Abstract
Lipid nanoparticles (LNPs) are key non-viral carriers for mRNA vaccines and therapeutics, but the inherent instability of mRNA necessitates sub-zero storage with cryoprotectants (CPAs) to prevent freeze-induced LNP aggregation and compromised mRNA delivery. Here we show that ice formation during freezing concentrates CPAs with LNPs in the remaining liquid—a phenomenon known as freeze concentration. This creates a steep concentration gradient of CPAs across the lipid membrane that drives passive CPAs diffusion into LNPs. By leveraging this process, we developed betaine-based CPAs that both preserve the stability of LNP and enter LNP during freeze-thaw. The incorporated betaine enhances endosomal escape and boosts mRNA delivery of LNP. In female mice, betaine-loaded LNPs elicit stronger humoral and cellular immune responses, providing dose-sparing advantages. These findings highlight freeze concentration as a promising LNP formulation strategy and underscore the role of CPA as active modulators of LNP structure and function.
Similar content being viewed by others
Introduction
Lipid nanoparticles (LNPs) have gained significant attention as non-viral vectors for messenger RNA (mRNA) delivery, particularly following the approval of the COVID-19 vaccines1,2,3. mRNA-LNPs are being extensively explored for applications in vaccines, protein replacement therapy, cancer immunotherapy, and CRISPR/Cas9 gene editing4,5,6,7. However, a key challenge in the clinical use of mRNA-LNPs is ensuring their long-term stability and mRNA delivery efficacy8,9,10. mRNA is highly susceptible to degradation via hydrolysis, oxidation, and enzymatic activity, necessitating storage at sub-zero temperatures to maintain stability11,12. However, freezing and thawing cycles introduce additional challenges to LNP formulations. Ice crystal formation and osmotic stress during freeze-thaw (F-T) processes can lead to fusion, aggregation, and leakage of encapsulated mRNA, significantly compromising the stability and mRNA delivery efficacy of LNPs13,14. Current strategies to mitigate the damaging effects of freezing typically focus on stabilizing LNPs with cryoprotectants (CPAs), such as sucrose, which is used in the storage of commercially approved mRNA vaccines like mRNA-1273 and BNT162b215,16. These cryoprotectants help preserve the integrity of LNPs during cryopreservation, enabling their storage at −20 °C or −70 °C for extended periods17.
An underexplored yet promising approach involves leveraging the freeze-thaw process itself to preserve stability and even enhance the delivery efficacy of LNPs18,19. During freezing, water undergoes a phase transition, forming ice and concentrating solutes in the remaining liquid phase, a phenomenon known as freeze concentration20,21. This process significantly alters the physicochemical environment around LNPs by increasing local solute concentrations and creating steep concentration gradients across the LNP membrane22,23. These gradients can drive the passive incorporation of small molecules—such as cryoprotectants—into LNPs through diffusion or transient membrane disruptions induced by the phase transition of lipids24. Freezing-induced content exchange between liposomes and surrounding solutions has been widely observed, and multiple F-T cycles have been used to fabricate drug-loaded liposomes with high loading efficiency25,26. These observations suggest that interactions between CPA and LNPs during F-T cycles may be more complex and dynamic than previously understood. This interplay offers an opportunity to not only mitigate freeze-induced damage but also to reformulate LNPs in ways that actively improve their delivery efficacy.
Herein, we show that freezing could be leveraged to enhance the mRNA delivery efficacy of LNP by incorporating functional molecules during the F-T process (Fig. 1a). Specifically, we focused on betaine, a zwitterionic cryoprotectant known for its ability to protect cellular structures during cryopreservation and its potential to enhance endosomal escape by protonating in the acidic environment of endosomes27,28,29. By incorporating betaine into LNPs during the F-T cycle, we aim to synergistically exploit its cryoprotective properties and its potential to improve endosomal escape, ultimately enhancing the structural integrity of LNPs and their mRNA delivery efficiency.
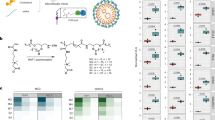
a Schematic illustration showing the proposed mechanism of freezing-induced incorporation of betaine into LNPs. b The hydrodynamic diameters and polydispersity index (PDI) of LNPs (n = 3 technical replicates). Data are shown as mean ± standard deviation (SD). c A representative Cryogenic transmission electron microscopy (Cryo-TEM) image of LNP. The scale bar is 100 nm. The TEM imaging experiment was repeated independently three times. d The mRNA encapsulation of LNPs (n = 3 technical replicates). Data are shown as mean ± SD. e The mLuc expression in DC2.4 cells after treatment with free mRNA, LNP, or freeze-thawed LNPs with different cryoprotectants (CPAs) (n = 4 technical replicates). Data are shown as mean ± SD. Statistical significance was analyzed by one-way analysis of variance (ANOVA) and Tukey’s multiple comparisons test. f A representative Cryo-TEM image of LNP+BT-CPA F-T. The scale bar is 100 nm. The TEM imaging experiment was repeated independently three times. g The mLuc expression in DC2.4 cells after treatment with free mRNA, LNP, or freeze-thawed LNP+BT-CPA with varying concentrations of betaine (n = 4 technical replicates). Data are shown as mean ± SD. h Representative IVIS images and i quantitative analysis of C57BL/6 mice following intramuscular injection of LNP or LNP+BT-CPA F-T at 4 and 24 h post-administration (n = 6 biologically independent samples). Data are shown as mean ± SD. Statistical significance was analyzed by unpaired two-tailed Student’s t-test. j Normalized mLuc expression in DC2.4 cells after treatment with LNPs formulated with ALC-0315, cKK-E12, and MC3 as ionizable lipids before and after F-T with BT-CPA (n = 4 technical replicates). Data are shown as mean ± SD. Statistical significance was analyzed by one-way ANOVA and Tukey’s multiple comparisons test. a was created in BioRender. Lu, X. (2025) https://BioRender.com/umtq3e5.
