Abstract
Cyclic GMP-AMP synthase (cGAS), a key mediator of the cGAS-STING DNA sensing pathway that triggers type-I interferon responses, plays a crucial role in innate immunity and has been implicated in the pathogenesis of various disease. Despite advances in the development of cGAS inhibitors, none have reached the market and there remains an unmet need for divergent chemical scaffolds with high selectivity, potency across species, and target-adaptive mechanisms of action to explore cGAS’s potential as a therapeutic target. Here we report the structural, biochemical, cellular, and mechanistic characterization of the XL series of allosteric inhibitors, designed to engage an innovative allosteric site near the activation loop of cGAS. Among them, XL-3156 and XL-3158 emerge as potent, selective, cross-species cGAS inhibitors that simultaneously occupy allosteric and orthosteric sites, stabilizing the activation loop in a closed, inactive conformation and thereby attenuating the cGAS-DNA interactions. Moreover, these allosteric inhibitors, also known as protein condensation inhibitors (PCIs), significantly suppress cGAS-DNA condensate formation, triggering a morphological transition from liquid-solid phase separation (LSPS) to liquid-liquid phase separation (LLPS) at the molecular level while eliminating LLPS in cells. The distinct mechanism of action enables PCIs to achieve synergistic effects in combination with orthosteric inhibitors. These results establish a mechanism-driven pharmacological strategy to inhibit cGAS through PCIs that modulate phase separation primarily by engagement of the allosteric site.
Similar content being viewed by others
Introduction
The innate immune system has evolved to dynamically detect both pathogen-associated molecular patterns and damage-associated molecular patterns through pattern recognition receptors such as Toll-like receptors and nucleotide oligomerization ___domain-like receptor1. As a key PRR for cytosolic double-stranded DNA (dsDNA), cyclic GMP-AMP synthase (cGAS) catalyzes the formation of cyclic GMP-AMP (cGAMP) upon dsDNA binding, initiating a signaling cascade that activates the interferon (IFN) pathway and host defense responses2,3. While crucial for pathogen defense, dysregulated cGAS activation is implicated in the pathogenesis of many diseases, including Aicardi-Goutières syndrome, autoimmune disorders, inflammatory conditions, neurodegenerative diseases, aging, and COVID-194,5,6,7,8. cGAS inactivation achieved genetically or pharmacologically consistently mitigates disease progression in diverse animal models in preclinical studies4,5,6,7,8. Notably, VENT-039, a small-molecule cGAS inhibitor with a currently confidential structure, has recently advanced to phase I clinical trials, underscoring the therapeutic promise of cGAS inhibition.
Previous efforts in identifying cGAS inhibitors have yielded several scaffolds, such as those of G15010,11,12, PF-0692821513, S314, and flavonoid15, which specifically bind to the catalytic ___domain of human cGAS (h-cGASCD). However, these inhibitors often manifest either species-specific inhibitory activity or insufficient cellular potency. Among these, only G150 demonstrates cellular human cGAS (h-cGAS) inhibition with an EC50 value of 1.96 μM, nearly 200-fold weaker than its enzymatic inhibition potency (IC50 = 0.01 μM)10. By contrast, compounds such as compound 316 selectively inhibit the enzymatic activity of mouse cGAS (m-cGAS) but not h-cGAS. Although RU.521 was originally designed as a m-cGAS inhibitor17, it exhibits cross-species potency by inhibiting both mouse and human cGAS activity at the cellular level18,19. In addition, X620, suramin21, CU-3222, compound C23, and aspirin24 have been reported to inhibit cGAS through various, yet-to-be-determined binding mechanisms, but lack the on-target specificity. Accordingly, there is a critical unmet need for the development of cGAS inhibitors with distinct chemical structures, cross-species potency, selectivity, and novel mechanisms of action to advance cGAS therapeutic target validation across multiple diseases.
Liquid-liquid phase separation (LLPS) has emerged as an important mechanism for arrangement and regulation of biomolecules in cells25. Recent studies have highlighted the importance of these biomolecular condensates as membrane-less organelles that can be pharmacologically modulated through different mechanisms. As reviewed by Patel et al., drugs can be classified as dissolvers (dissolve condensates), inducers (induce condensates), localizers (alter localization), or morphers (alter physiochemical properties) based on their effects on condensates, which provides alternative perspectives for drug development targeting phase separation26. Upon sensing cytosolic DNA, cGAS undergoes LLPS via multivalent interactions formed between cGAS and DNA27,28,29, which provide the structural basis for cGAS phase separation with unique features induced by species-specific variations28,30,31. Multiple factors, including nucleic acids27,32, proteins33,34,35,36,37, and metabolites such as spermine38,39 and fatty acids40, have been reported to regulate cGAS LLPS, offering an opportunity for the development of cGAS inhibitors by targeting the LLPS of cGAS. Though a class of cyclopeptides has been identified to inhibit cGAS phase separation by specifically binding to the DNA binding site of cGAS19, small molecule inhibitors that are rationally designed to disturb the LLPS of cGAS remain beyond reach.
In this work, we carry out a structure-based design of a distinct class of cGAS inhibitors by targeting an allosteric site to interact with the activation loop of cGAS, a region known to undergo conformational changes upon DNA binding28,30,41 and recognized as an allosteric inhibitor binding site. These allosteric inhibitors are able to inhibit the activity of both the recombinant and intracellular cGAS by preventing cGAS LLPS. It has been noted that cGAS can condense from ~10 nM to ~10 μM in cells upon LLPS27,29, which might account for the significantly lower cellular potency (EC50) of orthosteric inhibitors in contrast to their potent inhibitory activity against cGAS in enzymatic assays (IC50). Moreover, our inhibitors are found to block cGAS LLPS by directly preventing the formation of cGAS-DNA condensates, thereby termed protein condensation inhibitors (PCIs). Such a distinct mechanism allows PCIs to potently inhibit cellular cGAS at lower concentrations, demonstrating a superior inhibition potential compared to the orthosteric cGAS inhibitors. In addition, the in vivo efficacy of these compounds is evaluated in a cerulein-induced acute pancreatitis (AP) model, indicating their potential as drug candidates for the treatment of cGAS-related inflammatory diseases. Accordingly, the rational design and development of PCIs provide a mechanistic framework for enhancing the cellular efficacy of inhibitors targeting cGAS and other phase-separating proteins, highlighting the broad utility of this strategy.
Results
Identification of an allosteric site followed by the design of an allosteric inhibitor XL-3156
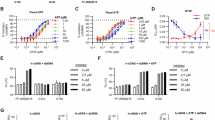
At the beginning of our cGAS inhibitor discovery project, PF-0692821513 and RU.52117 have been reported to specifically bind to the catalytic ___domain of h-cGAS and m-cGAS, respectively. To understand the ligand binding modes of different inhibitors and carry out the structure-based design of small-molecule inhibitors of cGAS, we determined high-resolution crystal structures of PF-06928215 and RU.521 in complex with the catalytic ___domain of human cGAS (h-cGASCD). The crystal structure of h-cGASCD bound with PF-06928215 was used as a model for virtual screening in our previous study14. A careful structure refinement on the h-cGASCD-RU.521 complex surprisingly unveiled that two enantiomers of RU.521 occupied two distinct sites with 50% occupancy each, reminiscent of butterfly wings. Their isobenzofuran-1(3H)-one moiety overlapped in the same spatial position (Fig. 1a and Supplementary Fig. 1a, b). The S-enantiomer of RU.521 ((S)-RU.521) fits well into the electron density at the classical inhibitor binding site (i.e., the active or orthosteric site) and is sandwiched between R376 and Y436. While the R-enantiomer of RU.521 ((R)-RU.521) is well positioned between S434 and V218 at a non-canonical ligand binding site, where it forms direct contacts with the activation loop (residues 210-220) (Fig. 1a). Conformational changes of the activation loop play a crucial role in cGAS-DNA interactions as the activation loop itself contributes to site A of dimeric h-cGASCD for DNA binding (Fig. 1b)28,30. In the open conformation, the activation loop forms direct contacts with DNA, whereas the closed conformation sterically prevents DNA engagement (Fig. 1c)28,41. Importantly, cGAS catalytic domains bind to DNA as dimers (Fig. 1b). When the activation loop is in the closed conformation, both cGAS monomers of the dimer induce steric clashes with DNA. Such a steric clash impacts two distinct DNA-binding sites, rather than a single one, significantly impairing the overall DNA binding capacity of the cGAS dimer. The co-crystal structure suggests that the direct interactions of (R)-RU.521 with the activation loop stabilize this loop in the closed conformational state, preventing cGAS activation and thereby blocking cGAS-DNA interactions28. Therefore, this non-canonical site which (R)-RU.521 engages is considered to be an allosteric site of cGAS.
a Left, an overall crystal structure of h-cGASCD in complex with RU.521; Right, an enlarged view of interactions between residues and two enantiomers of RU.521 (light blue (S)-RU.521 binds to the orthosteric site and light green (R)-RU.521 engages the allosteric site). b Superimposed structures of DNA-bound cGAS dimer to show three DNA-binding sites: A (green), B (red), and C (purple) (PDB IDs: 6EDB and 6CT9). The activation loop at site A is highlighted in two black boxes. c Superimposed activation loops of cGAS bound with DNA (green) and RU.521 (blue). Interactions between residues and XL-3156 (yellow, d) or G150 (purple, e). f Superimposed structures of RU.521 (light green & blue), G150 (purple), and XL-3156 (yellow) in complex with h-cGASCD. Cartoon representation of h-cGASCD omitted for clarity. The orthosteric site and allosteric site are highlighted in purple and yellow, respectively.
Inspired by the distinct binding mode of RU.521 with h-cGASCD, we successfully designed and synthesized an inhibitor, XL-3156, targeting both the orthosteric and allosteric sites. XL-3156 was prepared by reacting pyridinecarboxaldehyde with two molecules of 1-(1H-benzo[d]imidazol-2-yl)-3-methyl-1H-pyrazol-5-ol (the wings) (Supplementary Fig. 1a). To enhance the aqueous solubility of the compound, we prepared a hydrochloride salt of XL-3156 and its chemical structure determined by small-molecule single crystal X-ray diffraction analysis does contain two hydrochlorides per molecule. Most importantly, we solved a crystal structure of h-cGASCD in complex with XL-3156 (Fig. 1d and Supplementary Fig. 1b). For a direct comparison, we also determined a crystal structure of h-cGASCD in complex with G150, a typical orthosteric inhibitor (Fig. 1e and Supplementary Fig. 1b). As expected, XL-3156 perfectly achieves the integration of the binding modes of both R- and S-enantiomers of RU.521 and effectively engages the allosteric site as well as the orthosteric site. The allosteric site is ~11.7 Å away from the orthosteric site (Supplementary Fig. 1c) as shown in the co-crystal structure. By contrast, G150 binds solely to the orthosteric site in the co-crystal structure (Fig. 1f). The divergent binding modes of XL-3156, RU.521, and G150 to h-cGASCD revealed by co-crystal structures thereby establish a structural foundation underpinning their potentially different mechanisms of action.
In addition, we determined a crystal structure of RU.521 in complex with m-cGASCD in the absence of DNA. Unlike the co-crystal structure of h-cGASCD in complex RU.521, no electron density of the compound was found at the allosteric site of m-cGASCD, indicating that RU.521 only occupies the orthosteric site (Supplementary Fig. 1b, d). This structure is almost identical to a previously reported crystal structure of m-cGASCD bound with RU.521 but that was determined in the presence of DNA17. These two structures reveal that RU.521 is only able to occupy the orthosteric site of m-cGASCD, regardless of the conformational states of the activation loop induced by the presence or absence of DNA. Moreover, RU.521 is a covalent inhibitor of m-cGAS, as evidenced by the observed electron density between RU.521 and the murine-specific residue C419 (Supplementary Fig. 1b, d). To further demonstrate its covalent binding capability, we incubated RU.521 with glutathione (GSH) and detected the presence of an adduct by high resolution mass spectrometer, validating its ability to form a covalent bond with GSH (Supplementary Fig. 1e). Additionally, we investigated the reactivity of RU.521 with cysteine under various pH conditions and incubation times using HPLC analysis. These experiments revealed that RU.521 could covalently react with the thiol group under non-acidic conditions, demonstrating a rapid and reversible covalent reaction (Supplementary Fig. 1f). Based on the covalent bond observed in the co-crystal structure, we proposed a mechanism for the reaction between RU.521 and cysteine (Supplementary Fig. 1g). Overall, RU.521 is well characterized as a covalent orthosteric inhibitor of m-cGAS while it indeed can noncovalently occupy both the orthosteric and allosteric sites of h-cGAS, demonstrating its distinct mechanisms of action with cGAS from different species. Notably, although RU.521 has been widely used as a tool compound in numerous studies on m-cGAS, here we provide a systematic exploration of its covalent binding mechanism of inhibition on m-cGAS and identify a previously uncharacterized warhead targeting cysteine as well.
