was used to record the fluorescence signals in freely moving mice during seizure. In Fig. 7a 10-Hz 470/10-nm filtered light (20–30 µW) was used to excite HypnoS. A 10-Hz 410/10-nm filtered light (20–30 µW) served as a control channel and a 520/20-nm bandpass filter was used to collect the fluorescence signals.
The fluorescence intensity of HypnoS sensors excited by 410 nm and 470 nm was recorded as F410 and F470, respectively. The ∆R/R0 was calculated under the above conditions using the formula: ∆R/R0 = ∆(R470/R410)/(R470/R410). Baseline values were determined before the application of saline or KA.
Polysomnographic recording and analysis
The sleep–wake state of each animal was determined using the EEG and EMG recordings. The EEG and EMG signals were amplified (NL104A, Digitimer), filtered (NL125/6, Digitimer) at 0.5–100 Hz (EEG) and 30–500 Hz (EMG), and then digitized using a Power1401 digitizer (Cambridge Electronic Design). The Spike2 software program (Cambridge Electronic Design) was used for recording, with a sampling rate of 1000 Hz. The sleep–wake state was classified semi-automatically in 4-s epochs using AccuSleep81 and then validated manually using a custom-made MATLAB GUI. The wake state was defined as desynchronized EEG activity combined with high EMG activity. NREM sleep was defined as synchronized EEG activity with high-amplitude delta activity (0.5–4 Hz) and low EMG activity. REM sleep was defined as high-power theta frequencies (6–9 Hz) combined with low EMG activity.
Quantification and statistical analysis
Where appropriate, cells or animals were randomly assigned to either the control or experimental group. Imaging data were processed using ImageJ (1.52p) software (NIH) and custom-written MATLAB (R2020b) programs. Data were plotted using GraphPad Prism 8.4.3 (Dotmatics) or Adobe Illustrator CC. Except where indicated, all summary data are reported as the mean ± s.e.m. The SNR was calculated as the peak response divided by the s.d. of the baseline fluorescence fluctuation. All data were assumed to be distributed normally, and equal variances were formally tested. Differences were analyzed using the two-tailed Student’s t-test; *P < 0.05, **P < 0.01, ***P < 0.001, and n.s., not significant (P ≥ 0.05).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
PDB files used in this study are: 3WLD, 2AMX and 2PGF (https://www.rcsb.org/structure/3WLD, https://www.rcsb.org/structure/2AMX and https://www.rcsb.org/structure/2PGF). Source data are provided with this paper.
Code availability
The code used for mesoscopic imaging data analysis in this study is provided and is available at https://github.com/damiers/wave_analysis. The custom Arduino code for stimulation and two-photon imaging in Drosophila, and the custom MATLAB codes for fiber photometry data analysis in mice can be made available upon request. Requests will be fulfilled within 2 weeks.
References
Drury, A. N. & Szent-Gyorgyi, A. The physiological activity of adenine compounds with especial reference to their action upon the mammalian heart. J. Physiol. 68, 213–237 (1929).
Chen, J. F., Eltzschig, H. K. & Fredholm, B. B. Adenosine receptors as drug targets–what are the challenges? Nat. Rev. Drug Discov. 12, 265–286 (2013).
Dias, R. B., Rombo, D. M., Ribeiro, J. A., Henley, J. M. & Sebastiao, A. M. Adenosine: setting the stage for plasticity. Trends Neurosci. 36, 248–257 (2013).
Cunha, R. A. How does adenosine control neuronal dysfunction and neurodegeneration? J. Neurochem. 139, 1019–1055 (2016).
Theparambil, S. M. et al. Adenosine signalling to astrocytes coordinates brain metabolism and function. Nature 632, 139–146 (2024).
Lazarus, M., Chen, J. F., Huang, Z. L., Urade, Y. & Fredholm, B. B. Adenosine and Sleep. Handb. Exp. Pharmacol. 253, 359–381 (2019).