Results and discussion
To evaluate the efficacy of betaine as a CPA for LNP, we synthesized the LNP encapsulating firefly luciferase mRNA using the clinically approved mRNA-1273 formulation using a microfluidic device. Cryogenic transmission electron microscopy (Cryo-TEM) and dynamic light scattering (DLS) measurements confirmed the spherical morphology of the LNPs (Fig. 1b, c). Betaine at 25 mg/mL was added to the LNP solution, and the samples were subjected to two F-T cycles at −80 °C. LNPs in sucrose (87 mg/mL) or phosphate-buffered saline (PBS) were used as the positive and negative controls, respectively. As shown in Fig. 1b, d, e, LNPs in PBS without cryoprotectants exhibited significant aggregation and mRNA leakage, leading to a nearly complete loss of mRNA delivery efficiency in DC2.4 cells. In contrast, LNPs with sucrose maintained their hydrodynamic diameter, polydispersity index (PDI), mRNA encapsulation, and mRNA delivery efficacy. LNPs with betaine had a similar hydrodynamic diameter but slightly reduced mRNA encapsulation, indicating that betaine at 25 mg/mL preserves LNP stability as a cryoprotectant, but is less effective than sucrose at 87 mg/mL. However, despite the lower mRNA encapsulation, LNPs with betaine showed ~1.7-fold higher mRNA expression compared to fresh LNPs or LNPs with sucrose.
We then optimized the betaine formulation not only to enhance LNP stability during F-T cycles but also as a potential strategy to improve mRNA delivery efficacy. To improve the stability of LNP during F-T, trehalose, a commonly used cryoprotectant for stabilizing nanoparticles, was added to betaine in equal mass (both at 25 mg/mL), resulting in a combined cryoprotectant formulation (BT-CPA). LNPs+BT-CPA showed minimal changes in size and mRNA encapsulation after F-T cycles. Cryo-TEM imaging confirmed the intact spherical morphology of the LNPs (Fig. 1f). The mRNA delivery efficiency of LNPs+BT-CPA was ~2.4-fold higher than that of fresh LNPs and ~1.4-fold higher than that of LNPs with betaine alone (LNP+betaine). LNPs with trehalose alone showed similar mRNA delivery efficiency to that of fresh LNPs. These findings suggest that trehalose further enhances the stability of LNP+betaine during F-T, leading to greatly improved mRNA delivery efficiency. We then optimized the amount of betaine in the BT-CPA formulation to maximize mRNA delivery efficiency. A series of betaine concentrations were tested while keeping the trehalose concentration constant. As shown in Fig. 1g, increasing betaine from 10 mg/mL to 25 mg/mL improved mRNA delivery. However, further increases in betaine concentration, up to 75 mg/mL, did not further improve mRNA delivery. Consequently, we selected the BT-CPA formulation with 25 mg/mL betaine and 25 mg/mL trehalose for further investigation.
To validate the enhanced mRNA delivery efficacy of LNP+BT-CPA after F-T, these LNPs were intramuscularly injected into C57BL/6 mice, with freshly prepared LNP as a control. Figure 1h, i and Supplementary Fig. 1 show that LNP+BT-CPA after F-T exhibits approximately 2.3-fold and 1.7-fold higher mLuc expression than fresh LNP at 4 h and 24 h post-injection, respectively. We also tested the versatility of BT-CPA across different LNP formulations using ALC-0315, cKK-E12, or DLin-MC3-DMA (MC3) as ionizable lipids. The hydrodynamic diameters of LNPs were all below 200 nm with a narrow polydispersity index (PDI < 0.2, Supplementary Fig. 2). As shown in Fig. 1j, all LNPs with BT-CPA show significantly enhanced mRNA delivery efficiency following F-T. Collectively, these results indicate that BT-CPA not only maintains LNP stability after F-T but also serves as a general strategy to enhance the mRNA delivery efficacy of LNPs both in vitro and in vivo.
The enhanced mRNA delivery is likely due to betaine altering the functions of either LNPs or cells. Betaine is a well-known osmoprotectant that can be rapidly taken up by cells in large quantities30, potentially influencing cellular functions and subsequent mRNA expression. To test this, DC2.4 cells were treated with BT-CPA for 4 h before incubation with SM102-LNP. As shown in Fig. 2a, the mRNA expression in cells treated with BT-CPA (BT-CPA prime) or PBS was nearly identical, indicating that BT-CPA does not affect the cellular mRNA expression.
a The mLuc expression in DC2.4 cells after incubation with free mRNA, LNP, and LNP+BT-CPA following different treatments. The group of BT-CPA prime represents pretreating the cells with BT-CPA for 4 h before incubating with LNP. (n = 4 technical replicates). Data are shown as mean ± SD. Statistical significance was analyzed by one-way ANOVA and Tukey’s multiple comparisons test. b Proton nuclear magnetic resonance (1H-NMR) spectra of betaine, trehalose, and LNPs following different treatments. The characteristic peaks of trehalose and betaine were highlighted. Figure 2a was created in BioRender. Lu, X. (2025) https://BioRender.com/h45y717.
We then investigated the mechanism by which BT-CPA alters LNP functions during F-T cycles. LNP was incubated with BT-CPA for 4 h before being added to DC2.4 cells. As shown in Fig. 2a, this incubation did not affect mRNA delivery. However, after F-T, LNPs+BT-CPA showed significantly enhanced mRNA expression, indicating that the F-T process is critical for the improved mRNA delivery of LNPs+BT-CPA. Since betaine requires transport proteins to enter cells and cannot pass through lipid bilayers of LNP on its own31, it’s very likely that betaine enters LNP through freeze concentration. Freezing can induce mechanical stress from ice crystal formation, potentially disrupting the lipid bilayer and allowing content exchange, while thawing can lead to fusion and fission of vesicles32,33,34. This process might allow betaine to enter LNPs, altering their compositions and enhancing their mRNA delivery efficiency.
To determine if CPA was incorporated into LNPs during F-T, we used proton nuclear magnetic resonance (1H-NMR) to characterize the LNPs. LNP+BT-CPA after F-T was dialyzed against PBS to remove any BT-CPA outside the LNP, as BT-CPA would remain trapped inside the LNP due to its inability to cross lipid bilayers at room temperature. After dialysis, deuterium methanol was added for subsequent NMR analysis. Free betaine, trehalose, LNP, and LNP+BT-CPA without F-T were also characterized by 1H-NMR as controls. As shown in Fig. 2b and Supplementary Fig. 3–8, the dialyzed LNP+BT-CPA without F-T showed no proton peaks corresponding to betaine (3.28 ppm and 3.83 ppm) or trehalose (3.34, 3.51, 3.68, 3.75-3.85, and 5.13 ppm). In contrast, LNP+BT-CPA after F-T exhibits characteristic peaks for both betaine and trehalose, indicating their incorporation into LNPs after F-T. High-resolution mass-spectrometry measurements also confirmed the presence of betaine and trehalose in LNP+BT-CPA after F-T (Supplementary Fig. 9). The mRNA delivery efficiency of the dialyzed LNP+BT-CPA after F-T was also significantly higher than that of fresh LNP. These findings suggest that BT-CPA is incorporated into LNPs during F-T, leading to their enhanced mRNA delivery efficiency.