Disruption of cGAS-DNA interactions as well as phase separation by XL-3156
Given that the allosteric site is located in proximity to the activation loop that plays a crucial role in cGAS activation, we hypothesized that the binding of allosteric inhibitors might affect the interactions between h-cGASCD and DNA through interfering with the conformational changes of the activation loop. To test this hypothesis, an electrophoretic mobility shift assay (EMSA) was employed to assess the inhibition effect of the compound on the binding of DNA to cGAS. Incubation of Cy5-labeled 45 bp DNA (ISD45) with six different concentrations of h-cGASCD (serially diluted 2-fold from 1 μM) in the presence of an inhibitor or DMSO was carried out. In general, higher concentrations of h-cGASCD result in lower mobility of free DNA after electrophoretic separation. However, the addition of XL-3156 or RU.521 leads to retaining a higher amount of free DNA when incubated with h-cGASCD at 0.5 μM (lane 2) or 0.25 μM (lane 3) compared to the addition of DMSO or G150 (Fig. 2a). This observation confirms that XL-3156 and RU.521, but not G150, efficiently reduce the h-cGASCD-DNA interactions. Using this assay, it is also revealed that the allosteric inhibitors attenuate the interactions between DNA and full-length h-cGAS (h-cGASFL) but not the N-terminal ___domain of h-cGAS (Supplementary Fig. 2a, b). Importantly, to rule out the intercalation of inhibitors to DNA directly, we employed an acridine orange fluorescence polarization assay to show that no DNA intercalation occurred for any of the three inhibitors, even at concentrations as high as 100 μM (Supplementary Fig. 2c). Therefore, the engagement of the allosteric site by XL-3156 and RU.521 changes the conformation of the activation loop, thereby reducing the interactions of cGAS with DNA. Such an effect can not be achieved by G150 that only occupies the orthosteric site.
a Electrophoretic mobility shift assays (EMSA) of Cy5-labeled DNA (ISD45, 0.05 μM) incubated with h-cGASCD (lanes 1–6: 1, 0.5, 0.25, 0.125, 0.063, and 0.031 μM, respectively) and indicated compound (20 μM). The experiment was repeated three times independently. b Quantified fluorescence intensities of condensates formed by mixing Cy3-labeled h-cGASCD at indicated concentration with FAM-labeled DNA (ISD100, 0.25 μM) in buffer containing indicated concentration of NaCl and indicated compound (100 μM) in the FAM channel. Each data point represents the mean intensity from 5 different images. c Representative images of phase separation induced by mixing Cy3-labeled h-cGASCD (2 μM) with FAM-labeled DNA (ISD100, 0.25 μM) in buffer containing 150 mM NaCl and different concentrations of indicated compound (left). Green, FAM-labeled DNA (ISD100); Red, Cy3-labeled h-cGASCD. Scale bar, 20 μm. Quantification of fluorescence intensities in the images (right). Data represent the mean ± SD from five different images. Statistical significance was determined by an unpaired two-tailed t test. *p < 0.05, **p < 0.01, ***p < 0.005, and ****p < 0.001. d Turbidity analysis of h-cGASCD at indicated concentration with DNA (ISD100, 0.25 μM) in buffer containing indicated concentration of NaCl and indicated compound (100 μM), where the numbers indicate OD340 values of three independent experiments. e EC50 fitting curves and histogram of DNA condensation assays for DNA-h-cGASCD interactions. Cy3-labeled DNA (ISD100, 0.15 μM) was incubated with various concentrations of h-cGASCD (51–6560 nM, in two-fold increments) in the presence of 50 μM indicated compound. Data are presented as mean ± SD from three independent experiments. Statistical significance was determined by an unpaired two-tailed t test. *p < 0.05, **p < 0.01, ***p < 0.005, and ****p < 0.001. Source data is available with this manuscript as a Source Data file.
The interactions between cGAS and DNA are essential to the occurrence of phase separation. The inhibitor binding interferes with cGAS-DNA interactions, which is expected to affect the phase separation of h-cGASCD. To test this prediction, different concentrations of Cy3-labeled h-cGASCD were incubated with FAM-labeled 100-bp DNA (ISD100) in the presence of different inhibitors and various concentrations of salt (NaCl). After that, images of the Cy3 and FAM channels were captured using confocal microscopy, and the fluorescence intensity of condensates was determined to assess the extent of phase separation (Fig. 2b and Supplementary Fig. 3). In the absence of inhibitors, cGAS forms amyloid-like solid phase separation with DNA31. The intensity of this separation decreases with increased salt concentrations and decreased protein concentrations. Higher salt concentrations contribute to less phase separation due to an electrostatic interaction-driven mechanism27,31. In the compound-treated groups, both XL-3156 and RU.521 significantly reduce the phase separation of h-cGASCD with DNA, and the former is more effective compared to the latter. In contrast, the orthosteric inhibitor G150 at a concentration of 100 μM has no effect on the phase separation (Fig. 2b and Supplementary Fig. 3). Confocal microscopy images further showed that XL-3156 and RU.521 inhibit the phase separation of h-cGASCD with DNA in a dose-dependent manner (Fig. 2c). In addition, a turbidity assay measuring OD340 values of unlabeled protein and DNA reveals that the addition of RU.521 and XL-3156 in particular distinctly reduces the condensates formed by unlabeled h-cGASCD and DNA (Fig. 2d). G150 was not used as a reference compound in the turbidity assay due to its absorbance at 340 nm. After that, a DNA condensation assay was used to quantitatively assess the h-cGASCD-DNA condensation28. In this assay, we incubated fluorescently labeled DNA (i.e., Cy3-labeled ISD100) with different concentrations of unlabeled h-cGASCD in the presence of DMSO or inhibitors at room temperature for 20 min, centrifuged the mixture, and collected the supernatant for the measurement of fluorescence intensity, which corresponded to the amount of DNA that did not form condensates with cGAS and was used to calculate the EC50 values. As a result, the determined EC50 values of h-cGASCD-DNA condensation in the presence of 50 μM DMSO, XL-3156, RU.521, or G150 are 300 nM, 783 nM, 873 nM, or 348 nM, respectively (Fig. 2e). The significantly higher EC50 values of XL-3156 and RU.521 over that of DMSO or G150 indicate that these two inhibitors effectively impair the formation of h-cGASCD-DNA condensates.
Taken together, the use of XL-3156 and RU.521 significantly attenuates the phase separation of h-cGASCD by preventing the formation of condensates containing h-cGASCD and DNA. These two allosteric inhibitors are thus named PCIs.
A morphological shift from LSPS to LLPS induced by XL-3156
Intriguingly, a close examination of h-cGASCD-DNA condensates revealed that XL-3156 treatment leads to the formation of distinct circular liquid droplets, contrasting sharply with the irregular solid-like aggregates observed in both the DMSO- and G150-treated groups (Supplementary Fig. 3). Given the low quantity of condensates in the XL-3156 group, we reduced the salt concentration to 50 mM in order to increase cGAS-DNA interactions, thereby facilitating the condensate formation. Reducing the NaCl concentration resulted in a markedly increased number of condensates. Notably, in the XL-3156-treated group, circular liquid droplets form at low concentrations of h-cGASCD (1.25 μM and 2.5 μM), transforming into gel-like forms at higher concentrations of h-cGASCD (5 μM). In contrast, the DMSO-treated group consistently produces fibrous solid aggregates, regardless of changes in protein concentration (Fig. 3a). This demonstrates that XL-3156 induces a morphological shift from LSPS to LLPS or liquid-gel phase separation (LGPS) of h-cGASCD-DNA in vitro at the molecular level, suggesting that the use of XL-3156 reduces the degree of phase separation.
a Representative images of the phase separation induced by mixing Cy3-labeled h-cGASCD at indicated concentration with FAM-labeled DNA (ISD100, 0.25 μM) in a buffer containing 50 mM NaCl and DMSO or XL-3156 (100 μM). Green, FAM-labeled DNA; Red, Cy3-labeled h-cGASCD. Scale bar, 25 μm. b Fluorescence recovery after photobleaching (FRAP) of h-cGASCD-DNA condensates. The condensates of LLPS (liquid-liquid phase separation), LGPS (liquid-gel phase separation), and LSPS (liquid-solid phase separation) were formed after FAM-labeled DNA (ISD100, 0.25 μM) mixed with 2.5 μM h-cGASCD plus XL-3156, 5 μM h-cGASCD plus XL-3156, and 2.5 μM h-cGASCD plus DMSO, respectively, in a buffer containing 50 mM NaCl. Time 0 indicates the time of photobleaching. Data represent the mean ± SD of six independent experiments. Scale bar, 5 μm. c, Time-lapse micrographs of LLPS merging droplets that were formed as described in (a). Data are representative of at least 3 merging droplets. Scale bar, 2 μm. d 3D imaging of cGAS-DNA LSPS or LLPS condensates that were formed as described in (a). A side (left top) and top (left bottom) view and the view of isometric projection (right) of the condensates were presented. e Quantified fluorescence intensities of condensates formed by mixing Cy3-labeled h-cGASFL at indicated concentration with FAM-labeled DNA (ISD100, 0.25 μM) in buffer containing indicated concentration of NaCl and indicated compound (100 μM) in the FAM channel. Each data point represents the mean intensity from five different images. f Representative images of (e) with indicated marks. Scale bar, 25 μm. g Schematic of phase transition of cGAS-DNA condensation. Enhanced cGAS-DNA interactions promote the transition of soluble solution to LLPS, followed by the transition from LLPS to LSPS, e.g., enhancing the extent of phase separation. In contrast, elevated concentrations of NaCl and the allosteric inhibitors lead to attenuating cGAS-DNA interactions and thereby reducing the extent of phase separation. Source data is available with this manuscript as a Source Data file.
Subsequently, the fluorescence recovery after photobleaching (FRAP) and time-dependent imaging of condensation experiments show that the fluorescence recovery of h-cGASCD-DNA condensates after photobleaching is much faster in the presence of XL-3156 compared to DMSO (Fig. 3b, c). In addition, two circular droplets of h-cGASCD-DNA condensates can merge into one in the presence of XL-3156 (Fig. 3c, Supplementary Movie 1). These results confirm the LLPS nature of XL-3156-treated condensates. Interestingly, 3D images reveal that the LLPS condensates accumulate at the bottom of the sample well, distinct from the pervasive LSPS mesh, displaying a fluorescence intensity significantly higher than that of LSPS (Fig. 3d, Supplementary Movies 2, 3). Moreover, h-cGASFL together with DNA spontaneously forms LLPS in the presence of 225 mM NaCl but remains in an LSPS state at lower salt concentrations (Fig. 3e, f, Supplementary Fig. 4), aligning with the finding reported previously31. Treatment with XL-3156 markedly diminishes the degree of phase separation of h-cGASFL and shifts from LSPS to LLPS at 150 mM of NaCl. By comparison, RU.521 has a similar but weaker effect, whereas both G150 and DMSO fail to induce any significant changes. Together, these data demonstrate that the addition of XL-3156 prevents h-cGAS-DNA from forming LSPS and instead promotes the formation of LLPS condensates in vitro at the molecular level. Furthermore, the turbidimetric assays and DNA condensation assays confirmed that PCIs like XL-3156 significantly impeded the h-cGASFL-DNA condensation, whereas G150 had no such effect (Supplementary Fig. 5). These results are similar to those of h-cGASCD mentioned above.
On the basis of these findings, we propose that weaker cGAS-DNA interactions drive the spontaneous formation of LLPS condensates from a dissolved state, while stronger cGAS-DNA interactions favor the occurrence of LSPS in vitro at the molecular level. Unlike the nonspecific attenuation of electrostatic interactions caused by elevated NaCl concentrations, allosteric inhibitors, serving as PCIs, impair the cGAS-DNA interactions through occupying the specific sites so as to selectively modulate cGAS phase separation (Fig. 3g).