Garcia-Gil, M., Camici, M., Allegrini, S., Pesi, R. & Tozzi, M. G. Metabolic aspects of adenosine functions in the brain. Front. Pharm. 12, 672182 (2021).
Hasko, G., Linden, J., Cronstein, B. & Pacher, P. Adenosine receptors: therapeutic aspects for inflammatory and immune diseases. Nat. Rev. Drug Discov. 7, 759–770 (2008).
Dale, N. & Frenguelli, B. G. Release of adenosine and ATP during ischemia and epilepsy. Curr. Neuropharmacol. 7, 160–179 (2009).
Rho, J. M. & Boison, D. The metabolic basis of epilepsy. Nat. Rev. Neurol. 18, 333–347 (2022).
Ma, L. et al. Locomotion activates PKA through dopamine and adenosine in striatal neurons. Nature 611, 762–768 (2022).
Badimon, A. et al. Negative feedback control of neuronal activity by microglia. Nature 586, 417–423 (2020).
Lovatt, D. et al. Neuronal adenosine release, and not astrocytic ATP release, mediates feedback inhibition of excitatory activity. Proc. Natl. Acad. Sci. USA 109, 6265–6270 (2012).
Wu, Z. et al. Neuronal activity-induced, equilibrative nucleoside transporter-dependent, somatodendritic adenosine release revealed by a GRAB sensor. Proc. Natl. Acad. Sci. USA 120, e2212387120 (2023).
Wall, M. J. & Dale, N. Neuronal transporter and astrocytic ATP exocytosis underlie activity-dependent adenosine release in the hippocampus. J. Physiol. 591, 3853–3871 (2013).
Zhou, X. et al. 40 Hz light flickering promotes sleep through cortical adenosine signaling. Cell Res. 34, 214–231 (2024).
Sun, X. et al. 40 Hz light flickering facilitates the glymphatic flow via adenosine signaling in mice. Cell Discov. 10, 81 (2024).
Latini, S. & Pedata, F. J. J. o.n. Adenosine in the central nervous system: release mechanisms and extracellular concentrations. J. Neurochem. 79, 463–484 (2001).
Camici, M., Garcia-Gil, M. & Tozzi, M. The inside story of adenosine. Int. J. Mol. Sci. 19, 784 (2018).
Wu, Z. & Li, Y. New frontiers in probing the dynamics of purinergic transmitters in vivo. Neurosci. Res. 152, 35–43 (2020).
Nguyen, M. D. & Venton, B. J. Fast-scan cyclic voltammetry for the characterization of rapid adenosine release. Comput. Struct. Biotechnol. J. 13, 47–54 (2015).
Zhang, Z., Cheng, X., Zhao, Y. & Yang, Y. Lighting up live-cell and in vivo central carbon metabolism with genetically encoded fluorescent sensors. Annu. Rev. Anal. Chem. 13, 293–314 (2020).
Greenwald, E. C., Mehta, S. & Zhang, J. Genetically encoded fluorescent biosensors illuminate the spatiotemporal regulation of signaling networks. Chem. Rev. 118, 11707–11794 (2018).
Zhang, J., Campbell, R. E., Ting, A. Y. & Tsien, R. Y. Creating new fluorescent probes for cell biology. Nat. Rev. Mol. Cell Biol. 3, 906–918 (2002).
Peng, W. et al. Regulation of sleep homeostasis mediator adenosine by basal forebrain glutamatergic neurons. Science 369, eabb0556 (2020).
Dunwiddie, T. V. & Masino, S. A. The role and regulation of adenosine in the central nervous system. Annu. Rev. Neurosci. 24, 31–55 (2001).
Cunha, R. A. Heteroexchange of purines in the hippocampus: mixing-up or messing-up ATP and adenosine. Br. J. Pharmacol. 139, 473–474 (2003).
Larson, E. T. et al. Structures of substrate- and inhibitor-bound adenosine deaminase from a human malaria parasite show a dramatic conformational change and shed light on drug selectivity. J. Mol. Biol. 381, 975–988 (2008).