To evaluate the impact of freeze-thaw cycles on mRNA delivery efficacy, we compared 1, 2, and 6 F-T cycles. As shown in Supplementary Fig. 10, mRNA delivery efficiency of LNP gradually increased after 1 and 2 F-T cycles, with hydrodynamic diameters and encapsulation efficiencies remaining unchanged. However, performing 6 F-T cycles led to LNP aggregation and mRNA leakage, which ultimately reduced delivery efficacy. These results suggest that while betaine progressively enters the LNPs with each cycle, excessive F-T cycles may allow too much betaine to penetrate, causing structural damage to the LNPs. Therefore, we selected two freeze-thaw cycles for maintaining LNP integrity and maximizing mRNA delivery.
Many studies have demonstrated that small molecules can cross the phospholipid membrane of liposomes during the F-T process, leading to content exchange between liposomes and surrounding solutions35,36. This phenomenon has significant implications for regulatory-approved liposome and LNP formulations, which often use CPAs and undergo F-T cycles as part of their production or storage processes. Our findings indicate that CPAs can alter the composition and potentially the functionality of these liposomes or LNPs. Indeed, 1H-NMR analysis of the commercial mRNA-1273 formulation revealed that sucrose was incorporated into LNPs after F-T cycles (Fig. 2b). While the internalized sucrose did not affect the overall mRNA delivery efficacy of the approved LNP, this outcome might be fortuitous and specific to the benign properties of sugars like sucrose. However, this raises a critical issue: CPAs are not merely inert stabilizers—they can become integral components of the liposome or LNP, potentially altering their nanostructure, physicochemical properties, and biological function.
We then studied whether freezing is necessary to incorporate BT-CPA into LNPs. We added BT-CPA directly into the aqueous phase of mRNA during the synthesis of LNPs (Fig. 3a). If freezing were not necessary for incorporating BT-CPA, we would expect similar results regardless of the F-T process. After synthesis and dialysis, LNPs with BT-CPA were successfully formed with diameters of less than 200 nm and a PDI of less than 0.2 (Supplementary Fig. 11). However, the 1H-NMR analysis revealed minimal characteristic peaks of betaine (Fig. 3b, Supplementary Fig. 12), indicating that a negligible amount of BT-CPA was encapsulated in the LNPs. In vitro transfection results further demonstrated that the incorporation of BT-CPA during synthesis did not improve the mRNA delivery efficacy of LNPs (Fig. 3c). These findings suggest that BT-CPA has poor encapsulation efficiency when incorporated during synthesis, likely due to insufficient interactions between BT-CPA and the LNP components during formulation. We have also explored whether BT-CPA could enter LNP using ultrasound, which could induce membrane disruption and fusion. GelRed was added to the solution to monitor the internalization of small molecules during sonication. As shown in Supplementary Fig. 13, sonication facilitated GelRed internalization into LNPs, suggesting that ultrasound induced membrane disruption or potentially led to LNP reassembly. However, ultrasound-treated LNPs exhibited markedly reduced mRNA delivery efficacy. These findings indicate that although ultrasound can promote the incorporation of BT-CPA, it concurrently compromises LNP structural integrity and mRNA delivery performance. Collectively, these results indicate the critical role of freezing in facilitating the incorporation of BT-CPA into LNPs, likely through the content exchange mechanism observed during the freezing process.
a Schematic illustration of BT-CPA formulated LNP. BT-CPA was directly added into the aqueous phase containing mRNA during LNP fabrication. b 1H-NMR spectra of BT-CPA formulated LNP and dialyzed LNP+BT-CPA F-T that is subjected to F-T cycles from −15 °C to 20 °C. The characteristic peaks of betaine and trehalose were highlighted. c The mLuc expression in DC2.4 cells after treatment with free mRNA, LNP, BT-CPA formulated LNP, or LNP+BT-CPA F-T (n = 4 technical replicates). Data are shown as mean ± SD. d Differential scanning calorimetry (DSC) thermograms of PBS and LNP. e Representative fluorescence microscope images of Cy5-mCherry-loaded LNP+BT-CPA F-T in PBS buffer at −15 °C. The bright field image shows the formation of ice crystals. The scale bar is 50 µm. The imaging experiment was repeated independently at least three times. f Schematic illustration shows that as LNP freezes, lipid phase separation occurs, allowing the incorporation of highly concentrated solutes into the LNP. Figure 3a was created in BioRender. Lu, X. (2025) https://BioRender.com/i28r052. Figure 3f was created in BioRender. Lu, X. (2025) https://BioRender.com/v74ycml.
The content exchange between liposomes and surrounding solutions has been shown to occur during the phase transition of lipids and the freezing of water. Current mRNA vaccines, such as mRNA-1273 and BNT162b2, are stored at various sub-zero temperatures, including −20 °C and −70 °C. Recent data also show that LNP maintained stability in temperatures between −25 °C and −15 °C for up to two weeks37. While these storage conditions have been effective in maintaining vaccine stability, the exact timing of content exchange during F-T remains unclear. To investigate this, the phase transition temperature of LNP was measured using differential scanning calorimetry (DSC). As shown in Fig. 3d and Supplementary Fig. 14, LNP exhibited an additional exothermic peak at approximately −20 °C compared to PBS, indicating the phase transition temperature of LNP. At this temperature, the presence of frozen water severely restricts the transmembrane movement of solutes, suggesting that the incorporation of BT-CPA into the LNPs is not solely a result of cooling below the lipid phase transition temperature. Previous studies have indicated that freezing can also induce a fluid-to-gel phase transition, driven by the dehydration of water bound to the hydrophilic groups of phospholipids38,39. Therefore, the content exchange between LNP and the surrounding solution very likely occurs during the freezing of water, which induces membrane defects and phase transition of lipids.
To test this hypothesis, LNP+BT-CPA was subjected to F-T cycles ranging from −15 °C to 20 °C, resulting in the freezing of water without reaching the LNP phase transition temperature. 1H-NMR results showed the characteristic peaks of trehalose and betaine after F-T, indicating that content exchange occurs during the freezing stage, without the need to reach the phase transition temperature of the LNPs (Fig. 3b and Supplementary Fig. 15). During the freezing process, solutes are progressively concentrated and excluded into the grain boundary of ice crystals. Fluorescence microscopy imaging (Fig. 3e) confirmed that LNPs also accumulate within these grain boundaries. As freezing progresses, the concentrations of both LNP and solutes dramatically increase. When the surface of the LNP begins to freeze, lipid phase separation occurs, allowing highly concentrated solutes to enter the LNP structure while solutes inside the LNP diffuse outward along the concentration gradient (Fig. 3f).