PCIs inhibit phase separation via the activation loop of cGAS
The significant effects of XL-3156 and RU.521 on cGAS phase separation prompt us to investigate how the binding of these PCIs hinders the interaction of h-cGASCD with DNA, ultimately reducing the h-cGAS-DNA condensation. As mentioned above, the activation loop of cGAS is crucial for the recognition of DNA. A multiple sequence alignment reveals that the residues at this loop are highly conserved compared to other regions of cGAS (Fig. 4a, Supplementary Fig. 6a), suggesting that the activation loop are essential for the function of cGAS. Our co-crystal structures show that XL-3156 or RU.521 form hydrophobic, π-π stacking, and cation-π interactions with surrounding residues including V218 and K219 at the activation loop together with S434, S435, Y436, and R376 (Fig. 4b). To demonstrate the impact of these important residues on the binding of PCIs, we introduced several mutations and carried out the aforementioned DNA condensation assays with these mutants. The hydrophobic V218 was mutated to a hydrophilic amino acid E218, significantly decreasing the h-cGASCD(V218E)’s ability to form condensates with DNA and consequently nullifying the effects of two PCIs. The V218K mutation maintains h-cGASCD condensation but attenuates the efficacy of the PCIs (Fig. 4c, Supplementary Fig. 7a, b). By contrast, mutation of V218 to amino acids M and F that favor hydrophobic interactions enhances RU.521’s ability to disrupt h-cGASCD (V218M or V218F)-DNA condensates, while XL-3156’s efficacy remains unchanged (Fig. 4c, Supplementary Fig. 7c, d). Mutations at the orthosteric site (S435L or Y436A) also reduced the inhibitory effect of both PCIs on condensate formation (Fig. 4c, Supplementary Fig. 7e, f). Together, these results support the notion that the specific interactions of PCIs with key residues allow the PCIs to achieve the inhibition of the h-cGASCD-DNA condensation.
a Sequence alignment of residues around the activation loop of cGAS from Homo sapiens (human; Uniprot Q8N884), Rhesus macaque (Rhesus monkey; Uniprot A0A0F7J4C8), Mus musculus (mouse; Uniprot Q8C6L5), Gallus gallus (chicken; Uniprot A0A8V0X930), Xenopus tropicalis (xenopus; Uniprot A0A6I8R967), and Danio rerio (zebrafish; Uniprot F1QCP4). Residue numbers above the sequences are from h-cGAS. Blue triangles highlight the residues of the activation loop that interact directly with DNA, according to the crystal structure with PDB ID of 6CT9. The black underlines indicate the highly conserved residues of the activation loop. The alignment was visualized with the ENDscript server. b Key residues involved in the interactions with XL-3156. c EC50 values determined by the DNA condensation assays for WT h-cGASCD and various mutants incubated with different compounds (50 μM). The residues at the activation loop (V218) and orthosteric site (S435 and Y436) that interact with XL-3156 were mutated. The Δloop refers to the substitution of YYEHVK with SG. Data are presented as mean ± SD from three independent experiments. d Distance variation between the Cα atoms of S213 at the activation loop and K414 at the helix (red) represents conformational states of the activation loop in DNA- (cyan) and ligand-bound (green) forms. Violin plots display the Cα-Cα distance distributions between S213 and K414 of h-cGASCD in an apo state and in complex with XL-3156, and G150 calculated based on 1-μs MD simulation trajectories. Violin plot elements: white dot (median), bounds of box (25th-75th percentiles, IQR = Interquartile Range), whiskers (1.5 × IQR from box edges). Data from 1 μs production phase trajectories (n = 500,000 frames/group, 2 ps sampling interval). Source data is available with this manuscript as a Source Data file.
Furthermore, a truncation of the activation loop by replacing six residues (YYEHVK) with SG (Δloop) results in the EC50 of condensation for Δloop h-cGASCD (306 nM) similar to that for the wild-type h-cGASCD (300 nM), although no enzymatic activity was detected for Δloop h-cGASCD in the pyrophosphatase-coupled assay (PPI assay) (Supplementary Fig. 7i). While our structural analysis indicates that the closed conformation of the activation loop creates a steric hindrance to DNA binding, the truncation of the loop eliminates such a steric hindrance and has no significant impact on cGAS-DNA condensation or phase separation. Notably, such a truncation leads two PCIs to almost have no impact on the phase separation of the Δloop h-cGASCD (Fig. 4c, Supplementary Fig. 7g). These data, combined with the co-crystal structures, reveal that the PCIs are not able to exert the inhibitory effect on the h-cGAS-DNA condensation without the help of the activation loop. Specifically, the closed conformation of the loop stabilized by the engagement of PCIs serves as the steric hindrance to DNA binding, which is crucial for inhibiting phase separation.
In addition, molecular dynamics (MD) simulations were utilized to examine conformational changes of the activation loop in the presence and absence of inhibitors. Stable root-mean-square deviation (RMSD) values during the 1 μs MD trajectory confirm system equilibrium and unchanged ligand binding modes (Supplementary Fig. 8). To well characterize the activation loop conformation, we monitored the distance between the Cα atoms of S213 at the flexible activation loop and K414 at a stable helix. This distance increases when the loop adopts a closed conformation. It is revealed that XL-3156-bound protein maintains a greater S213-K414 distance compared to both apo and G150-bound states, indicating that XL-3156 stabilizes the closed conformation of the activation loop (Fig. 4d, Supplementary Fig. 9a). By the different method, this provides support for the conclusion inferred from the co-crystal structures and various biochemical assays.
Taken together, the conformational states of the activation loop are of utmost importance for cGAS phase separation while the allosteric PCIs lock the closed conformation of the loop to exert the inhibiting effect on the cGAS-DNA condensates formation.
Structure-based hit-to-lead optimization
After confirming that XL-3156 effectively inhibits cGAS-DNA phase separation, we conducted structure-based inhibitor design and optimization. The compounds were systematically evaluated using a combination of the PPI assay, condensation assay, and THP-1 dual reporter gene assay for cellular activity assessment.
Guided by the co-crystal structure of h-cGASCD bound with XL-3156, which shows two basic amino acids (K362 and R376) nearby the pyridine moiety, we first introduced a carboxyl group at the ortho position of the six-membered aromatic ring connected to the tertiary carbon, yielding XL-3117. This modification significantly improved the inhibitory activity against the enzymatic activity (IC50 = 5.5 μM) and cGAS-DNA condensation (EC50 = 0.4 μM) (Supplementary Fig. 10 and Supplementary Table 3). However, XL-3117 had poor cellular potency, most likely due to limited membrane permeability caused by its high polarity. Employing a bioisosteric replacement strategy, we substituted the NH moiety in the benzimidazole scaffold with a sulfur atom, thereby generating a benzothiazole pharmacophore in compound XL-3119. Furthermore, building upon the well-documented efficacy of fluorination in small-molecule drug optimization42, we carried out 4,6-difluoro substitution on the benzothiazole pharmacophore, yielding compound XL-3123 (Supplementary Table 3). The solved co-crystal structure of h-cGASCD in complex with XL-3123 confirms the rationale of our design strategy. As expected, XL-3123 utilizes a ligand binding mode identical to that of XL-3156 and its carboxyl group forms the desired salt bridge interactions with residues K362 and R376 (Fig. 5a, b). Unfortunately, while the molecular potency of XL-3119 and XL-3123 are similar to and slightly less than that of XL-3117, respectively, neither compound shows satisfactory cellular potency (Supplementary Table 3).
a Key residues of h-cGASCD involved in the interactions with XL-3123 in the co-crystal structure. b 2Fo–Fc electron density map of XL-3123 in the co-crystal structure with h-cGASCD, contoured at 1.0 σ level. c The RMSFs of residues (203–232) of h-cGASCD in complex with XL-3156 or XL-3158 calculated based on 1-μs MD simulation trajectories. d Time-dependent numbers of hydrogen bonds between h-cGASCD and XL-3156 or XL-3158 during the 1-μs simulation. e Noncovalent interaction energies between h-cGASCD and XL-3156 or XL-3158 during the 1-μs simulation (EP-I, vdW: vdW energy, EP-I, Electr.: electrostatic energy and EP-I, total: the sum of vdW energy and electrostatic energy). Energies are in unit of kcal/mol. Violin plot elements: white dot (median), bounds of box (25th-75th percentiles), whiskers (1.5 × IQR from box edges). Data derived from 400 ns production phase (600-1000 ns) trajectories (n = 200,000 frames/group, 2 ps sampling interval). Source data is available with this manuscript as a Source Data file.
Subsequently, we explored the substitution of the carboxyl group with a trifluoromethyl group at the ortho-, meta-, and para- positions of the benzene ring, resulting in compounds XL-3157, XL-3159, and XL-3158, respectively (Supplementary Table 3). All three compounds demonstrated both potent h-cGAS inhibition at the molecular level and effective cellular activity, with XL-3158 overall exhibiting the most favorable profiles (Supplementary Table 3, Supplementary Fig. 10). The microscale thermophoresis (MST) binding assay revealed XL-3158’s binding affinity of h-cGASCD with a Kd of 7.19 μM (Supplementary Fig. 10c). Additionally, the condensation assay results confirmed that all compounds in this series (including XL-3117, XL-3119, XL-3123, XL-3157, XL-3159, and XL-3158) effectively inhibit cGAS-DNA condensate formation.
Despite many attempts, the co-crystal structure of h-cGASCD in complex with XL-3158 has not yet been determined, most likely because the compound is not sufficiently soluble for X-ray protein crystallography study. Alternatively, we utilized molecular docking and MD simulations to investigate its binding mode with h-cGASCD, with the help of the co-crystal structure of h-cGASCD-XL-3156. The modeled complex structure suggests that the binding mode of XL-3158 is similar to that of XL-3156 or XL-3123 shown in the co-crystal structures (Supplementary Fig. 9b, c). This consistent binding mode across multiple compounds demonstrates the conserved interaction pattern between the XL series inhibitor and h-cGAS. In addition, the follow-up 1-μs MD simulation of the docked complex structure demonstrated the ability of XL-3158 to stabilize the closed conformation of the activation loop better than that of XL-3156 (Fig. 5c). Comparative analyses of XL-3158 and XL-3156 showed that XL-3158 established more stable hydrogen bonding (Fig. 5d and Supplementary Fig. 9d, e) and stronger electrostatic interactions with h-cGAS while maintaining comparable van der Waals interactions (Fig. 5e). These results provide molecular basis underlying the improved binding affinity of XL-3158 relative to XL-3156, primarily contributed by enhanced hydrogen bonding and electrostatic interactions with h-cGAS.
PCIs inhibit cGAS phase separation in cells
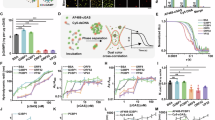
To determine whether the XL series compounds are able to act as PCIs to inhibit cGAS in the cellular environment, we used the representative XL-3158 to investigate its impact on cGAS phase separation in cells. We generated m-cGAS-knockout L929 cells followed by stably transducing of tetracycline-inducible (Tet-on) EGFP-h-cGASFL or EGFP (as a control) using lentiviral infection (Supplementary Fig. 11). To visualize the interaction between h-cGASFL and DNA, we transfected the cells with Cy5-labeled ISD100 followed by confocal microscopy studies. In Tet-on EGFP-h-cGASFL cells, we observed colocalization of h-cGASFL and DNA, suggesting the possible presence of phase-separated condensates (i.e., puncta in the image shown in Fig. 6a). In contrast, no colocalization was found in the control cells stably expressing EGFP only, confirming the specificity of the h-cGASFL-DNA interaction (Supplementary Fig. 11). Treatment with 20 μM XL-3158 resulted in a significant reduction in the number of puncta in the cells, while G150 did not have such an effect (Fig. 6b). Considering RU.521 at 20 μM was found to impair the complex formation of DNA with the transfection regent by visualizing labeled DNA, it was not included in this experiment. Furthermore, FRAP and time-dependent imaging confirmed the liquid nature of the condensates in cells (Fig. 6c, d), demonstrating the occurrence of LLPS with h-cGASFL without the treatment of inhibitors, which is different from the LSPS of h-cGAS (both h-cGASCD and h-cGASFL) in in vitro phase separation assays at the molecular level (Fig. 3a). These results together demonstrate that XL-3158 inhibits the LLPS of h-cGASFL with DNA in the cellular environment.
a Left: representative live-cell images of cGAS-DNA puncta formation in L929 cells stably expressing Tet-on EGFP-h-cGASFL, transfected with Cy5-labeled ISD100 (DNA), and treated with indicated compound (20 μM), n = 30. The first column shows full-field views with scale bars representing 20 μm. The other columns are enlarged views of the areas marked by white boxes in the first column. Green, EGFP-h-cGASFL; magenta, Cy5-labeled DNA. Scale bars, 10 μm. Right: quantified fluorescence intensities of different channels along a line perpendicular to one of the arrows in (left). The y-axis and x-axis represent the fluorescence intensity and the distance along the line, respectively. b Violin plot representation of numbers of h-cGASFL-DNA puncta counted from (a). Data are presented as mean ± SD from three independent experiments, with each experiment analyzing 10 different images. Statistical significance was determined by an unpaired two-tailed t test. ***p < 0.005. c FRAP of h-cGASFL-DNA puncta in cells. The puncta were formed as described in (a). Data represent the mean ± SD of five independent experiments. Scale bar, 2 μm. d Time-lapse micrographs of h-cGASFL-DNA merging puncta formed as described in (a). Data are representative of at least 3 merging puncta. Scale bar, 0.5 μm. Source data is available with this manuscript as a Source Data file.