Sun, F. et al. A genetically encoded fluorescent sensor enables rapid and specific detection of dopamine in flies, fish, and mice. Cell 174, 481–496.e419 (2018).
Marvin, J. S., Schreiter, E. R., Echevarría, I. M. & Looger, L. L. A genetically encoded, high-signal-to-noise maltose sensor. Proteins 79, 3025–3036 (2011).
Zhuo, Y. et al. Improved green and red GRAB sensors for monitoring dopaminergic activity in vivo. Nat. Methods 21, 680–691 (2024).
Li, X. et al. Ultrasensitive sensors reveal the spatiotemporal landscape of lactate metabolism in physiology and disease. Cell Metab. 35, 200–211.e209 (2023).
Traut, T. W. Physiological concentrations of purines and pyrimidines. Mol. Cell Biochem. 140, 1–22 (1994).
Jaruwat, A. et al. Crystal structure of Plasmodium falciparum adenosine deaminase reveals a novel binding pocket for inosine. Arch. Biochem. Biophys. 667, 6–13 (2019).
Cambronne, X. A. et al. Biosensor reveals multiple sources for mitochondrial NAD⁺. Science 352, 1474–1477 (2016).
Liu, B. et al. GlutaR: a high-performance fluorescent protein-based sensor for spatiotemporal monitoring of glutamine dynamics in vivo. Angew. Chem. Int. Ed Engl. 64, e202416608 (2024).
Tantama, M., Martinez-Francois, J. R., Mongeon, R. & Yellen, G. Imaging energy status in live cells with a fluorescent biosensor of the intracellular ATP-to-ADP ratio. Nat. Commun. 4, 2550 (2013).
Berg, J., Hung, Y. P. & Yellen, G. A genetically encoded fluorescent reporter of ATP:ADP ratio. Nat. Methods 6, 161–166 (2009).
Porkka-Heiskanen, T. & Kalinchuk, A. V. Adenosine, energy metabolism and sleep homeostasis. Sleep. Med. Rev. 15, 123–135 (2011).
Boison, D. & Yegutkin, G. G. Adenosine metabolism: emerging concepts for cancer therapy. Cancer Cell 36, 582–596 (2019).
Parpura, V. et al. Glutamate-mediated astrocyte-neuron signalling. Nature 369, 744–747 (1994).
Lee, C. J. et al. Astrocytic control of synaptic NMDA receptors. J. Physiol. 581, 1057–1081 (2007).
Rangaraju, V., Calloway, N. & Ryan, T. A. Activity-driven local ATP synthesis is required for synaptic function. Cell 156, 825–835 (2014).
Mann, K., Deny, S., Ganguli, S. & Clandinin, T. R. Coupling of activity, metabolism and behaviour across the Drosophila brain. Nature 593, 244–248 (2021).
Klapoetke, N. C. et al. Independent optical excitation of distinct neural populations. Nat. Methods 11, 338–346 (2014).
Latini, S. & Pedata, F. Adenosine in the central nervous system: release mechanisms and extracellular concentrations. J. Neurochem. 79, 463–484 (2001).
Boison, D. Adenosine kinase, epilepsy and stroke: mechanisms and therapies. Trends Pharmacol. Sci. 27, 652–658 (2006).
Boison, D. Adenosine dysfunction in epilepsy. Glia 60, 1234–1243 (2012).
Van Gompel, J. J. et al. Increased cortical extracellular adenosine correlates with seizure termination. Epilepsia 55, 233–244 (2014).
Raol, Y. H. & Brooks-Kayal, A. R. Experimental models of seizures and epilepsies. Prog. Mol. Biol. Transl. Sci. 105, 57–82 (2012).
Dana, H. et al. Sensitive red protein calcium indicators for imaging neural activity. Elife 5, e12727 (2016).
Hamodi, A. S., Martinez Sabino, A., Fitzgerald, N. D., Moschou, D. & Crair, M. C. Transverse sinus injections drive robust whole-brain expression of transgenes. Elife 9, e53639 (2020).