To further demonstrate that LNPs undergo content exchange with surrounding solutions during F-T cycles and to rule out the possibility that BT-CPA may merely adsorb onto the LNP surface, we incubated LNP+BT-CPA with GelRed, a fluorescent nucleic acid stain that increases in fluorescence intensity when it binds to mRNA40 (Fig. 4a). As shown in Fig. 4b, the LNP solution without GelRed exhibited no fluorescence, and GelRed alone showed weak fluorescence. When GelRed was added to the LNP solution, it binds to unencapsulated mRNA, resulting in increased fluorescence. After F-T cycles, the fluorescence of GelRed further increases, suggesting that GelRed enters LNP and binds to mRNA. To rule out the possibility that the increased fluorescence of GelRed was due to mRNA leakage during F-T, we subjected LNP+BT-CPA to F-T cycles first and then incubated the formulation with GelRed. The fluorescent intensities of LNP solutions before and after F-T are nearly identical, suggesting that there is no significant mRNA leakage from the LNPs. These findings are consistent with the mRNA encapsulation data (Fig. 1d) and indicate that large hydrophilic molecules like mRNA cannot pass through the lipid bilayer during the F-T process.
a Schematic illustration of freeze-thawed LNP with GelRed. After freeze-thaw, GelRed enters LNP and binds with mRNA to emit fluorescence. b Fluorescence intensity of LNP, GelRed, LNP+GelRed, LNP+BT-CPA F-T+GelRed (GelRed was added after F-T), and LNP+BT-CPA+GelRed F-T (GelRed was added before F-T) (n = 3 technical replicates). Data are shown as mean ± SD. Statistical significance was analyzed by one-way ANOVA and Tukey’s multiple comparisons test. c Fluorescence intensity of LNP+BT-CPA+GelRed F-T, temperature decreasing rate with 1, 2.5, 5, 10, 15, 20 °C/min (n = 3 technical replicates). Data are shown as mean ± SD. d Schematic illustration of freeze-thawed fluorescein-loaded LNP. The encapsulated fluorescein in LNPs leaked out after F-T. e The concentration of encapsulated fluorescein in LNP before and after F-T. (n = 3 technical replicates). Data are shown as mean ± SD. Statistical significance was analyzed by unpaired two-tailed Student’s t-test. Figures 4a and 4d were created in BioRender. Lu, X. (2025) https://BioRender.com/d64j818.
We next investigated the effect of the cooling rate on the content exchange process. The cooling rate is known to influence the phase transition of lipids and the formation of ice crystals41, which can impact the extent of solute infiltration into LNPs. We hypothesized that slower cooling rates might allow more time for the phase separation of lipids, facilitating greater solute exchange between LNPs and the surrounding solution. As shown in Fig. 4c, the fluorescence intensity of GelRed decreased progressively with increasing cooling rate. These results suggest that slower cooling rates facilitate the exchange of solutes between LNPs and the surrounding solution, potentially due to an extended phase separation period of lipids, which provides additional time for solute infiltration into LNPs.
The diffusion of small hydrophilic molecules from LNP to surrounding solutions was also investigated by using fluorescein as the model molecule (Fig. 4d). Fluorescein was added to the aqueous phase during LNP synthesis. After LNP fabrication and dialysis to remove free fluorescein, a small portion of fluorescein was shown to be encapsulated in the LNPs after dialysis. The encapsulation efficiency is very low (<0.01%) due to the weak interaction between fluorescein and lipids. These F-LNPs were then incubated with BT-CPA and subjected to F-T cycles. The leaked fluorescein was isolated through centrifugal filters and quantified. Figure 4e shows that approximately 43% of the fluorescein diffuses out from the LNPs. These results indicate that LNPs undergo content exchange with the external solution during the F-T process.
We next investigate the mechanism by which BT-CPA enhances the delivery efficacy of LNPs by evaluating the cellular uptake and endosomal escape efficiencies of SM102-LNP+BT-CPA after F-T cycles. Cyanine 5-labeled mRNA encoding mCherry (Cy5-mCherry) was encapsulated in the SM102-LNP to enable fluorescent tracking and imaging. SM102-LNPs were incubated with BT-CPA, subjected to F-T cycles, and dialyzed against PBS to obtain BT-CPA-incorporated SM102-LNPs (BT-CPA-LNP). DC2.4 cells were incubated with BT-CPA-LNP and analyzed by flow cytometry and confocal laser-scanning microscopy (CLSM). Free Cy5-mCherry and SM102-LNP were used as controls. Flow cytometry analysis showed nearly identical cellular uptake ability between freshly prepared SM102-LNP and BT-CPA-LNP, suggesting that the incorporation of BT-CPA does not improve the cellular uptake ability of LNP (Fig. 5a). CLSM was employed to further assess the endosomal escape efficiency of the LNPs by staining the nucleus and lysosomes using Hoechst (shown as blue) and LysoTracker (shown as green). The fluorescence of Cy5-mCherry was shown as red. As shown in Fig. 5b, cells treated with BT-CPA-LNP exhibited fewer overlapping between signals of Cy5 and lysosome compared to SM102-LNP. Quantitative analysis using Pearson’s correlation coefficient (PCC) between signals of Cy5 and LysoTracker also shows that the BT-CPA-LNP group has a significantly lower PCC than freshly prepared LNPs without CPA (Fig. 5c). These results suggest that the incorporation of CPA enhances the endosomal escape capability of the LNPs.
a Flow cytometry analysis of DC2.4 cells treated with free Cy5-mCherry, LNP, or LNP+BT-CPA F-T. (n = 3 technical replicates) Data are shown as mean ± SD. Statistical significance was analyzed by one-way ANOVA and Tukey’s multiple comparisons test. ns, not significant. b Representative CLSM images of DC2.4 cells treated with LNP or LNP+BT-CPA F-T encapsulating Cy5-mCherry. The nuclei and lysosomes of cells were stained with Hoechst and LysoTracker, respectively. The scale bar is 20 µm. c Pearson’s correlation coefficient of (b) was quantified using ImageJ (n = 3 technical replicates). Data are shown as mean ± SD. Statistical significance was analyzed by unpaired two-tailed Student’s t-test. d Schematic illustration of the membrane fusion assay between LNP and the model endosome. e The fluorescence intensity of the model endosome was monitored after adding different amounts of LNPs. f The proposed mechanism through which betaine enhances the endosomal escape of LNP. g Normalized mLuc expression in DC2.4 cells after treatment with different LNPs (n = 3 technical replicates). Data are shown as mean ± SD. Statistical significance was analyzed by one-way ANOVA and Tukey’s multiple comparisons test. h The viability of DC2.4 cells after incubation with varying concentrations of sucrose or BT-CPA (n = 3 technical replicates). Data are shown as mean ± SD. Figure 5 d and 5 f were created in BioRender. Lu, X. (2025) https://BioRender.com/q10z184.