Enzymatic and cellular potency of PCIs targeting cGAS
The revealed capability of allosteric inhibitors to disrupt cGAS phase separation at the cellular level prompts us to further investigate their impact on cGAS enzymatic activity in the cellular conditions. Using an IFN luciferase reporter cell assay (IFN-Luc, THP-1 dual cells), we determined the cellular potency of the allosteric inhibitor XL-3158 and the orthosteric inhibitor G150 for comparison. In the enzymatic PPI assay, XL-3158 and G150 exhibit an IC50 value of 11.1 μM and 0.06 μM against h-cGAS, respectively, suggesting that the inhibitory activity of G150 is more potent than that of XL-3158 at the molecular level. However, when evaluated by the cellular assay, XL-3158 and G150 surprisingly show an EC50 value of 11.85 μM and 9.78 μM, respectively (Fig. 7a–c). These data demonstrate that both inhibitors have similar cellular potency against cGAS despite the much lower molecular potency of XL-3158 compared to G150. In this context, the DNA condensation assay is necessary to purposefully evaluate the propensity of these small-molecule inhibitors to inhibit the condensation of cGAS with DNA. The determined EC50 value of h-cGASCD-DNA condensation in the presence of XL-3156 (783 nM) is much higher than that of G150 (348 nM) (Fig. 2e).
a, b Dose-dependent curves for inhibition of two compounds against h-cGASFL enzymatic activity at the molecular level measured by the PPI assay and IFN-luciferase activity assay in THP-1 Dual cells induced by HT-DNA. c IC50, EC50, IC50/EC50 ratio, and CC50 values of three compounds. d A schematic representation of the mechanism by which allosteric inhibitors effectively inhibit cGAS cellular activity. Blue circles represent LLPS droplets. The different positions of the fulcra in the two balances represent distinct leverage effects, reflecting varying efficacies of different types of cGAS inhibitors. e Synergistic effects of XL-3158 and G150 on inhibition of HT-DNA-induced IFN-luciferase activity in THP-1 Dual cells. Compounds (XL-3158: G150) were tested at molar ratios of 1:10, 1:5, 1:2.5, 1:1, 2.5:1, 5:1, and 10:1. Combination Index (CI) values were determined at the 90% effective dose (ED90) using CompuSyn software. CI < 1 indicates synergistic effects. f A schematic diagram of the DNA-induced IFN pathway activation. Inhibition of the IFN pathway is activated by different inducers in the presence of various compounds. g Quantification of IFNB-1 mRNA levels by qPCR, demonstrating the effects of compounds on the IFN pathway activation in THP-1 Dual cells induced by HT-DNA for 4 h. Statistical significance was determined by an unpaired two-tailed t test. **p < 0.01, ***p < 0.005, and ****p < 0.001. h Western blot analysis showing the effect of different concentrations of XL-3158 on HT-DNA-induced activation of TBK-1 and IRF3 phosphorylation in THP-1 Dual cells. Quantification of IFNB-1 mRNA levels by qPCR, demonstrating the effects of compounds on the IFN pathway activation in THP-1 Dual cells induced by Poly(I:C) for 4 h (i) and cGAMP for 4 h ( j). Statistical significance was determined by an unpaired two-tailed t test. *p < 0.05, ***p < 0.005, and ****p < 0.001. Representative western blot analysis demonstrating the effects of different compounds on TBK-1 phosphorylation in THP-1 Dual cells activated by Poly(I:C) for 4 h (k) and cGAMP for 2 h (l). Data are presented as mean ± SD from three independent experiments for all panels except d, f, h, k, l. Source data is available with this manuscript as a Source Data file.
Consequently, the question arises: how does XL-3158 achieve high cellular potency even though it does show weak inhibition of cGAS in the enzymatic assay at the molecular level, while G150 shows the opposite pattern? To exclude the influence of confounding factors and provide a quantitative comparison, we calculated the ratio of each compound’s enzymatic IC50 to its cellular EC50. Notably, G150 exhibits a ratio of 0.6%, whereas XL-3156 demonstrated a substantially higher ratio of 93.7%. This stark contrast can be well explained by our proposed model illustrated in Fig. 7d. It illustrates that in cells without phase separation, the concentration of cGAS is around 10 nM, where the orthosteric inhibitors can effectively suppress its activity. When DNA enters the cell, it induces cGAS allosteric activation followed by the occurrence of LLPS. The condensation in LLPS droplets increases local cGAS concentration to 10 µM27,29 in cells, facilitating the catalysis of substrate synthesis, a high concentration of the orthosteric inhibitor is thereby required to effectively inhibit concentrated cGAS in the condensates. In contrast, by inhibiting phase separation and preventing protein condensation, allosteric inhibitors at a low concentration can yield good inhibition of cGAS, thereby showing superior cellular inhibition potency compared to orthosteric inhibitors. Accordingly, the different positions of the fulcra in the two balances in Fig. 7d represent distinct leverage effects, reflecting varying efficacies of different types of cGAS inhibitors.
Since PCIs could maintain the intracellular concentration of free cGAS at a lower level while G150 could potently inhibit the free cGAS, it is speculated that the combination of PCIs with G150 might have a synergistic effect. To test this, we evaluated the inhibitory activity of XL-3158 and G150 in different combination ratios. The results show that there is a significant synergistic effect when the proportion of XL-3158 is higher, and the effect is significantly enhanced as the proportion of XL-3158 increases (Fig. 7e, Supplementary Fig. 12a). In contrast, when G150 is the dominant component, the synergistic effect is greatly diminished. These data firmly support our proposal shown in Fig. 7d that XL-3158 inhibits cGAS phase separation in cells by disrupting the formation of condensates, thus maintaining free cGAS at low concentrations and consequently allowing G150 to effectively inhibit the enzyme.
In addition, we employed quantitative PCR (qPCR) and western blot (WB) to measure the mRNA expression of IFN-1β and the phosphorylation levels of downstream pathways induced by HT-DNA to confirm that XL-3158 effectively suppressed the downstream signaling of the cGAS pathway (Fig. 7f–h, Supplementary Fig. 12b). On the other hand, instead of HT-DNA, we activated the pathway by using cGAMP, the downstream of cGAS, and RNA (Poly(I:C)) that activates the IFN pathway via another mechanism. As expected, both XL-3158 and G150 had no effect on the pathway in these two cases. By comparison, a STING inhibitor H151 and a TBK-1 inhibitor GSK8612 exhibited significant inhibition on the pathway activated by cGAMP and Poly(I:C), respectively (Fig. 7i–l, Supplementary Fig. 12b). The determined CC50 values for XL-3158 and G150 were 77.55 μM and 62.17 μM, respectively, demonstrating acceptable cytotoxicity (Fig. 7c, Supplementary Fig. 12c). These results reveal the specific inhibition of the DNA-induced activation of cGAS as well as the whole pathway by XL-3158, highlighting the excellent on-target selectivity of this allosteric inhibitor.
Given the high conservation of the activation loop and the limitation of species-specific inhibitory activity of existing inhibitors, we utilized recombinant m-cGAS and RAW-ISG cells to investigate the inhibitory activity of allosteric inhibitors against cGAS in murine species. The resulting IC50 and EC50 values of XL-3158 are 2.19 μM and 2.67 μM, respectively, demonstrating that XL-3158 effectively inhibits m-cGAS at both the molecular and cellular levels. Since G150 shows no inhibitory activity against m-cGAS10, we selected RU.521, a well-established m-cGAS inhibitor17, as a positive control. In comparison, the IC50 and EC50 values of RU.521 are 0.37 μM and 2.62 μM, respectively (Fig. 7c, Supplementary Fig. 13a, b). And both compounds have the similar CC50 values, 56.79 μM for XL-3158 and 70.06 μM for RU.521 (Fig. 7c, Supplementary Fig. 13c). The much lower IC50 value of RU.521 over XL-3158 is ascribed to the covalent binding of RU.521 to the orthosteric site of m-cGAS revealed by the aforementioned co-crystal structure. Nevertheless, the calculated ratio of each compound’s enzymatic IC50 to its cellular EC50 is 81.9% for XL-3158 and 14.1% for RU.521, again suggesting a better inhibitory efficiency of XL-3158 in mouse cells compared to RU.521, a covalent orthosteric inhibitor of m-cGAS (Fig. 7c). Furthermore, we employed WB and qPCR analyses to verify the efficacy and specificity of XL-3158 in mouse cells (Supplementary Fig. 13d–h). The results consistently demonstrate that XL-3158 inhibits the DNA-activated pathway, while showing no effect on the Poly(I:C)-activated pathway. Accordingly, these findings provide additional evidence for the broad utility of XL-3158 as a potent and selective inhibitor of cGAS-mediated signaling across different species of cells.
XL-3158 attenuates cerulein-induced acute pancreatitis
It has been reported in a previous study43 that the cGAS-STING pathway plays a critical role in the progression of acute pancreatitis (AP) while genetic knockout of cGAS significantly alleviates the disease progression. Therefore, we investigated whether XL-3158 could ameliorate cerulein-induced AP in vivo. We first assessed the in vivo pharmacokinetic properties of XL-3158 in mice (Supplementary Table 7). The results show a favorable oral bioavailability (F% = 64.2) and half-life (t1/2 = 4.07 h) in mice. According to the good pharmacokinetic parameters of XL-3158, we evaluated the therapeutic effects of oral administration of XL-3158 at a dose of 25 mg/kg in a cerulein-induced mouse model of AP (Fig. 8a). The results show that injection of cerulein successfully induced AP in mice, while XL-3158 treatment significantly attenuated pancreatic injury compared to the DMSO-treated group. Oral administration of XL-3158 reduced the pancreas-to-body weight ratio (Fig. 8b). Histological examinations of pancreatic sections by Hematoxylin and Eosin (H&E) staining reveal that the model group exhibited substantial edema, inflammatory cell infiltration, and parenchymal necrosis. These pathological changes were remarkably ameliorated by the treatment of XL-3158 (Fig. 8c, d). Moreover, compared to the model group, XL-3158-treated mice showed significantly decreased serum amylase and lipase activities, as well as reduced levels of inflammatory cytokines TNF-α and IL-6 in serum (Fig. 8e–h). Collectively, these findings suggest that XL-3158 holds therapeutic potential for cerulein-induced AP.
a Schematic illustration of the experimental protocol. Mice were intraperitoneally injected with cerulein (50 μg/kg) or saline (Vehicle) at 1-h intervals for a total of 7 injections. XL-3158 (25 mg/kg) or vehicle was administered by oral gavage at 0 h and 5 h after the first cerulein injection. Mice were euthanized 12 h after the last cerulein injection. b Pancreas-to-body weight ratio of mice from vehicle (saline), cerulein, and cerulein + XL-3158 groups. c Histopathological scores based on five parameters: edema, inflammatory infiltration, fat necrosis, parenchymal necrosis, and hemorrhages from different groups. d Representative H&E staining of pancreatic sections from different groups. Black, red, and green arrows indicate edema, inflammatory infiltration, and parenchymal necrosis, respectively. Scale bar = 200 µm. Relative serum amylase (e) and lipase (f) activity in different groups. Serum levels of TNF-α (g) and IL-6 (h) in different groups of mice were measured by ELISA. All quantitative data (b, c, e–h) are shown as scatter plots with horizontal lines indicating mean ± SD. For b, c, e, f: n = 10 biological replicates (individual mice) for vehicle and XL-3158 groups; n = 9 for cerulein group; g, h: n = 6 biological replicates (individual mice per group). Statistical significance determined by an unpaired two-tailed t test. *p < 0.05, **p < 0.01, ***p < 0.005, ****p < 0.001. Source data is available with this manuscript as a Source Data file.