Dong, A. et al. A fluorescent sensor for spatiotemporally resolved imaging of endocannabinoid dynamics in vivo. Nat. Biotechnol. 40, 787–798 (2022).
Deng, F. et al. Improved green and red GRAB sensors for monitoring spatiotemporal serotonin release in vivo. Nat. Methods 21, 692–702 (2024).
Farrell, J. S. et al. In vivo assessment of mechanisms underlying the neurovascular basis of postictal amnesia. Sci. Rep. 10, 14992 (2020).
Xu, M. et al. Basal forebrain circuit for sleep-wake control. Nat. Neurosci. 18, 1641–1647 (2015).
Simpson, E. H. et al. Lights, fiber, action! A primer on in vivo fiber photometry. Neuron 112, 718–739 (2024).
Carswell, H. V., Graham, D. I. & Stone, T. W. Kainate-evoked release of adenosine from the hippocampus of the anaesthetised rat: possible involvement of free radicals. J. Neurochem. 68, 240–247 (1997).
Weltha, L., Reemmer, J. & Boison, D. The role of adenosine in epilepsy. Brain Res. Bull. 151, 46–54 (2019).
Porkka-Heiskanen, T. et al. Adenosine: a mediator of the sleep-inducing effects of prolonged wakefulness. Science 276, 1265–1268 (1997).
Porkka-Heiskanen, T., Strecker, R. E. & McCarley, R. W. Brain site-specificity of extracellular adenosine concentration changes during sleep deprivation and spontaneous sleep: an in vivo microdialysis study. Neuroscience 99, 507–517 (2000).
Basheer, R., Strecker, R. E., Thakkar, M. M. & McCarley, R. W. Adenosine and sleep-wake regulation. Prog. Neurobiol. 73, 379–396 (2004).
Greene, R. W., Bjorness, T. E. & Suzuki, A. The adenosine-mediated, neuronal-glial, homeostatic sleep response. Curr. Opin. Neurobiol. 44, 236–242 (2017).
Peng, W. et al. Adenosine-independent regulation of the sleep-wake cycle by astrocyte activity. Cell Discov. 9, 16 (2023).
Werner, A. et al. Determination of nucleotides, nucleosides and nucleobases in cells of different complexity by reversed-phase and ion-pair high-performance liquid chromatography. J. Chromatogr. 421, 257–265 (1987).
Arnold, S. T. & Cysyk, R. L. Adenosine export from the liver: oxygen dependency. Am. J. Physiol. 251, G34–G39 (1986).
Fredholm, B. B. Analysis of purines. Life Sci. 41, 837–840 (1987).
Cunha, R. A. Adenosine as a neuromodulator and as a homeostatic regulator in the nervous system: different roles, different sources and different receptors. Neurochem. Int. 38, 107–125 (2001).
Zhao, Y. et al. An expanded palette of genetically encoded Ca²⁺ indicators. Science 333, 1888–1891 (2011).
Qian, Y. et al. A genetically encoded near-infrared fluorescent calcium ion indicator. Nat. methods 16, 171–174 (2019).
Deo, C. et al. The HaloTag as a general scaffold for far-red tunable chemigenetic indicators. Nat. Chem. Biol. 17, 718–723 (2021).
Deng, B. et al. Chemoconnectomics: mapping chemical transmission in Drosophila. Neuron 101, 876–893.e874 (2019).
Chen, W., Zhang, Y., Yeo, W. S., Bae, T. & Ji, Q. Rapid and efficient genome editing in Staphylococcus aureus by using an engineered CRISPR/Cas9 system. J. Am. Chem. Soc. 139, 3790–3795 (2017).
Zhang, M. et al. Monitoring the dynamic regulation of the mitochondrial GTP-to-GDP ratio with a genetically encoded fluorescent biosensor. Angew. Chem. Int Ed. Engl. 61, e202201266 (2022).
Wang, L. et al. A high-performance genetically encoded fluorescent indicator for in vivo cAMP imaging. Nat. Commun. 13, 5363 (2022).