In the acidic environment of the endosome, the carboxylic acid group of betaine becomes protonated, converting the zwitterionic betaine molecule into a positively charged species. This positive charge can interact with the negatively charged endosomal membrane, leading to enhanced membrane fusion. Previous studies have also demonstrated that zwitterionic polymers exhibit improved endosomal escape through similar mechanisms29. The membrane fusion ability of LNPs was further investigated using a model endosome containing two fluorescent dye-labeled lipids, NBD-PE and Rhod-PE42 (Fig. 5d). The fluorescence signal of NBD was quenched by PE through fluorescence resonance energy transfer (FRET). Upon fusion of the LNP with the model endosome, the fluorescence of NBD increases due to the greater distance between NBD and Rhod. As shown in Fig. 5e, BT-CPA-LNP exhibited higher NBD fluorescence intensity compared to fresh LNPs, indicating its enhanced membrane fusion capability. These results suggest that the incorporated betaine increases the endosomal escape ability of LNPs, resulting in their increased mRNA delivery efficacy (Fig. 5f).
In addition to the enhanced mRNA delivery efficacy, the long-term storage of mRNA-LNP is also important for CPA. We tested the mRNA expression efficacy of LNP after storage at −80 °C for 4 and 6 months. Figure 5g showed that LNP+BT-CPA exhibited consistently enhanced mRNA expression after storage for over a period of 6 months. Both betaine and trehalose showed excellent profiles in a variety of biological applications. We do not anticipate high toxicity associated with BT-CPA. Indeed, cytotoxicity evaluation in DC2.4 cells showed that BT-CPA is not cytotoxic at all tested concentrations (Fig. 5h).
The enhanced mRNA expression of LNPs stored with BT-CPA could offer dose-sparing benefits. To evaluate the efficacy of LNP+BT-CPA after F-T as a vaccine, mRNA encoding a model antigen ovalbumin (mOVA) was encapsulated into LNPs. C57BL/6 mice were intramuscularly injected with either freshly prepared LNP or freeze-thawed LNP+BT-CPA at weeks 0 and 2, at doses of 0.1 µg or 1 µg mOVA per injection (Fig. 6a). The serum was collected in week 3 after the first injection for subsequent analysis of OVA-specific IgG antibody titers via enzyme-linked immunosorbent assay (ELISA). As shown in Fig. 6b, LNP+BT-CPA F-T induced higher OVA-specific antibody responses than freshly prepared LNP at both doses. The OVA-specific CD4 and CD8 T cell responses in the spleens of mice were further evaluated via an enzyme-linked immunosorbent spot (ELISpot) assay. LNP+BT-CPA F-T also generates higher Interferon-gamma (IFN-γ) producing CD4 and CD8 T cells than freshly prepared LNP (Fig. 6c, d, and Supplementary Fig. 16). These results indicate that BT-CPA enhances both the humoral and cellular immune responses of the mRNA-LNP vaccine after storage under freezing conditions, thus providing dose-sparing advantages.
a Schematic of vaccination regimen using C57BL/6 mice. b OVA-specific IgG response in the serum from 0.1 µg and 1 µg of mOVA-LNP vaccinated mice were analyzed by ELISA (n = 4 biologically independent samples). Data are shown as mean ± SD. c Optimal images of IFN-γ-spot-forming cells via ELISpot assay. Splenocytes were isolated 3 weeks post prime shot and stimulated by CD4 or CD8 peptide. d Quantitative analysis of IFN-γ spots of ELISpot assay stimulated with CD4 or CD8 peptide (n = 4 biologically independent samples). Data are shown as mean ± SD. Figure 6a was created in BioRender. Lu, X. (2025) https://BioRender.com/xdgkl76.
In conclusion, our study demonstrates that the combined cryoprotectant formulation of betaine and trehalose (BT-CPA) not only stabilizes lipid nanoparticles (LNPs) during freeze-thaw cycles but also enhances mRNA delivery. By leveraging the freezing process to incorporate betaine into LNPs, driven by freezing-induced concentration and lipid phase separation, we significantly improve their structural integrity, endosomal escape ability, and mRNA expression efficiency. These findings underscore the potential of using freezing to enhance LNP functionality by incorporating functional molecules during the freeze-thaw process, opening possibilities for the development of cryoprotectants that can improve the efficacy of mRNA-based therapeutics. Additionally, our results highlight the need for a more comprehensive understanding of the interactions between cryoprotectants and LNPs during freeze-thaw cycles. The chemical properties of cryoprotectants, such as charge, polarity, and functional activity, may profoundly influence nanoparticle behavior—affecting stability, payload release, and cellular uptake. Moving forward, cryoprotectants should be evaluated not only for their ability to stabilize nanoparticles but also for their potential role as active modulators of nanoparticle function, providing a perspective on formulation strategies for mRNA delivery.
Methods
Synthesis of Lipid nanoparticles
Lipid nanoparticles (LNPs) were synthesized by mixing the lipids and mRNA in a microfluidic chip. Ionizable lipids SM-102 (Sinopeg), ALC-0315 (Sinopeg), cKK-E12 (WuXi AppTec), or MC3 (Sinopeg), DSPC (Avanti Polar Lipids), cholesterol (Sigma–Aldrich), and DMG-PEG2000 (Avanti Polar Lipids) were dissolved in ethanol. The mLuc-mRNA (Firefly luciferase mRNA, APExBIO) or OVA-mRNA was dissolved in 50 mM citrate buffer (pH 4, Biorigin). mOVA were synthesized using T7 RNA polymerase-mediated transcription from a linearized DNA template (GenScript). The ethanol phase and aqueous phase were mixed with a volume ratio of 3:1 using syringe pumps (Harvard Apparatus). The mixture was dialyzed (MWCO = 12 k-14 kDa, Biorigin) against 1× phosphate-buffered saline (PBS, Sigma) at 4 °C for over 4 h. The hydrodynamic diameter and polydispersity index (PDI) of LNP were measured at 25 °C using a Zetasizer Nano ZSP (Malvern Instruments). The mRNA encapsulation efficiency was measured using Quant-iT RiboGreen RNA assay (Invitrogen according to the manufacturer’s instructions.