Discussion
The cGAS-STING pathway plays a central role in innate immunity, while aberrant activation of this pathway has been implicated in various autoimmune and inflammatory diseases. However, the lack of highly selective cGAS inhibitors with in vivo efficacy has hindered the exploration of the therapeutic potential targeting this pathway. In the present study, we introduce a strategy to inhibit cGAS by modulating its phase separation with DNA which is a striking feature of cGAS. A highly conserved allosteric site formed by residues nearby and at the activation loop of cGAS, a loop critical for DNA sensing and cGAS activation, has been identified. It is taken as an allosteric site because engagement of this site by our inhibitors stabilizes the inactive conformation of the activation loop, thereby preventing the conformational transition required for cGAS activation. Such a mechanism has been well-established in other targets, even though it may partially overlap with the substrate-binding region in the active site (e.g., DFG loop in kinase and sodium ion site in GPCRs44,45). Additionally, the cross-species conservation of this allosteric site not only confirms its evolutionary importance in cGAS function, but also crucially bridges preclinical findings to human therapeutic contexts through conserved pharmacodynamic responses.
Upon the discovery of the allosteric site, a series of inhibitors that occupy both the allosteric site and the orthosteric site have been designed, synthesized, and well characterized by X-ray crystallography and various biochemical assays. The comprehensive mechanistic studies have further revealed that these allosteric inhibitors work by locking the activation loop in a closed conformation, thereby attenuating cGAS-DNA interactions as well as the formation of cGAS-DNA condensates, and are thus termed PCIs. This mode of action distinguishes PCIs from traditional orthosteric inhibitors like G150 and provides an inhibitor that inhibits cGAS activity by modulating its phase separation. Interestingly, a recent study has reported that a class of cyclic peptide inhibitors can bind to cGAS with an open conformation of the activation loop, attenuating phase separation through competitive DNA binding19, while our PCIs employ a distinct mechanism to hinder DNA binding by stabilizing the closed conformation of this loop. These findings complementarily demonstrate the critical roles of the flexible activation loop in interfering with DNA binding and suggest multiple strategies for therapeutic intervention.
The compounds involved in this study include G150, RU.521, and our recently developed PCIs (i.e., XL series compounds represented by XL-3158), which exhibit distinctly different binding modes and inhibitory mechanisms. G150 binds potently to the less conserved orthosteric site, while RU.521 shows a species-dependent mode of action, forming a covalent bond with m-cGAS but noncovalent binding to either the orthosteric or the allosteric site of h-cGAS. By contrast, our PCIs engage both the orthosteric and allosteric sites and in particular retain effective interactions with the conserved allosteric site, resulting in broad cross-species inhibitory activity against cGAS. From a drug development perspective, the cross-species activity of the compounds like XL-3158 is particularly valuable as it enables reliable preclinical studies in animal models, potentially facilitating the translation of the preclinical findings into clinical trials. Furthermore, the distinct binding modes of these inhibitors correlate with their differential effects on cGAS activity and phase separation. While G150 primarily affects the enzymatic activity through orthosteric inhibition, our PCIs modulate both enzymatic activity and phase separation by stabilizing the closed conformation of the activation loop. In this regard, not only the typical enzymatic assay but also the DNA condensation assay have to be utilized to comprehensively assess the in vitro inhibitory activity of small-molecule inhibitors against cGAS. This dual mechanism of action, primarily facilitated by the engagement of the allosteric site, underscores the potential of our PCIs as more comprehensive and broadly applicable cGAS inhibitors, representing a significant advancement in cGAS inhibition strategies.
Our imaging studies reveal an intriguing property of PCIs: an ability to transform LSPS into LLPS by attenuating the overall phase separation of cGAS-DNA in vitro at the molecular level. This phenomenon is reminiscent of previous reports showing that increased salt concentrations weaken cGAS-DNA electrostatic interactions, leading to an LSPS to LLPS transition31. However, unlike the non-specific effects caused by NaCl, our subsequent site-directed mutagenesis experiments demonstrate that PCIs inhibit cGAS-DNA phase separation specifically through interacting with the allosteric site as well as the activation loop. Notably, despite the lower potency shown in biochemical assays compared to orthosteric inhibitors, XL-3158 exhibits comparable potency in cellular assays. We propose that by inhibiting LLPS and preventing condensation in cellular environments, PCIs can efficiently reduce condensed cGAS and achieve a high cellular inhibition efficiency. This finding underscores the therapeutic potential of targeting protein condensation and highlights the importance of evaluating inhibitors in cellular contexts. Importantly, the strategy of developing PCIs to enhance the cellular efficacy of inhibitors may apply to other proteins that undergo phase separation, opening up broader avenues for drug discovery and development. This study not only establishes a previously unrecognized mechanism for cGAS inhibition but also highlights the therapeutic potential of targeting phase separation dynamics for modulating protein functionality.
Methods
Ethics
All animal experiments and protocols were approved by the Animal Ethics Committee of the Shanghai Institute of Materia Medica (Approval No. [2020-0042]).
Commercial compounds
G150 and RU.521 were purchased from InvivoGen. H151 was obtained from TopScience. GSK8612 was acquired from Bidepharm. All compounds were dissolved in dimethyl sulfoxide (DMSO) and stored at −80 °C until use.
Chemical synthesis
Synthetic schemes, detailed procedures, and characterization of XL series compounds were provided in the Supplementary Information.
Expression and purification of cGAS proteins
The nucleic acid sequence of h-cGASFL (amino acids 1-522, Uniprot ID: Q8N884), h-cGASCD (amino acids 157-522), N ___domain of human cGAS (h-cGASN (amino acids 1-157)), and m-cGASCD (amino acids 147-507, Uniprot ID: Q8C6L5) were synthesized at GenScript. They were cloned into a pET-15b vector with 6×His-SUMO2 at the N-terminal respectively according to previously published methods14. Between the SUMO2 and the N-terminal of m-cGASCD, a serine residue was introduced as a linker to facilitate the removal of the 6×His-SUMO2 tag.
The recombinant fusion proteins were expressed in E. coli strain Rosetta 2 (DE3) cells (Sigma-Aldrich) or BL21 (DE3) pLysS (Beyotime Biotechnology). Cells were cultured in 2x YT media at 37 °C until reaching an OD600 of 0.8–1.0. After that, the culture temperature was reduced to 17 °C, and the protein expression was induced by the addition of 0.4 mM IPTG. Harvesting of cells was carried out 8–10 h after induction.
Cell pellets were disrupted using a high-pressure homogenizer in lysis buffer (20 mM HEPES, 400 mM NaCl, pH 7.5). Then the lysates were centrifugated at 47,850 × g for 30 min, and the supernatant was loaded on a manually packed Ni-NTA affinity column (GE Healthcare). The column was washed with a wash buffer (20 mM HEPES, 1 M NaCl, 50 mM imidazole, pH 7.5), and the recombinant protein was eluted with an elution buffer (20 mM HEPES, 400 mM NaCl, 300 mM imidazole, pH 7.5).
To remove the 6×His-SUMO2 tag, human SENP2 protease was added to the eluted protein and incubated at 4 °C for 4 h. Untagged cGAS was further purified using Heparin columns (GE Healthcare), followed by gel filtration with a size exclusion chromatography (HiLoad 16/600 Superdex 200 pg, GE Healthcare). The gel filtration buffer used for h-cGAS includes 20 mM HEPES, 250 mM KCl, 1 mM TCEP, pH 7.5, while the buffer used for m-cGAS includes 20 mM Tris, 300 mM NaCl, 1 mM DTT, pH 7.5. cGAS mutants were purified with the same protocol as wild-type (WT) cGAS.
Crystallization and structure determination
Crystals of h-cGASCD were obtained by the hanging-drop vapor diffusion method at 4 °C. The protein concentration used was 8–9 mg/mL, and the drops were set up with 1.5 μL of protein plus 1.5 μL of reservoir solution containing 17.5–19% (v/v) PEG 3350 and 0.2 M ammonium citrate (pH 7–7.5). To obtain crystals of h-cGASCD in complex with a compound, apo crystals were soaked overnight in the mother liquor supplemented with 10 mM of the compound. Crystals were flash-frozen in liquid nitrogen using the mother liquor supplemented with 20% glycerol as cryoprotectant.
Crystals of m-cGASCD were obtained by the hanging-drop vapor diffusion method at 4 °C. The protein concentration used was 7–12 mg/mL, and the drops were set up with 1.5 μL of protein plus 1.5 μL of reservoir solution containing 16–23% (v/v) PEG 3350 and 0.1 M HEPES (pH 7.5–7.8). To obtain crystals of m-cGASCD in complex with RU.521, apo crystals were soaked overnight in the mother liquor supplemented with 5 mM of RU.521. Crystals were flash-frozen in liquid nitrogen using the mother liquor supplemented with 20% glycerol as a cryoprotectant.
Diffraction data were collected at 100 K on the beamlines BL17U1, BL18U1, and BL19U1 at the Shanghai Synchrotron Radiation Facility (SSRF) and processed using the HKL3000 software package46. The structures were solved by molecular replacement using the program CCP4 (v7.0.078)47 with the structure of PDB IDs 4LEV for h-cGASCD and 4K8V for m-cGASCD as a search model48,49, respectively. Refinement was performed using PHENIX (v1.17.1–3660)50, and the program Coot (v0.8.9.2)51 was used to fit the compound and water molecules into the initial Fo-Fc map. Data collection and refinement statistics for five determined crystal structures are shown in Supplementary Table 1.
Analysis of RU.521 reactivity with cysteine
The reactivity of RU.521 with cysteine was investigated under various pH conditions and incubation times using HPLC analysis. RU.521 (a final concentration of 1 mM) was incubated with cysteine (a final concentration of 20 mM) in 500 mM Tris buffer at pH 5, 6, 7, 8, and 9. The reaction medium consisted of a 1:1 mixture of double-distilled water and methanol. Reactions were monitored at two time points: 5 min and 30 min. HPLC traces were collected and analyzed on an Agilent 1260 Infinity system. Chromatographic separation was performed using a GL Science C18 Superb column (5 μm, 4.6 mm × 250 mm). The mobile phase consisted of water containing 0.1% trifluoroacetic acid (TFA) as mobile phase A and acetonitrile containing 0.1% TFA as mobile phase B. The gradient elution was programmed as follows: 10% B to 100% B over 15 min, 100% B for 5 min, then 100% B to 10% B over 5 min. The flow rate was set at 1.0 mL/min, and the detection wavelength was 254 nm. The column temperature was maintained at 25 °C throughout the analysis.
Electrophoretic mobility shift assay (EMSA)
Cy5-labeled 45 bps DNA (ISD45) (0.05 μM) was incubated with h-cGASCD, h-cGASFL or h-cGASN at a gradient of concentrations (0.031–1 μM for h-cGASCD and h-cGASN, 0.016–0.5 μM for h-cGASFL) in 30 μL reaction buffer (20 mM Tris-HCl pH 7.5, 150 mM NaCl) with or without the compound. The incubation was carried out at room temperature for 20 min. After incubation, the reactions were mixed with 3x native DNA loading buffer (20 mM HEPES, pH 7.2, 150 mM NaCl, 30% sucrose, 0.01% bromophenol blue) and subsequently separated on a 1% agarose gel using 0.5x TBE buffer as a running buffer. The sequences of ISD45 were listed in the Supplementary Table 2. Gels were scanned with the Typhoon Fla-9500 imager (GE Healthcare) and visualized using ImageJ. Data are represented as means ± SD of three independent experiments.
DNA intercalation assay
dsDNA intercalation was assessed for compounds using a high-throughput fluorescence polarization (FP) assay following the protocol developed at the Broad Institute (National Center for Biotechnology Information. PubChem BioAssay Database; AID 504727, https://pubchem.ncbi.nlm.nih.gov/bioassay/504727). Acridine orange, a known DNA intercalator, was used as a fluorescent probe, while mitoxantrone served as a positive control. The assay was performed in a 384-well format with 30 μL reaction buffer per well. The buffer consists of 10 mM HEPES (pH 7.5), 1 mM EDTA, and 100 mM NaCl. To set up the assay, 6 μg/mL HT-DNA, 50 nM acridine orange, and 100 μM compound (final concentrations) were dispensed in 30 μL buffer per well in black opaque 384-well plates (Corning). Mitoxantrone at a final concentration of 10 μM was used as a positive control. The plates were incubated at room temperature in the dark for 20 min. Fluorescence polarization was measured using a plate reader (BioTek synergy 4) with a 480 nm excitation filter, 535 nm S and P emission filters, and an FP module. The FP values were directly calculated by the Gen5 software (3.33, BioTek). Data are represented as means ± SD of three independent experiments.
h-cGAS labeling
The purified protein was incubated overnight with Cy3 NHS ester (APExBIO) at a molar ratio of 1:8 at 4 °C. The labeled protein was then purified by gel filtration with size exclusion chromatography (HiLoad 16/600 Superdex 200 pg, GE Healthcare). The purified labeled protein was quantified by Bradford assay and frozen in liquid nitrogen. For imaging, the labeled protein was mixed with unlabeled protein at a ratio of 1:9.