Kao, Y. H. et al. Targeting ENT1 and adenosine tone for the treatment of Huntington’s disease. Hum. Mol. Genet. 26, 467–478 (2017).
Cantu, D. A. et al. EZcalcium: open-source toolbox for analysis of calcium imaging data. Front. Neural Circuits 14, 25 (2020).
Werley, C. A., Chien, M. P. & Cohen, A. E. Ultrawidefield microscope for high-speed fluorescence imaging and targeted optogenetic stimulation. Biomed. Opt. Express 8, 5794–5813 (2017).
Zhuang, C. et al. Real-time brain-wide multi-planar microscopy for simultaneous cortex and hippocampus imaging at the cellular resolution in mice. Biomed. Opt. Express 12, 1858–1868 (2021).
Jackson, J., Karnani, M. M., Zemelman, B. V., Burdakov, D. & Lee, A. K. Inhibitory control of prefrontal cortex by the claustrum. Neuron 99, 1029–1039.e1024 (2018).
Barger, Z., Frye, C. G., Liu, D., Dan, Y. & Bouchard, K. E. Robust, automated sleep scoring by a compact neural network with distributional shift correction. PLoS ONE 14, e0224642 (2019).
Acknowledgements
This work was supported by the National Basic Research Foundation of China (no. 2022YFC3400600 to J.W.), the National Natural Science Foundation of China (nos. 22277001 and 91853107 to J.W.; 32422031 to Z.W.), the National Key R&D Program of China (no. 2022YFE0108700 to Y.L.) and STI2030-Major Projects (2022ZD0208300 to Z.W.). J.W. also thanks the Youth Thousand-Talents Program of China for support. The authors thank the Large-scale Instrument Platform of the State Key Laboratory of Natural and Biomimetic Drugs, School of Pharmaceutical Sciences, Peking University, for instrument and technical support. The authors thank Ms. Liqin Fu at the National Center for Protein Sciences, Peking University, and Dr. Lei Su at Institute of Microbiology, Chinese Academy of Sciences, for technical support with confocal imaging. Z.W. is supported by the Young Elite Scientists Sponsorship Program by Beijing Association for Science and Technology. We also thank Drs. Jiang-fan Chen, Lei Ma and Hui Dong for useful suggestions of the manuscript.
Author information
Authors and Affiliations
Contributions
J.W. (corresponding author), Z.W. and Yulong L. conceived the project and designed the experiments; J.W. (corresponding author), Z.W., Yulong L., Q.W., Z.B. and L.W. wrote the manuscript; Q.W. and Y.H. developed and optimized the sensors; Q.W. and Y.H. performed the experiments related to characterizing the sensors in vitro with the help of Z.W. and W.L.; Q.W. and Z.W. performed the experiments related to characterizing the sensors in cultured neurons and astrocytes; L.W. and Q.W. performed the surgery and two-photon imaging experiments related to the validation of the sensors in acute brain slices; Q.W. and Y.W. performed the two-photon imaging experiments in transgenic flies; Z.W. and Z.B. performed the surgery and the mesoscopic imaging in head-fixed mice with the help of F.D., P.Z. S.D. and C.L.; Z.W. and Z.B. performed the surgery and in vivo two-photon imaging experiments in head-fixed mice with the help of Y.Z.; Q.W., Z.M., Yumo L. and J.W.(Capital Medical University, Beijing, China) performed the fiber photometry recordings in freely moving mice under the supervision of Z.W.; All authors contributed to the data interpretation and analysis.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Katalin Török, who co-reviewed with Olga Kopach; and Michael Lazarus for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wei, Q., Bai, Z., Wang, L. et al. A high-performance fluorescent sensor spatiotemporally reveals cell-type specific regulation of intracellular adenosine in vivo. Nat Commun 16, 4245 (2025). https://doi.org/10.1038/s41467-025-59530-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-59530-7
This article is cited by
-
State of the art indicators for imaging purinergic dynamics in vitro and in vivo
Purinergic Signalling (2025)