Freeze-thaw treatment to LNP
The cryoprotectant solution was prepared by dissolving trehalose (w/v) and betaine in the aqueous buffer of 10 mM sodium citrate and 20 mM Tris with the final pH adjusted to 7.4. The solution was then filtered through a 0.22 μm membrane to remove all particulates. Cryoprotectant solution was then mixed with LNP solution at room temperature in a 1:1 (v/v) ratio. For the freezing process, the cryoprotectant-LNP mixture was placed in a NalgeneTM Cryo Freezing Container, which provided controlled cooling at a rate of approximately 1 °C/min. The samples were maintained within the freezing container for at least 2 h in a −80 °C freezer. Then the frozen LNP suspensions were thawed at room temperature. The thawed solutions were then gently mixed to restore homogeneity before subsequent analysis.
Evaluation of mRNA expression in vitro
DC2.4 cells were incubated in a 37 °C humidified incubator with 5% CO2. 2 × 105 DC2.4 cells per well were seeded in each well of a 96-well plate (Merck). The cells were cultured in 200 µL of Roswell Park Memorial Institute (RPMI) medium 1640 basic (Gibco) supplemented with 10% fetal bovine serum (FBS, Gibco) and 1% penicillin-streptomycin (Gibco). When the cell confluence reached 50%, LNPs encapsulating Fluc-mRNA were added to the 96-well plate. The plate was incubated for 24 h before the detection of luciferase expression using the One-Lite Luciferase Assay System (Vazyme) according to the manufacturer’s instructions. The bioluminescence was measured using a multimode microplate reader (Synergy H1, BioTek).
Evaluation of mRNA expression in vivo
All animal experiments were approved by the Peking University Institutional Animal Care and Use Committee (PUIRB-LA2023156). The studies, study design, results, and findings are not specific towards any sex or gender. The animals were maintained in a controlled environment with a 12 h light/dark cycle. The ambient temperature was maintained at 22–24 °C, and the humidity level was kept at 40 ~ 60%. Eight-week-old female C57BL/6 mice were used to evaluate the mRNA expression in vivo. LNP and LNP+BT-CPA F-T containing 2 µg of mLuc were intramuscularly injected into the left thigh of each mouse (n = 6). Potassium (S)-2-(6-hydroxybenzo[d]thiazol-2-yl)-4, 5-dihydrothiazole-4-carboxylate (Bidepharm) in 200 µL of PBS (20 mg/mL) was intraperitoneally injected into each mouse at 4 and 24 h post-injection. The mice were then imaged using the IVIS Lumina system (Perkin Elmer).
1H NMR
1H NMR spectra were recorded using a Bruker NEO 700-NMR spectrometer operating at 700 MHz at room temperature. Betaine and trehalose were dissolved into methanol-D4 (Innochem) before scanning. All samples containing LNPs were dialyzed against PBS before scanning. 50 µL of the sample solution (in PBS) was mixed with 450 µL of methanol-D4, and the LNPs were then disrupted using an ultrasound sonicator. Chemical shifts are given on the parts per million scale (ppm). The center of the methanol-D4 (3.310 ppm) signals was used as the reference. All 1H NMR data were analyzed using the MestReNova software.
Differential Scanning Calorimetry (DSC)
DSC measurements of PBS or LNP solutions were performed on a calorimeter (PE DSC8500) equipped with liquid nitrogen under an argon atmosphere. The samples were first cooled from 20 °C to −80 °C at a rate of 5 °C min−1. After holding at −80 °C for 2 min, the samples were heated from −80 °C to 80 °C at a rate of 2.5 °C/min. The heat flow and temperature were calibrated before the experiments. All the DSC curves were normalized against sample weight. Phase transition and ice melting temperatures were obtained from the onset point of exothermic peaks during heating procedures.
Fluorescence microscopy
2.5 µL of the Cy5-mCherry@LNP+BT-CPA solution was deposited onto a silicon substrate and covered with a 12 mm diameter circular cover glass. The sample was placed on a cryo-stage (Instec mk2000, America) for temperature control. The temperature was decreased to −80 °C at a rate of 1 °C/min, held for 2 min, and subsequently ramped to −15 °C at a rate of 20 °C/min. Fluorescence and bright field microscopy images were captured at −15 °C using a Nikon optical microscope (LV100ND, Japan) equipped with a CCD camera (DS-Ri2).
LNP freeze-thaw with Gelred
The LNP solution with BT-CPA was mixed with Ultra GelRed 10,000× (GR501-01, Vazyme) at a 1000:1 (V/V) ratio. After F-T, the mixture was added to a black 96-well plate. The fluorescence of GelRed was measured using a multimode microplate reader (Synergy H1, BioTek) with excitation at 300 nm and emission at 613 nm.
Cellular uptake in vitro
DC2.4 cells were incubated in a 37 °C humidified incubator with 5% CO2. 5 × 104 DC2.4 cells per well were seeded in a 48-well plate (Merck) which were cultured in 200 µL RPMI medium 1640 basic supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. When the cell confluence reached 65%, LNPs encapsulating 100 ng Cy5-DNA per well were added into the 48-well plate to incubate for 3 h. Then the medium was removed. Cells were washed with PBS before adding 100 µL of 0.25% Trypsin-EDTA (Gibco) to digest cells. 100 µL medium was then added to each well. The cells were washed by PBS 3 times before being analyzed by a flow cytometer (Beckman Coulter).
Endosomal vesicle assay
Endosomal vesicles were prepared by mixing the ethanol phase containing 42% DOPE, 17% DOPC, 13% LBPA, 28% cholesterol, 1.5% Rhod-PE, and 2% NBD-PE with PBS (pH 5.5). The mixture was dialyzed using a dialysis membrane (MWCO = 12–14 kDa) against PBS (pH 5.5) at 4 °C for over 4 h. The successful synthesis of vesicles was confirmed by dynamic light scattering measurements. 2 mL of endosomal vesicles (110 nM of NBD-PE) were added to a fluorescence cuvette before sequentially adding 2 μL of LNPs containing 1 µg of mLuc. The fluorescence of the mixed solution was recorded after each addition of LNP by a Cary Eclipse fluorescence spectrometer (Ex/Em = 465/540 nm).
Confocal microscopy
DC2.4 cells were incubated in a 37 °C humidified incubator with 5% CO2. 2 × 105 DC2.4 cells per well were seeded in a glass bottom dish (Cellvis). The cells were cultured in RPMI medium 1640 basic supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin for 12 h. LNPs encapsulating Cy5-DNA were incubated with cells at a Cy5 concentration of 100 nM for 3 h. The medium was removed before being washed by PBS 3 times. The cell nucleus and lysosomes were stained with Hoechst (Gibco) and LysoTracker green (Beyotime) according to the manufacturer’s instructions. 4% paraformaldehyde was used to fix the cells. Cells were imaged by an LSM 880 confocal laser scanning microscope (Zeiss).