In vitro phase separation assays and image collection at the molecular level
In vitro phase-separated droplet formation was analyzed by imaging reactions in a 384-well glass-bottomed plate (Cellvis). Phase separation was induced by mixing Cy3 labeled-protein and FAM labeled-DNA (ISD100) (Sangon Biotech) in buffer (20 mM Tris-HCl pH 7.5, 1 mg/mL BSA, 1 mM TCEP) with various salt concentrations at 25 °C in a total reaction volume of 30 μL. Each imaging experiment was performed using a molar ratio of 10% labeled protein or DNA. The details of incubation time and specific protein, DNA, and salt concentrations are indicated in each Figure legend and the corresponding Figure schematics. The sequences of ISD100 were listed in the Supplementary Table 2.
Microscopy images were acquired at 25 °C using a Leica TCS SP8 confocal microscope (Leica Application Suite X 4.5.0) with an oil immersion 63x/1.4 numerical aperture objective lens (HCX PL APO; Leica Microsystems). Fluorescence images of FAM and Cy3 were acquired with excitation at 488 nm (emission at 498–549 nm) and 552 nm (emission at 562–637 nm), respectively. The formation of phase separation was evaluated by measuring the integrated density (IntDen) of 488 channels per image using FIJI. Briefly, unprocessed fluorescence microscopy images were analyzed with FIJI (ImageJ 1.53t) using the following batch process: run(“16-bit”), setThreshold(10, 65535, “raw”), run(“Convert to Mask”), and run(“Measure”). Data are represented as means ± SD of three independent experiments.
For 3D reconstruction, confocal microscopy images were acquired with a z-interval of 0.5 µm, and a total of 40 images along the z-axis were recorded.
Fluorescence recovery after photobleaching (FRAP)
FRAP analysis was performed using fluorescently labeled components (Cy3-cGAS, FAM-ISD100), and microscopy images were collected using a confocal microscope (Leica TCS SP8; Leica Microsystems). For FRAP analysis of cGAS-DNA phase separation, photobleaching of the FAM signal in selected droplets was performed with 100% maximum laser power of a 488 nm laser. The recovery was immediately recorded at an interval of 1.5 s for ~150 s. The results were processed using the Image J plugin, briefly, by running “create spectrum jru”, “combine all trajectories jru”, “normalize trajectories jru”, “batch FRAP fit jru” and “average trajectories jru” to get the results. Data are represented as means ± SD of six independent experiments.
Time-lapse microscopy
For time-lapse microscopy, droplet formation was induced as indicated in Figure legends, and fluorescence images were taken at intervals of 30 s with an inverted fluorescence microscope (ZEN 3.7, Zeiss LSM900; ZEISS Microscopy). Fluorescence images of FAM and Cy3 were acquired with excitation at 493 nm (emission at 498–554 nm) and 553 nm (emission at 565–639 nm), respectively.
Turbidity measurements
Phase separation was induced by mixing protein and DNA (ISD100) with various salt concentrations the same as the “In vitro phase separation assays and image collection at the molecular level” part. The absorbance values were read by the nanodrop (Kaiao K5600) at 340 nm wavelength. The sequences of ISD100 were listed in the Supplementary Table 2. Data are represented as means ± SD of three independent experiments.
Sequence alignment of cGAS from different species
To analyze the sequence conservation of cGAS across different species, the amino acid sequences were obtained from the NCBI Gene database. The accession numbers of the sequences used in this study are as follows: Homo sapiens (human; Uniprot Q8N884), Rhesus macaque (Rhesus monkey; Uniprot A0A0F7J4C8), Mus musculus (mouse; Uniprot Q8C6L5), Gallus gallus (chicken; Uniprot A0A8V0X930), Xenopus tropicalis (xenopus; Uniprot A0A6I8R967), and Danio rerio (zebrafish; Uniprot F1QCP4)
Multiple sequence alignment was performed using CLUSTAL 2.152 with default parameters. The aligned sequences were then visualized using ESPript 3.053, which highlights the conserved residues and regions among the sequences.
DNA condensation assay with different cGAS constructs
Cy3-ISD100 was incubated with WT h-cGASCD, indicated mutants h-cGASCD, and WT h-cGASFL at a gradient of concentrations in 60 μL reaction buffer (20 mM Tris-HCl pH 7.5, 150 mM NaCl) with or without the compound at room temperature for 30 min. After incubation, mixtures were centrifuged at 13,000 × g for 10 min. 10 μL supernatant was transferred to a black 384 wells plate. The Cy3 fluorescence intensity of each supernatant was quantified on the Biotek synergy 4 plate reader using 540 nm excitation and 585 nm emission wavelengths. The EC50s were obtained by plotting the fluorescence intensity values of Cy3 in the supernatant against cGAS protein concentrations using GraphPad Prism 9. The sequences of ISD100 were listed in the Supplementary Table 2. Data are represented as means ± SD of three independent experiments.
Pyrophosphatase (PPiase)-coupled activity assay
The pyrophosphatase-coupled (PPiase-coupled) assay for monitoring the enzymatic activity of h-cGASFL or m-cGASCD in vitro was performed following the protocol used in our previous publication with some modifications15. The final concentrations of cGAS, E. coli pyrophosphatase, ATP, GTP, and HT-DNA in the assay were 200 nM, 50 nM, 0.2 mM, 0.2 mM, and 8 μg/mL, respectively, in 40 μL of reaction buffer (10 mM HEPES, 140 mM NaCl, 0.01% Tween-20, 5 mM MgCl2, pH 7.5). The experiments were performed in 384-well transparent polypropylene plates. After 90 min incubation at room temperature, the reaction was stopped by adding 40 μL of a solution containing 40 mM ethylenediaminetetraacetic acid mixed with an equal amount of malachite green-molybdate solution, followed by an additional 10 min incubation before measuring the absorbance at 620 nm. The inhibition ratio of the tested compound was calculated using the following formula:
Inhibition% = [1 − (Absinhibitor – Abswithout cGAS)/(AbsDMSO − Abswithout cGAS)] × 100%
Data are represented as means ± SD of three independent experiments.
Microscale thermophoresis (MST) binding assay
h-cGASCD were fluorescently labeled using an amine-reactive 2nd Generation NHS conjugation kit (MO-L011, NanoTemper) following manufacturer’s protocol. For MST measurements, serial dilutions of the compound were incubated with 100 nM labeled protein in assay buffer (20 mM HEPES, 250 mM KCl, 1 mM TCEP, pH 7.5) for 20 min at 25 °C. The reaction mixtures were loaded into capillaries (MO-K022, NanoTemper) and analyzed using a Monolith X instrument (NanoTemper) under standardized conditions: excitation power at 50% with medium infrared laser intensity. Thermophoretic traces were recorded and processed through MO. Control software (v2.6.6.24191) to calculate dissociation constants (Kd). Three independent experiments were performed.
Inhibitory activity measurement of XL series compounds on WT h-cGASCD-DNA condensates
h-cGASCD at a concentration of 300 nM was incubated with varying concentrations of inhibitor for 10 min. After the incubation period, 25 nM of Cy3-ISD100 was added to the reaction mixture and incubated for an additional 20 min. The reaction mixture was then centrifuged at 15682 × g for 5 min to separate protein-DNA complexes from unbound Cy3-DNA. The Cy3 fluorescence intensity (Cy3int) of each supernatant was quantified on the Biotek synergy 4 plate reader using 540 nm excitation and 585 nm emission wavelengths. The inhibition ratio of the tested compound was calculated using the following formula:
Inhibition% = (Cy3intinhibitor – Cy3intDMSO)/(Cy3intwithout cGAS – Cy3intDMSO) × 100%
Molecular docking and molecular dynamics (MD) simulations
Molecular docking was conducted utilizing Glide54 within the Schrödinger software suite, employing the crystal structure of h-cGASCD in complex with XL-3156 (PDB ID: 9J2W) as the receptor. The receptor structure was preprocessed using the Protein Preparation Wizard55, which facilitated the addition of hydrogen atoms and missing residue side chains. The OPLS3 force field56 was used for minimization of the overall structure. Three-dimensional (3D) conformations of XL-3158 were yielded using LigPrep. The docking grid was centered on the centroid of XL-3156 and generated using the Receptor Grid Generation to define the ligand-binding site. An exclude volume constraint was implemented at coordinates (x = 146.0, y = 1.2, z = 1.2, radius = 3.4) to restrict ligand sampling in undesired regions. During the subsequent docking calculations, the excluded volume penalties were configured to Large with standard precision (SP) settings. The highest-scoring docked pose was subsequently utilized as the initial model for the molecular dynamics (MD) simulation.
All-atom MD simulations were performed using AMBER 20 package57 to investigate the conformational changes of the activation loop. The crystal structures of h-cGASCD in complex with XL-3156 (PDB ID: 9J2W), G150 (PDB ID: 9J2Y), the apo structure of h-cGASCD (PDB ID: 4LEV), and the docked structure of h-cGASCD in complex with XL-3158 were used for setting up the model systems submitted to MD simulations.
The parameters of XL-3156, XL-3158, and G150 were calculated using the B3LYP method at the 6-31 G* level with Gaussian 09. The restricted electrostatic potential was generated by Antechamber58 and assigned the general AMBER force field59. For the protein, we mutated the selenomethionine to methionine (4LEV), fixed the missing side chains and loops in the Schrödinger suite (9J2W and 9J2Y), and used the ff14SB force field60. The parameters for the zinc ion and its four coordinating residues (H390, C396, C397, and C404) were built by MCPB.py61 and Gaussian 09 with the same method mentioned above. Next, the protein or protein-ligand complex was solvated with a 10 Å TIP3P water box and neutralized by adding counterions (Cl− or Na+). Detailed system parameters are provided in the Supplementary Table 4. Two successive energy minimizations were carried out using the steepest descent and conjugate gradient algorithm in the Sander module. The system was gradually heated from 0 to 100 and 100 to 300 K separately in the NVT ensemble over 5 and 100 ps, respectively, with a smaller restriction on protein and ligand. Then, the system was equilibrated at 300 K and 1 atm without any restriction. Periodic boundary conditions were utilized, and the SHAKE algorithm62 was applied to constrain the hydrogen atoms. The long-range electrostatic interactions were calculated using the particle mesh Ewald (PME) method63.
Finally, a 1 μs productive MD trajectory was generated for each model at a constant temperature and pressure using the PMEMD module64 implemented in AMBER 20. The coordinates of the protein or the protein-ligand complex in the MD trajectory were saved every 10 ps and used for the data analysis with the cpptraj module. The RMSF value of residues was calculated based on the trajectory of the last 400 ns. The change in electrostatic and van der Waals interaction energy between the ligand and h-cGASCD were calculated by the linear interaction energy (LIE) approach65.
Cell cultures
HEK-293 (ATCC) and RAW-ISG (InvivoGen) cells were cultured in DMEM media (GIBCO) supplemented with 10% fetal bovine serum (GIBCO) at 37 °C and 5% CO2. L929 (ATCC) cells and modified L929 were cultured in MEM media (GIBCO) supplemented with 10% fetal bovine serum (GIBCO) at 37 °C and 5% CO2. THP-1 Dual cells (InvivoGen) were cultured in RPMI 1640 media (GIBCO) supplemented with 10% heat-inactivated fetal bovine serum (GIBCO) at 37 °C and 5% CO2.
Lentivirus packaging
For lentivirus packaging, HEK-293 cells were co-transfected with the modified pLVX-Puro (Addgene), psPAX2 (Addgene), and pMD2.G (Addgene). 36 h post-transfection, the lentivirus-containing supernatant was harvested and filtered through a 0.45 μm polyvinylidene fluoride filter (AISIMO). The collected virus was then concentrated using a virus concentration kit (Beyotime Biotechnology) according to the manufacturer’s instructions and used for infection.
Cell lines generated for this study
To generate the m-cGAS KO L929 (L929-m-cGAS−/−) cell line, 1 × 105 L929 cells were transfected with 28 pM sgRNA (Genscript), 8.4 pM TrueCut Cas9 Protein (Invitrogen) with Lipofectamine CRISPRMAX Cas9 (Invitrogen) as manual in a 24-well plate. The sgRNAs used for qPCR in this study were listed in Supplementary Table 5. 72 h post-transfection, the cells were seeded into 96-well plates at a density of 0.5 cell/well. Two weeks later, monoclonal L929- m-cGAS−/− cells were selected via western blotting using an antibody against m-cGAS (Cell Signaling Technology).
