Abstract
The ureter is the outlet for urine produced by the kidney. Recent advances in stem cell biology have enabled the in vitro generation of kidney organoids from pluripotent stem cells (PSCs), but they lack the ureter, which hinders the smooth drainage of urine. By mimicking the in vivo developmental process of ureteral stromal progenitors (SPs) from the posterior intermediate mesoderm, we report here in vitro induction protocols for ureteral SPs from mouse and human PSCs. When the induced SPs were combined with ureteral epithelia derived from mouse embryos, the elongated, multilayered ureteral structure was reconstituted, exhibiting peristaltic constriction. Some of the pathological features associated with the loss of Tbx18/TBX18 in mice and humans can be modeled using induced SPs lacking functional TBX18. Furthermore, the ureter-like spherical organoids entirely derived from mouse and human PSCs can also be generated by combining the induced SPs and the epithelial progenitor, the ureteric bud. Therefore, our induction protocols for ureteral SPs will be useful for the elucidation of ureteral development and diseases, and will be an important step towards functional kidney organoids with urine flow.
Similar content being viewed by others
Introduction
The kidney is a vital organ in the human body responsible for maintaining homeostasis. It fulfills a variety of functions, including the regulation of water volume and ion balance, the excretion of waste products, hematopoiesis, and bone metabolism. For the kidney to perform its function, it requires not only the kidney itself, but also the ureter, which is the outlet for the urine produced by the kidney. Recent advances in stem cell biology have enabled the generation of kidney organoids from pluripotent stem cells (PSCs)1,2,3,4,5,6,7,8,9,10,11,12. However, no kidney organoids with ureters or even ureteral organoids have been generated, which is one of the hurdles to overcome for the future application of organoids in transplant treatment.
The ureter is a straight tube that allows efficient transport of urine from the kidney to the bladder by unidirectional peristalsis. The ureter is composed of two distinct tissue compartments: the ureteral epithelium and the surrounding ureteral mesenchyme. The ureteral epithelium, also known as the urothelium, is a specialized epithelium that is highly extensible and forms a tight seal around the lumen. It is further divided into three cell layers, from the inside to the outside: superficial cells, intermediate cells, and basal cells13,14,15 (Supplementary Fig. 1a). The ureteral mesenchyme also consists of three cell layers: the inner lamina propria, the tunica muscularis with contractile smooth muscle cells (SMCs), and the outer tunica adventitia that anchors the ureter to the body wall16,17 (Supplementary Fig. 1a). Given that the ureter develops as a result of the interaction between the epithelium and the surrounding mesenchyme16,18, a logical approach to generate the ureter from PSCs would be to induce the progenitors of these tissues and combine them to elicit the interactions necessary for ureter development. While induction protocols for the progenitor of the ureteral epithelium, ureteric buds (UBs), have been established6,7,8,9,11,12, the progenitor of the ureteral mesenchyme has yet to be induced from PSCs. Stromal progenitors (SPs) of the mouse ureter surround the distal UB stalk at embryonic day (E) 11.5 and around the ureter from E12.5 onwards, and differentiate through interactions with the adjacent ureteral epithelia19,20. In the E11.5 kidney, SPs of the ureter, which are marked by Tbx18 expression, are already segregated from those of the kidney, which express Foxd110,20. Since the SP of the ureter is located in a ventral position with respect to that of the kidney, the SP of the ureter is referred to as the ventral SP, in contrast to the SP of the kidney, which is referred to as the dorsal SP (Supplementary Fig. 1b).
In this work, we establish induction protocols for ventral SPs from PSCs using the reverse induction approach, which has been previously established in studies of nephron progenitor (NP), UB, and dorsal SP inductions1,6,10. Initially, we identified the optimal conditions for the induction of ventral SPs from the posterior intermediate mesoderm (IM) in E9.5 mouse embryos. Subsequently, the same conditions were applied to establish induction protocols for ventral SPs from mouse and human PSCs via posterior IM. The induced ventral SPs were then combined with mouse embryonic ureteral epithelia to reconstitute the elongated, multilayered structures of the ureter with peristaltic constrictions. Some of the pathological features associated with the loss of Tbx18/TBX18 in mice and humans can be modeled using induced SPs lacking functional TBX18. Furthermore, the ureter-like spherical organoids derived entirely from mouse and human PSCs were generated by combining the induced ventral SPs and the induced UBs.