CCK-8 cytotoxicity assay
The cytotoxicity of LNPs was evaluated by CCK-8 assays. DC2.4 cells were seeded into 96-well plates (2 × 104 cells per well) in RPMI medium 1640. When DCs were 50% confluent, cells were incubated with different concentrations of sucrose and CPA for 24 h. The supernatant was aspirated before adding 100 µL of fresh RPMI medium 1640 and 10 µL of CCK-8 solution (Yeasen). Cells were incubated for 1 h at 37 °C. The 96-well plate was equilibrated to room temperature for 30 min. The absorbance at 450 nm was measured using a multimode microplate reader (Synergy H1, BioTek). Cell viability was calculated according to the manufacturer’s instructions.
Immunization and ELISA
Female C57BL/6 mice (6–8 weeks old) received intramuscular injections of mOVA@LNP or mOVA@LNP+CPA F-T on days 0 and 14. Each dose contained 0.1 µg or 1 µg mOVA. Serum samples were collected from the submandibular (facial) vein for ELISA analysis. Briefly, 5 µg/mL of OVA protein (Sigma) in 50 mM Na2CO3 was coated onto high binding 96-well plates (100 µL per well, Merck) and incubated at 4 °C overnight. Coating buffers were aspirated before adding 150 μL of blocking buffer (1× PBS with 0.05% Tween−20, 1% BSA, and 0.02% NaN3). The plate was incubated for 2 h at 37 °C before being washed three times with 300 μL of PBST buffer (1× PBS with 0.05% Tween−20). 100 μL of diluted serum sample was added to each well. The plates were incubated for 1 h at 37 °C. The ELISA plates were washed three times with 300 μL of PBST before adding 100 μL of anti-mouse IgG-HRP (RAS060-C04, Acro) and incubated for 1 h at 37 °C. The ELISA plates were washed three times with 300 μL of PBST before adding 100 μL of 1-Step Ultra TMB-ELISA (Thermo Scientific). The reaction was stopped with 100 μL of 2 M sulfuric acid after 8–12 min incubation. The absorbance of each well at 450 nm was read using a microplate reader.
ELISpot assay
The pre-coated strip plate (3321-4AST−2, Mabtech) was washed four times with sterile 1× PBS (200 μL per well) and conditioned with RPMI 1640 medium (200 μL per well) for 30 min at room temperature. Splenocytes were prepared from immunized mice and incubated (4 × 105 cells per well) in RPMI 1640 medium (200 μL per well) supplemented with 10% FBS, 1% penicillin–streptomycin, 1% HEPES pH 7.3, 1% MEM non-essential amino acids 1% sodium-pyruvate, and 0.1% 2-mercaptoethanol. Cells were stimulated with 10 µg/mL SIINFEKL peptide and ISQAVHAAHAEINEAGR peptide (GL Biochem) for restimulation of OVA-specific CD4+ and CD8+ T cells, respectively, for 24 h. Spots were visualized with streptavidin-alkaline phosphatase (3321-4AST-2, MabTech) and BCIP/NBT substrate (3321−4AST-2, MabTech) followed by incubation with biotin-conjugated anti-IFN-γ antibody (3321-4AST-2, MabTech). The number of spots was counted visually.
Statistics and reproducibility
All statistical analyses were performed using the GraphPad Prism software package (PRISM 8.2.1; GraphPad Software). Technical or biological replicates were used in all experiments unless otherwise stated. Data are presented as means ± SD. One-way analysis of variance (ANOVA) was used when there were multiple comparisons. Student’s t-test was used for single comparisons. The specific statistical methods are indicated in the figure legends. Data were significantly different if P < 0.05.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All the data generated in this study are provided in the Article, Supplementary Information, Source Data file, and deposited in the figshare database under accession code [https://doi.org/10.6084/m9.figshare.28774901]. Source data is available for Figs. 1–6 and Supplementary Figs. 1–16 in the associated source data file. Source data are provided in this paper.
References
Paunovska, K., Loughrey, D. & Dahlman, J. E. Drug delivery systems for RNA therapeutics. Nat. Rev. Genet. 23, 265–280 (2022).
Huang, X. et al. The landscape of mRNA nanomedicine. Nat. Med. 28, 2273–2287 (2022).
Han, X. et al. An ionizable lipid toolbox for RNA delivery. Nat. Commun. 12, 7233 (2021).
Cullis, P. R. & Hope, M. J. Lipid nanoparticle systems for enabling gene therapies. Mol. Ther. 25, 1467–1475 (2017).
Hajj, K. A. & Whitehead, K. A. Tools for translation: non-viral materials for therapeutic mRNA delivery. Nat. Rev. Mater. 2, 17056 (2017).
Pardi, N., Hogan, M. J., Porter, F. W. & Weissman, D. mRNA vaccines-a new era in vaccinology. Nat. Rev. Drug Discov. 17, 261–279 (2018).
Liu, J. et al. Fast and efficient CRISPR/Cas9 genome editing in vivo enabled by bioreducible lipid and messenger RNA nanoparticles. Adv. Mater. 31, 1902575 (2019).
Hou, X., Zaks, T., Langer, R. & Dong, Y. Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 6, 1078 (2021).
Kon, E., Elia, U. & Peer, D. Principles for designing an optimal mRNA lipid nanoparticle vaccine. Curr. Opin. Biotechnol. 73, 329–336 (2020).
Holm, M. R. & Poland, G. A. Critical aspects of packaging, storage, preparation, and administration of mRNA and adenovirus-vectored COVID-19 vaccines for optimal efficacy. Vaccine 39, 457–459 (2021).
Li, Y. & Breaker, R. R. Kinetics of RNA degradation by specific base catalysis of transesterification involving the 2γ-hydroxyl group. J. Am. Chem. Soc. 121, 5364–5372 (1999).
Chheda, U. et al. Factors Affecting Stability of RNA – Temperature, Length, Concentration, pH, and Buffering Species. J. Pharm. Sci. 113, 377–385 (2024).
Kim, B. et al. Optimization of storage conditions for lipid nanoparticle-formulated self-replicating RNA vaccines. J. Control. Release 353, 241–253 (2023).
Qin, B. et al. How does temperature play a role in the storage of extracellular vesicles?. J. Cell. Physiol. 235, 7663–7680 (2020).
Uddin, M. N. & Roni, M. A. Challenges of storage and stability of mrna-based covid-19 vaccines. Vaccines 9, 1033 (2021).