To generate the doxycycline-inducible EGFP-h-cGAS of L929-cGAS−/−, L929-cGAS−/− cells were infected with the lentivirus encoding the corresponding protein with 5 μg/mL polybrene (Beyotime Biotechnology). After 24 h post-infection, 2–5 μg/mL puromycin (Beyotime Biotechnology) was added to select the resistant cells. After 5 days, the expression of EGFP-h-cGAS was induced by adding 1 μg/mL doxycycline hyclate (Beyotime Biotechnology) for 24 h.
Confocal microscopy and live-cell imaging
For imaging experiments, cells were seeded onto glass-bottom confocal dishes (Cellvis) pre-coated with poly-D-lysine (Sigma) at a density of 1 × 105 cells per dish and were induced with 1 μg/mL doxycycline. After 24 h, Cy5-labeled ISD100 (Sangon Biotech) was transfected into the cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Two hours post-transfection, cells were washed three times with PBS and incubated in phenol red-free DMEM complete medium containing 10% FBS for imaging. The sequences of ISD100 were listed in the Supplementary Table 2.
Confocal imaging was performed using an inverted fluorescence microscope (Zeiss LSM900; ZEISS Microscopy) with two channels: Cy5 for DNA (excitation/emission wavelengths: 649/666 nm) and GFP for cGAS (excitation/emission wavelengths: 488/510 nm). Images were acquired using a 63×/1.4 NA oil immersion objective.
For compound-treatment groups, cells were incubated with the specified concentration of the compound for 2 h before transfection.
Luciferase reporter assay for THP1-Dual and RAW-ISG cells
THP1-Lucia ISG cells (InvivoGen) or RAW-ISG cells (InvivoGen) were seeded into a 24-well plate at a density of 5 × 105 cells per well and incubated with compounds for the indicated time. The cells were then transfected with HT-DNA using Lipofectamine 2000 (Invitrogen) at the indicated titer and incubated for 24 h. The supernatant was collected, and 20 μL of the supernatant from each well was mixed with 50 μL of QUANTI-Luc Luciferase reagent (InvivoGen). Luciferase luminescence was immediately detected using a microplate reader (BioTek Synergy H1) with an integration time of 1 second per well. Data are represented as means ± SD of three independent experiments.
Cell viability assay
Cell viability was assessed using the Cell Counting Kit-8 (CCK-8) assay (YEASEN Biotechnology) according to the manufacturer’s instructions. Briefly, cells were seeded into 96-well plates at a density of 1 × 105 cells per well and treated with the indicated compound for 24 h. Following treatment, 10 μL of CCK-8 solution was added to each well, and the plates were incubated at 37 °C for 1–4 h. The absorbance at 450 nm was measured using a microplate reader (BioTek Synergy H1). Cell viability was calculated as a percentage relative to the untreated control cells. Data are represented as means ± SD of three independent experiments.
Synergistic Effect of XL-3158 and G150 in THP-1 Dual Cells
The synergistic effect of XL-3158 and G150 was evaluated using a luciferase reporter assay in THP-1 Dual cells. XL-3158 and G150 were combined at molar ratios of 10:1, 5:1, 2.5:1, 1:1, 1:2.5, 1:5, and 1:10, with a final concentration of 20 μM in total. The combined compounds were then serially diluted in a 2-fold manner to obtain five concentrations. As controls, XL-3158 and G150 were individually prepared at the same concentrations as the combined treatments.
THP-1 Dual cells were seeded, treated with compounds, and transfected with HT-DNA as described above. Luciferase activity was measured according to the same protocol.
The combination index (CI) values at effective doses of 90%, 75%, and 50% (ED90, ED75, and ED50) were calculated using CompuSyn software (1.0, ComboSyn, Inc., Paramus, NJ, USA). CI values < 1, =1, and >1 indicate synergistic, additive, and antagonistic effects, respectively. Data are presented as means ± SD from three independent experiments.
Real-time quantitative PCR
THP-1 Dual and RAW-ISG cells were seeded into 12-well plates 1 day prior to the experiment. THP-1 Dual cells were induced with PMA for 24 h. The cells were then stimulated with either HT-DNA (Sigma), 2'3’-cGAMP (MedChemExpress), or Poly(I:C) (MedChemExpress) after treatment with XL-3158, G150, H151, or GSK8612. Total RNA was isolated from the cells incubated with the compound for 4 h using TRIeasy LS Total RNA Extraction Reagent (YEASEN Biotechnology) according to the manufacturer’s instructions. mRNA was reverse transcribed using Hifair III 1st Strand cDNA Synthesis SuperMix (YEASEN Biotechnology). Real-time PCR was performed using Hieff UNICON Universal Blue qPCR SYBR Green Master Mix (YEASEN Biotechnology) on the QuantGene 9600 (Bioer Technology). Samples were run in triplicate, and the target CT values were normalized to GAPDH CT values. Relative quantification results were directly exported from the GENE-9660 software (1.0.13, Bioer Technology). The primers used for qPCR in this study are listed in Supplementary Table 6. Data are presented as means ± SD from three independent experiments.
Antibodies
Rabbit anti-TBK1 (CST, 3504; 1:1000), rabbit anti-phospho-TBK1 (CST, 5483; 1:1000), rabbit anti-IRF3 (CST, 4302; 1:1000), rabbit anti-phospho-IRF3 (CST, 4947; 1:1000), Mouse anti-GAPDH (Beyotime, AF0006; 1:1000). Anti-rabbit IgG, HRP-linked Antibody (Beyotime, A0208; 1:10000), Anti-mouse IgG, HRP-linked Antibody (Beyotime, A0216; 1:10000).
Immunoblotting (Western blot)
Total cell lysates were prepared in RIPA lysis buffer (Beyotime Biotechnology) supplemented with 1x protease inhibitor cocktail (MedChemExpress) and 1x phosphatase inhibitor cocktail (Beyotime Biotechnology) according to the manufacturer’s instructions. The samples were separated by SDS-PAGE and transferred to PVDF membranes (Bio-Rad). The membranes were incubated with the indicated primary antibodies at 4 °C overnight, followed by incubation with HRP-conjugated anti-rabbit IgG antibody (Beyotime Biotechnology) or anti-mouse IgG antibody (Beyotime Biotechnology) at room temperature for 1 h. The signal was visualized using the Tanon ECL Western Blotting Substrate (Tanon Biotech) and detected using the Tanon 4600 imaging system (Tanon Biotech). Data are presented as means ± SD from three independent experiments.
In vivo PK study of XL-3158 in ICR mouse
Male ICR mice (18–22 g, n = 9) were housed under controlled environmental conditions (temperature 18–29 °C, relative humidity 30–70%) with a 12-h light/dark cycle. Sex was not considered as a variable in this study; gender selection followed established protocols from literature to align with standard practices for pharmacokinetic studies. Animals were maintained on standard rodent diet with free access to water. Prior to drug administration, mice were fasted for 12 h and remained fasting for 2 h post-administration. XL-3158 was administered via oral gavage (50 mg/kg in DMSO/0.5% HPMC, 5/95, v/v) at 10 mL/kg, intraperitoneal injection (10 mg/kg in DMSO/EtOH/PEG300/0.9% NaCl, 5/5/40/50, v/v/v/v) at 5 mL/kg, or intravenous injection (5 mg/kg in DMSO/EtOH/PEG300/0.9% NaCl, 5/5/40/50, v/v/v/v) at 5 mL/kg.
Induction of acute pancreatitis and assessment of pancreatic severity
Female C57BL/6J mice (6–8 weeks old) were used to generate an AP model. Sex was not considered as a variable in this study; gender selection followed established protocols from literature to align with standard practices for cerulein-induced pancreatitis models43. Mice were fasted for 12–24 h, then AP was induced by intraperitoneal (i.p.) injection of cerulein (50 µg/kg) administered seven times at 1-h intervals. XL-3158 (25 mg/kg) was administered via oral gavage at two time points: concurrently with the first and sixth cerulein injections. Twelve hours after the last cerulein injection, blood and pancreatic tissue samples were collected. Serum was isolated from whole blood by centrifugation at 3000 × g for 15 min at 4 °C and stored at −80 °C. Pancreatic tissues were fixed in 4% paraformaldehyde for histological examination.
Serum amylase activity
Serum amylase activity was measured according to previously published methods66. Briefly, the reaction was performed in a buffer containing 50 mM MES (pH 6.1), 300 mM NaCl, 5 mM CaCl2, and 450 mM KSCN, with 2.25 mM 2-chloro-4-nitrophenyl-α-d-maltotrioside (CNP-G3) (Sigma-Aldrich) as substrate. The reaction mixture (200 µL) containing 2 µL serum per well was monitored at 405 nm for 30 min in kinetic mode. The average amylase activity for each group was calculated based on the absorbance changes.
Serum lipase activity
Serum lipase activity was measured according to previously published methods67. The assay system consisted of two buffers: Buffer 1 containing 41 mM Tris-HCl (pH 8.4), 1 µg/mL colipase, 1.8 mM sodium deoxycholate, and 7.2 mM sodium taurodeoxycholate; Buffer 2 containing 1.6 mM tartaric acid (pH 4.0), 0.24 mM 1,2-o-dilauryl-rac-glycero-3-glutaric acid-(6’-methylresorufin) ester (DGGR) (Sigma-Aldrich), and 0.1 mM CaCl2. For the assay, 2 µL of serum was mixed with 180 µL of Buffer 1 and 18 µL of Buffer 2. The reaction was monitored at 580 nm for 30 min in kinetic mode. The average lipase activity for each group was calculated based on the absorbance changes.
H&E staining
Tissue specimens were fixed in 4% paraformaldehyde, dehydrated, and embedded in paraffin. Sections (5 µm thickness) were cut and stained with hematoxylin and eosin (H&E) according to standard protocols. The stained sections were examined under a light microscope. Histological images were captured using a Nikon Eclipse Ci-L light microscope (Nikon). Histopathological scoring was performed based on five parameters: edema, inflammatory infiltration, fat necrosis, parenchymal necrosis, and hemorrhages.
ELISA analysis of serum cytokines
Serum levels of TNF-α and IL-6 were determined using commercial enzyme-linked immunosorbent assay (ELISA) kits (TNF-α, MultiSciences, Cat. #EK282EGA; IL-6, MultiSciences, Cat. #EK206HS) according to the manufacturer’s instructions. For each experimental group, six mice were selected using a random number generator and assigned individual identification numbers. Serum samples were thawed on ice and diluted according to the kit instructions. All samples were analyzed in triplicate. Standard curves were generated using the recombinant cytokine standards provided with the kits. The optical density was measured at 450 nm with wavelength correction at 570 nm using a microplate reader (specify model if desired). Cytokine concentrations were calculated using the standard curves, and final results were expressed as pg/mL.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The sequence of h-cGASFL used in this study is available in the uniprot under Uniprot ID: Q8N884. The sequence of m-cGASCD used in this study is available in the uniprot under Uniprot ID: Q8C6L5 [https://www.uniprot.org/uniprotkb/ Q8C6L5/entry]. The sequence of m-cGASCD used in this study is available in the uniprot under Uniprot ID: Q8C6L5 [https://www.uniprot.org/uniprotkb/ Q8C6L5/entry]. Crystallographic data for the structures reported in this study have been deposited at the Cambridge Crystallographic Data Centre, under deposition numbers CCDC 2442513 (XL-3156). Copies of the data can be obtained free of charge via https://www.ccdc.cam.ac.uk/structures/ (https://doi.org/10.5517/ccdc.csd.cc2mzmr0). The full CheckCif validation report (including absence of A-/B-level alerts) is provided as a supplementary file (CheckCif_Report_CCDC_2442513.pdf). Further details on structural analysis and atomic coordinates are included in the Supplementary Information. The atomic coordinates and structure factors generated in this study have been deposited into the Protein Data Bank under accession codes 9J2X (h-cGASCD in complex with RU.521), 9J2W (h-cGASCD in complex with XL-3156), 9J2Y (h-cGASCD in complex with G150), 9LIO (h-cGASCD in complex with XL-3123), 9J2Z and (m-cGASCD in complex with RU.521). The Molecular Dynamics Simulations checklist is incorporated into the Supplementary Information as a Summary Table 8. The MD simulation data used in this study are available in the Zenodo database under accession code 15307131. Source data are provided with this paper as a Source Data file. Source data are provided with this paper.