Results
Induction of ventral SPs from E9.5 posterior IM
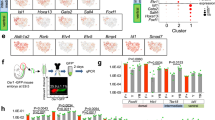
We first performed single-cell RNA sequencing (scRNA-seq) of in vivo mouse embryonic kidneys with ureters (E11.5, 13.5, and 15.5). The resulting merged UMAP plots showed multiple clusters for different cell types in the kidney, including NPs (Six2+), stroma (Pdgfra+), UBs (tip; Ret+, stalk; Shh+), and endothelial cells (ECs) (Pecam1+) (Supplementary Fig. 1c). To further characterize the stromal cells, we extracted them to identify representative markers of ventral SPs. The stromal cells were divided into multiple clusters, including ventral SPs (cluster 6), dorsal SPs (clusters 3, 4, 5, 10), cortical stromal cells (cluster 2), medullary stromal cells (cluster 7), differentiating cells (clusters 9, 11), and Top2a+ proliferating cells (clusters 0, 1, 8) (Fig. 1a). We here selected cluster 4 as the dorsal SP cluster because of the high expression of Ntn1, which is known to be restricted to the dorsal side of the renal stroma21,22 (Supplementary Fig. 1d). We then selected several representative markers in the ventral SPs compared to the dorsal SPs (Fig. 1b, Supplementary Fig 1d, e, Supplementary Data 1). These include not only known genes such as Tbx1819,20, Gata223, and Foxf124, but also unknown genes such as Plcxd3, ligp1, Sntg1, Col9a2, and Syt1. Notably, we found that Itga8 is strongly expressed in the population of ventral SP (Fig. 1b, Supplementary Fig. 1d). We also found that genes associated with BMP signaling activity (e.g., Bmp4, Smad7)25 were more highly expressed in ventral SPs, while those associated with retinoic acid (RA) signaling activity (e.g. Rarb)26 were widely expressed (Fig. 1c), which is consistent with previous report of BMP4-dependent dorsoventral patterning of the renal stroma10,27.
a–c scRNA-seq analysis of the stromal cells of in vivo mouse embryonic kidneys (E11.5, 13.5 and 15.5). a UMAP plots showing twelve clusters, with three representing Top2a+ proliferating cells. Cluster 6 is indicated as the ventral SP cluster. b Representative genes in ventral and dorsal SPs shown as dot plots. c Dot plots showing RA and BMP signal-related genes. d The origin of ventral SPs, shown by immunostaining of the kidney at E11.5. The expression of TBX18 (green) and tomato (derived from E9.5 Osr1+ cells) overlapped surrounding the UB stalk. NPs and UBs are indicated by blue and red arrows, respectively. The experiments were repeated twice with similar results. Scale bars: 50 μm. e Schematic diagram of isolation of posterior IM cells from E9.5 wild-type mouse embryos, followed by the 3 days culture and sorting the PDGFRA+ stromal cells for qPCR analysis. f Expression of ventral and dorsal SP genes after the culture, analyzed by qPCR (n = 3 biologically independent experiments). BMP4 upregulates ventral SP genes in a dose-dependent manner, whereas downregulates dorsal SP genes. Relative mRNA expression levels normalized to β-actin gene expression are shown as mean ± SD. Dunnett’s multiple comparison test (two-sided) was performed. The source data are provided as a Source Data file. B0: BMP4 (0 ng/ml); B1: BMP4 (1 ng/ml); B10: BMP4 (10 ng/ml); Ref: Tbx18-GFP+/PDGFRA+ ventral SPs harvested at E13.5 (non-cultured; presented as a reference). g Flow cytometric analysis before (upper) and after (lower) the culture in the ventral SP induction from the E9.5 posterior IM. The source data are provided as a Source Data file.
We next performed lineage tracing analysis to determine the origin of the ureteral (i.e. ventral) SPs. Osr1 is a transcription factor that is expressed in the posterior IM at E9.51,28. Upon administration of tamoxifen to Osr1CreER; R26tdTomato mice at E9.5, TBX18+ ventral SPs surrounding the UB stalk at E11.5 were positive for tdTomato, indicating that ventral SPs are derived from the posterior IM at E9.5 (Fig. 1d). NPs, but not UBs, were positive for tdTomato (Fig. 1d), consistent with our previous reports1,29.