Kamiya, M. et al. Stability Study of mRNA-Lipid nanoparticles exposed to various conditions based on the evaluation between physicochemical properties and their relation with protein expression ability. Pharmaceutics 14, 2357 (2022).
Crommelin, D. J. A., Anchordoquy, T. J., Volkin, D. B., Jiskoot, W. & Mastrobattista, E. Addressing the cold reality of mRNA vaccine stability. J. Pharm. Sci. 110, 997–1001 (2021).
Patel, S. et al. Naturally-occurring cholesterol analogues in lipid nanoparticles induce polymorphic shape and enhance intracellular delivery of mRNA. Nat. Commun. 11, 983 (2020).
Álvarez-Benedicto, E. et al. Optimization of phospholipid chemistry for improved lipid nanoparticle (LNP) delivery of messenger RNA (mRNA). Biomater. Sci. 10, 549–559 (2022).
DeGrandpre, E. L., DeGrandpre, M. D., Colman, B. P. & Valett, H. M. Observations of river solute concentrations during ice formation. ACS ES T Water 1, 1695–1701 (2021).
Siow, L. F., Rades, T. & Lim, M. H. Characterizing the freezing behavior of liposomes as a tool to understand the cryopreservation procedures. Cryobiology 55, 210–221 (2007).
Peter, B., Levrier, A. & Schwille, P. Spatiotemporal propagation of a minimal catalytic RNA network in GUV protocells by temperature cycling and phase separation. Angew. Chem. Int. Ed. 62, e202218507 (2023).
Vuist, J. E., Boom, R. M. & Schutyser, M. A. I. Solute inclusion and freezing rate during progressive freeze concentration of sucrose and maltodextrin solutions. Dry. Technol. 39, 1285–1293 (2021).
Wolkers, W. F. et al. Factors affecting the membrane permeability barrier function of cells during preservation technologies. Langmuir 35, 7520–7528 (2019).
Costa, A. P., Xu, X. & Burgess, D. J. Freeze-anneal-thaw cycling of unilamellar liposomes: Effect on encapsulation efficiency. Pharm. Res. 31, 97–103 (2014).
Koide, H. et al. One-step encapsulation of siRNA between lipid-layers of multi-layer polycation liposomes by lipoplex freeze-thawing. J. Control. Release 228, 1–8 (2016).
Mori, N., Ishihara, M., Tasaki, H., Sankai, T. & Otsuki, J. The effect of betaine for mouse sperm cryopreservation. Cryobiology 106, 157–159 (2022).
Li, Y. et al. Enhanced endosomal/lysosomal escape by distearoyl phosphoethanolamine- polycarboxybetaine lipid for systemic delivery of siRNA. J. Control. Release 176, 104–114 (2014).
Peng, H. et al. PH-sensitive zwitterionic polycarboxybetaine as a potential non-viral vector for small interfering RNA delivery. RSC Adv. 10, 45059–45066 (2020).
Yang, J. et al. Natural zwitterionic betaine enables cells to survive ultrarapid cryopreservation. Sci. Rep. 6, 37458 (2016).
Petronini, P. G., De Angelis, E., Borghetti, A. F. & Wheeler, K. P. Osmotically inducible uptake of betaine via amino acid transport system A in SV-3T3 cells. Biochem. J. 300, 45–50 (1994).
Steponkus, P. L. & Lynch, D. V. Freeze/thaw-induced destabilization of the plasma membrane and the effects of cold acclimation. J. Bioenerg. Biomembr. 21, 21–41 (1989).
Peter, B. & Schwille, P. Elucidating the mechanism of Freeze-Thaw driven content mixing between protocells. ChemSystemsChem 5, e202300008 (2023).
Litschel, T. et al. Freeze-thaw cycles induce content exchange between cell-sized lipid vesicles. N. J. Phys. 20, 055008 (2018).
Tsuji, G., Fujii, S., Sunami, T. & Yomo, T. Sustainable proliferation of liposomes compatible with inner RNA replication. Proc. Natl. Acad. Sci. USA 113, 590–595 (2016).
Salibi, E., Peter, B., Schwille, P. & Mutschler, H. Periodic temperature changes drive the proliferation of self-replicating RNAs in vesicle populations. Nat. Commun. 14, 1222 (2023).
Schoenmaker, L. et al. mRNA-lipid nanoparticle COVID-19 vaccines: structure and stability. Int. J. Pharm. 601, 120586 (2021).
Akhoondi, M., Oldenhof, H., Sieme, H. & Wolkers, W. F. Freezing-induced removal of water from phospholipid head groups in biomembranes. Biomed. Spectrosc. Imaging 1, 293–302 (2012).
Drobnis, E. Z. et al. Cold shock damage is due to lipid phase transitions in cell membranes: A demonstration using sperm as a model. J. Exp. Zool. 265, 432–437 (1993).
Crisafuli, F. A. P., Ramos, E. B. & Rocha, M. S. Characterizing the interaction between DNA and GelRed fluorescent stain. Eur. Biophys. J. 44, 1–7 (2015).
Wolkers, W. F., Balasubramanian, S. K., Ongstad, E. L., Zec, H. C. & Bischof, J. C. Effects of freezing on membranes and proteins in LNCaP prostate tumor cells. Biochim. Biophys. Acta - Biomembr. 1768, 728–736 (2007).
Miao, L. et al. Synergistic lipid compositions for albumin receptor mediated delivery of mRNA to the liver. Nat. Commun. 11, 2424 (2020).
Acknowledgements
This research was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences No.XDB0960100 to X.L., the National Natural Science Foundation of China (No. 22175188 to X.L. T2293762 and T2293760 to J.W., 22472188 to Z.L.), International Partnership Program of the Chinese Academy of Sciences No. 027GJHZ2023179GC to X.L., and start-up funding from the Institute of Chemistry, Chinese Academy of Sciences to X.L.
Author information
Authors and Affiliations
Contributions
X.C., X.Z., Z.L., X.L., and J.W. conceived the idea, analyzed the data, and wrote the manuscript. X.C., X.Z., K.T., and H.H. performed experiments. All authors contributed to the discussion and editing of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
X.C., X.Z., Z.L., X.L., and J.W. are inventors on a patent application (No. CN2023115331792) held by the Institute of Chemistry Chinese Academy of Sciences that covers the design and applications of BT-CPA reported in this study. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Cheng, X., Zheng, X., Tao, K. et al. Freezing induced incorporation of betaine in lipid nanoparticles enhances mRNA delivery. Nat Commun 16, 4700 (2025). https://doi.org/10.1038/s41467-025-60040-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-60040-9