References
Grivennikov, S. I., Greten, F. R. & Karin, M. Immunity, inflammation, and cancer. Cell 140, 883–899 (2010).
Sun, L., Wu, J., Du, F., Chen, X. & Chen, Z. J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791 (2013).
Wu, J. et al. Cyclic GMP-AMP Is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339, 826–830 (2013).
Crow, Y. J. & Manel, N. Aicardi-Goutières syndrome and the type I interferonopathies. Nat. Rev. Immunol. 15, 429–440 (2015).
Gao, D. et al. Activation of cyclic GMP-AMP synthase by self-DNA causes autoimmune diseases. Proc. Natl. Acad. Sci. USA 112, E5699–E5705 (2015).
Gulen, M. F. et al. cGAS–STING drives ageing-related inflammation and neurodegeneration. Nature 620, 374–380 (2023).
Domizio, J. D. et al. The cGAS-STING pathway drives type I IFN immunopathology in COVID-19. Nature 603, 145–151 (2022).
Hu, M.-M. & Shu, H.-B. Innate immune response to cytoplasmic DNA: mechanisms and diseases. Annu. Rev. Immunol. 38, 79–98 (2020).
Mullard, A. Biotechs step on cGAS for autoimmune diseases. Nat. Rev. Drug Discov. 22, 939–941 (2023).
Lama, L. et al. Development of human cGAS-specific small-molecule inhibitors for repression of dsDNA-triggered interferon expression. Nat. Commun. 10, 2261 (2019).
Tan, J. et al. Synthesis and pharmacological evaluation of tetrahydro-γ-carboline derivatives as potent anti-inflammatory agents targeting cyclic GMP–AMP synthase. J. Med. Chem. 64, 7667–7690 (2021).
Chen, M. et al. Design, synthesis, and pharmacological evaluation of spiro[carbazole-3,3′-pyrrolidine] derivatives as cGAS inhibitors for treatment of acute lung injury. J. Med. Chem. 67, 6268–6291 (2024).
Hall, J. et al. Discovery of PF-06928215 as a high affinity inhibitor of cGAS enabled by a novel fluorescence polarization assay. PLoS ONE 12, e0184843 (2017).
Zhao, W. et al. In silico screening-based discovery of novel inhibitors of human cyclic GMP–AMP synthase: a cross-validation study of molecular docking and experimental testing. J. Chem. Inf. Model. 60, 3265–3276 (2020).
Li, J. et al. Discovery of novel cGAS inhibitors based on natural flavonoids. Bioorg. Chem. 140, 106802 (2023).
Song, J. et al. Discovery and characterization of a novel cGAS covalent inhibitor for the treatment of inflammatory bowel disease. Acta Pharmacol. Sin. 44, 791–800 (2023).
Vincent, J. et al. Small molecule inhibition of cGAS reduces interferon expression in primary macrophages from autoimmune mice. Nat. Commun. 8, 750 (2017).
Wiser, C., Kim, B., Vincent, J. & Ascano, M. Small molecule inhibition of human cGAS reduces total cGAMP output and cytokine expression in cells. Sci. Rep. 10, 7604 (2020).
Wang, X. et al. Development of cyclopeptide inhibitors of cGAS targeting protein-DNA interaction and phase separation. Nat. Commun. 14, 6132 (2023).
An, J. et al. Inhibition of Cyclic GMP-AMP synthase using a novel antimalarial drug derivative in Trex1-deficient mice. Arthritis Rheumatol. 70, 1807–1819 (2018).
Wang, M., Sooreshjani, M. A., Mikek, C., Opoku-Temeng, C. & Sintim, H. O. Suramin potently inhibits cGAMP synthase, cGAS, in THP1 cells to modulate IFN-β levels. Future Med. Chem. 10, 1301–1317 (2018).
Padilla-Salinas, R. et al. Discovery of small-molecule cyclic GMP-AMP synthase inhibitors. J. Org. Chem. 85, 1579–1600 (2020).
Lai, J. et al. Compound C reducing interferon expression by inhibiting cGAMP accumulation. Front. Pharmacol. 11, 88 (2020).
Dai, J. et al. Acetylation blocks cGAS activity and inhibits self-DNA-induced autoimmunity. Cell 176, 1447–1460.e14 (2019).
Banani, S. F., Lee, H. O., Hyman, A. A. & Rosen, M. K. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298 (2017).
Patel, A. et al. Principles and functions of condensate modifying drugs. Front. Mol. Biosci. 9, 1007744 (2022).
Du, M. & Chen, Z. J. DNA-induced liquid phase condensation of cGAS activates innate immune signaling. Science 361, 704–709 (2018).
Xie, W. et al. Human cGAS catalytic ___domain has an additional DNA-binding interface that enhances enzymatic activity and liquid-phase condensation. Proc. Natl. Acad. Sci. USA 116, 11946–11955 (2019).
Yao, Y., Wang, W. & Chen, C. Mechanisms of phase-separation-mediated cGAS activation revealed by dcFCCS. PNAS Nexus 1, pgac109 (2022).
Zhou, W. et al. Structure of the human cGAS–DNA complex reveals enhanced control of immune surveillance. Cell 174, 300–311.e11 (2018).
Zhou, W., Mohr, L., Maciejowski, J. & Kranzusch, P. J. cGAS phase separation inhibits TREX1-mediated DNA degradation and enhances cytosolic DNA sensing. Mol. Cell 81, 739–755.e7 (2021).
Chen, S., Rong, M., Lv, Y., Zhu, D. & Xiang, Y. Regulation of cGAS activity by RNA-modulated phase separation. EMBO Rep. 24, e51800 (2023).
Bhowmik, D. et al. Cooperative DNA binding mediated by KicGAS/ORF52 oligomerization allows inhibition of DNA-induced phase separation and activation of cGAS. Nucleic Acids Res. 49, 9389–9403 (2021).
Xu, G. et al. Viral tegument proteins restrict cGAS-DNA phase separation to mediate immune evasion. Mol. Cell 81, 2823–2837.e9 (2021).
Zhao, M. et al. The stress granule protein G3BP1 promotes pre-condensation of cGAS to allow rapid responses to DNA. EMBO Rep. 23, e53166 (2021).
Shi, C. et al. USP15 promotes cGAS activation through deubiquitylation and liquid condensation. Nucleic Acids Res. 50, 11093–11108 (2022).
Tao, X. et al. Ku proteins promote DNA binding and condensation of cyclic GMP-AMP synthase. Cell Rep. 40, 111310 (2022).
Wang, L. et al. Spermine enhances antiviral and anticancer responses by stabilizing DNA binding with the DNA sensor cGAS. Immunity 56, 272–288.e7 (2023).
Zhao, C. et al. Polyamine metabolism controls B-to-Z DNA transition to orchestrate DNA sensor cGAS activity. Immunity 56, 2508–2522.e6 (2023).
Wang, L. et al. Oleic acid dissolves cGAS–DNA phase separation to inhibit immune surveillance. Adv. Sci. 10, 2206820 (2023).
Zhang, X. et al. The cytosolic DNA sensor cGAS forms an oligomeric complex with DNA and undergoes switch-like conformational changes in the activation loop. Cell Rep. 6, 421–430 (2014).
Meanwell, N. A. Fluorine and fluorinated motifs in the design and application of bioisosteres for drug design. J. Med. Chem. 61, 5822–5880 (2018).
Zhao, Q., Wei, Y., Pandol, S. J., Li, L. & Habtezion, A. STING signaling promotes inflammation in experimental acute. Pancreat. Gastroenterol. 154, 1822–1835.e2 (2018).
Roskoski, R. Classification of small molecule protein kinase inhibitors based upon the structures of their drug-enzyme complexes. Pharmacol. Res. 103, 26–48 (2016).
Changeux, J.-P. & Christopoulos, A. Allosteric modulation as a unifying mechanism for receptor function and regulation. Cell 166, 1084–1102 (2016).
Minor, W., Cymborowski, M., Otwinowski, Z. & Chruszcz, M. HKL-3000: the integration of data reduction and structure solution-from diffraction images to an initial model in minutes. Acta Crystallogr. D. Biol. Crystallogr. 62, 859–866 (2006).
Collaborative Computational Project, Number 4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994).
Li, X. et al. Cyclic GMP-AMP synthase is activated by double-stranded DNA-induced oligomerization. Immunity 39, 1019–1031 (2013).
Gao, P. et al. Cyclic [G(2′,5′)pA(3′,5′)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell 153, 1094–1107 (2013).
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D. Biol. Crystallogr. 58, 1948–1954 (2002).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 (2004).
Larkin, M. A. et al. Clustal W and clustal X version 2.0. Bioinformatics23, 2947–2948 (2007).
Robert, X. & Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 (2014).
Halgren, T. A. et al. Glide: A new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J. Med. Chem. 47, 1750–1759 (2004).
Sastry, G. M., Adzhigirey, M., Day, T., Annabhimoju, R. & Sherman, W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 27, 221–234 (2013).
Harder, E. et al. OPLS3: A force field providing broad coverage of drug-like small molecules and proteins. J. Chem. Theory Comput 12, 281–296 (2016).
Case, D. A. et al. AmberTools. J. Chem. Inf. Model. 63, 6183–6191 (2023).
Wang, J., Wang, W., Kollman, P. A. & Case, D. A. Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph. Model. 25, 247–260 (2006).
Wang, J., Wolf, R. M., Caldwell, J. W., Kollman, P. A. & Case, D. A. Development and testing of a general amber force field. J. Comput. Chem. 25, 1157–1174 (2004).
Maier, J. A. et al. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 11, 3696–3713 (2015).
Li, P. & Merz, K. M. Jr. MCPB.py: a Python based metal center parameter builder. J. Chem. Inf. Model. 56, 599–604 (2016).
Lippert, R. A. et al. A common, avoidable source of error in molecular dynamics integrators. J. Chem. Phys. 126, 046101 (2007).
Cerutti, D. S., Duke, R. E., Darden, T. A. & Lybrand, T. P. Staggered mesh Ewald: an extension of the smooth particle-mesh Ewald method adding great versatility. J. Chem. Theory Comput. 5, 2322–2338 (2009).
Salomon-Ferrer, R. et al. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh Ewald. J. Chem. Theory Comput. 9, 3878–3888 (2013).
Åqvist, J., Luzhkov, V. B. & Brandsdal, B. O. Ligand binding affinities from MD simulations. Acc. Chem. Res. 35, 358–365 (2002).
Gella, F. J., Gubern, G., Vidal, R. & Canalias, F. Determination of total and pancreatic alpha-amylase in human serum with 2-chloro-4-nitrophenyl-alpha-D-maltotrioside as substrate. Clin. Chim. Acta Int. J. Clin. Chem. 259, 147–160 (1997).
Panteghini, M., Bonora, R. & Pagani, F. Measurement of pancreatic lipase activity in serum by a kinetic colorimetric assay using a new chromogenic substrate. Ann. Clin. Biochem. Int. J. Lab. Med. 38, 365–370 (2001).
Acknowledgements
We thank the Institutional Technology Service Center of Shanghai Institute of Materia Medica for providing the instrumental platform, Ms. Haixin Huang for guidance on laser confocal fluorescence microscopy imaging, and the staff from beamlines BL02U1, BL10U2, and BL18U1 at Shanghai Synchrotron Radiation Facility for assistance in data collection. This work was supported by the National Key R&D Program of China (No. 2023YFF1205104) to Y.X., the Strategic Priority Research Program of the Chinese Academy of Sciences (No. XDB0830000) to Y.X., and Research Program of Shanghai Institute of Materia Medica, Chinese Academy of Sciences (No. SIMM0320231006) to Y.X.
Author information
Authors and Affiliations
Contributions
Y.X. and W.Z. conceived the study and designed experiments. G.C., W.Z., and J.H. performed synthesis experiments. W.Z., X.S., H.D., M.L., and J.L. conducted biological assays and crystallography experiments. M.X. performed molecular dynamics simulations. W.Z., X.S., L.X., Z.Q., X.H., and W.L. executed animal studies. X.S., H.H., H.S., Q.S., and H.S. assisted with data analysis. Y.X. supervised project strategy, methodology, and funding acquisition. Y.X. and W.Z. drafted the manuscript with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Junmin Quan, Yinghao Wu, and the other, anonymous, reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhao, W., Chen, G., He, J. et al. De novo design of protein condensation inhibitors by targeting an allosteric site of cGAS. Nat Commun 16, 5140 (2025). https://doi.org/10.1038/s41467-025-60297-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-60297-0