Since ventral SPs are derived from the posterior IM (Fig. 1d), we sought an optimal condition to induce ventral SPs from the posterior IM. As we previously reported10, we isolated the posterior IM as a ROBO2+/PDGFRA+ fraction from E9.5 wild-type mice, cultured for three days, and sorted the PDGFRA+ stromal cells for quantitative PCR (qPCR) analysis (Fig. 1e). We have previously demonstrated that inhibition of BMP signaling, together with addition of RA and FGF9 during the culture, induces dorsal SPs10. Thus, to induce ventral SPs, we tested the effects of BMP4 in the presence of RA and FGF9. The expression levels of Tbx18, which has been demonstrated to act upstream of BMP430, and Gata2, which was dependent on RA23, were already comparable to endogenous levels even in the absence of BMP4. In contrast, BMP4 upregulated the expression levels of other ventral SP markers in a dose-dependent manner, whereas dorsal SP markers were downregulated (Fig. 1f). This result indicates that BMP4 reduces the expression of dorsal (i.e. renal) stromal genes and that SPs are shifted towards a ventral (i.e. ureteral) fate, as previously reported30. We determined the optimal concentration of BMP4 (10 ng/ml) for ventral SP induction, as almost all markers reached levels equivalent to the in vivo reference samples (Tbx18-GFP+/PDGFRA+ ventral SPs harvested at E13.5). Additionally, we found that BMP4 improved the percentage of the PDGFRA+ fraction by flow cytometry, suggesting that BMP4 efficiently induces the SP lineage instead of the NP lineage (ITGA8+/PDGFRA−) from E9.5 posterior IM (Fig. 1g and Supplementary Fig. 1f, g). In conclusion, the combination of RA, FGF9, and BMP4 is effective in inducing ventral SPs from E9.5 posterior IM.
Induction of ventral SPs from mouse ESCs
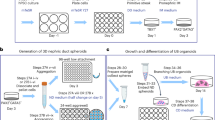
Next, we investigated whether ventral SPs could be induced from mouse embryonic stem cells (ESCs). We have previously established a protocol to induce dorsal SPs from mouse ESCs via the posterior IM, which can be isolated based on the expression of ROBO2 and PDGFRA1,10,29. The addition of SHH is essential for the final step in the mouse ESC induction protocol10, and the sorted posterior IM cells stimulated with RA, FGF9, SHH, and LDN193189 (LDN), a BMP receptor antagonist, give rise to dorsal SPs. Therefore, to induce ventral SPs, we adopted the same protocol to induce the posterior IM from mouse ESCs, and the sorted cells were subjected to the similar conditions except for the BMP signaling: we added BMP4 instead of LDN193189 (Fig. 2a, b), based on the results in Fig. 1. As in mouse embryos, the addition of BMP4 shifted mouse ESC-derived SPs to a ventral fate (Supplementary Fig. 2a). Furthermore, gene expression levels of ventral SP markers were higher in the presence of BMP4 than in the presence of LDN, while dorsal SP markers were higher in the presence of LDN (Fig. 2c). This trend was also observed when using a different mouse ESC line, suggesting the robustness of our protocols (Supplementary Fig. 2b). In addition, the induced ventral and dorsal SPs (ventral and dorsal iSPs) showed different FACS patterns. While both protocols for dorsal and ventral SPs induced more than 90% of PDGFRA+ cells, ventral SPs were strongly positive for ITGA8, whereas dorsal SPs were weakly positive (Fig. 2d). This is consistent with the expression pattern of Itga8 in the stromal cells in vivo (Fig. 1b, Supplementary Fig. 1d).
a Ventral SP induction protocol from mouse ESCs. A: ActivinA (D2-D3; 1 ng/ml, D5.5-6.5; 10 ng/ml); C3: CHIR (3 μM); C10: CHIR (10 μM); Y: Y27632 (10 μM); R: RA (D5.5-6.5; 100 nM, D6.5-9.5; 10 nM); F: FGF9 (10 ng/ml); S: SHH-N (1 μg/ml); B: BMP4 (D5.5-6.5; 3 ng/ml, D6.5-9.5; 10 ng/ml). b Schematic diagram of the induction of two types of SPs from mouse ESCs. c Comparison of gene expression levels between LDN and BMP4 in the final step of the SP induction from mouse ESCs. Ref: Tbx18-GFP+/PDGFRA+ ventral SPs harvested at E13.5 (non-cultured; presented as a reference). Data are shown as mean ± SD (n = 3 biologically independent experiments). Two-sided Student’s t-test was performed. The source data are provided as a Source Data file. d Flow cytometric analysis of two types of SP inductions between LDN and BMP4 from mouse ESCs at day 9.5. e, f scRNA-seq analysis of mouse ESC-derived dorsal and ventral iSPs, and stromal cells in E13.5 mouse embryonic kidney. e UMAP plots and the expression of representative ventral SP genes in dorsal and ventral iSPs from mouse ESCs, and stromal cells in E13.5 mouse embryonic kidney. f Unbiased hierarchical clustering analysis of the two types of induced stroma (viSP ventral induced SP, diSP dorsal induced SP) from mouse ESCs, and E13.5 embryonic stroma.
Next, we compared the gene expression of ventral and dorsal iSP with mouse embryonic stromal cells in vivo (E13.5) by scRNA-seq analysis (Supplementary Fig. 2c). In the UMAP plots, the stromal cells were divided into nine clusters (Fig. 2e). Based on the expression patterns of specific marker genes in mouse embryos, cluster 2 (Foxd1, Crabp1, Ntn1) and cluster 4 (Tbx18, Syt1, Sntg1, Iigp1, Plcxd3, Gata2, Col9a2, Foxf1) were shown to represent the dorsal SPs and ventral SPs, respectively (Fig. 2e, Supplementary Figs. 2d, 3). Cluster 3 represented the differentiating cortical (Ace2, Clca3a1) and medullary (Alx1, Wnt4) stromal cells (Supplementary Figs. 2e, 3). Notably, the ventral iSPs contained cluster 4 but not cluster 2, whereas the dorsal iSPs contained cluster 2 but not cluster 4 (Fig. 2e, Supplementary Fig. 2f), suggesting the differential induction of the two types of SPs. An unbiased hierarchical clustering analysis of these on-target clusters showed that ventral (Tbx18, Gata2, Plcxd3, Iigp1, Foxf1, Sntg1, Col9a2, Syt1) and dorsal iSPs (Foxd1, Ntn1, Igfbp6, Adamts18) had specific gene expressions that were similar to their counterparts in vivo (Fig. 2f, Supplementary Fig. 2g, Supplementary Data 2, 3). In addition, the expression of signaling-related genes, such as Fgf731,32and Bmp424,27, which are important for ureter development in vivo, was also observed in ventral iSPs (Fig. 2f, Supplementary Fig. 2g, Supplementary Data 2, 3). However, the BMP4-treated ventral iSP sample also contained other clusters. Cluster 1, which was present only in the ventral iSP sample, expressed not only ventral SP genes but also splanchnic mesodermal genes (Gata4, Hand2, Tbx20) (Fig. 2e, Supplementary Figs. 2d, h, 3). Clusters 0 and 5 were likely to represent proliferating dorsal SPs in vivo, since they expressed cell cycle markers (Top2a, Fbxo5, Esco2) and dorsal SP markers (Supplementary Figs. 2d, 3). However, these clusters in the ventral iSP sample also expressed Tbx18 (Fig. 2e, Supplementary Figs. 2d, 3). Indeed, most of the cells in the BMP4-treated sample expressed Tbx18 (90.5%). Other ventral SP genes were also expressed in most of the induced cells (Iigp1: 73.7%; Sntg1: 81.4%; Syt1: 90.6%) (Fig. 2e, Supplementary Figs. 2d, 3). These results suggest that the current protocol induced cells expressing representative ventral SP marker genes, although many cells co-expressed other lineage markers. While there is room for improvement in the protocol, cells similar to ventral SPs were induced from mouse ESCs in vitro.
Ureter reconstitution by assembly of ventral iSPs with embryonic ureteral epithelia
We next investigated whether ventral iSPs derived from mouse ESCs could fulfill the functionality of ventral SPs in vivo, i.e. to induce ureteral development by interacting with the ureteral epithelia. For this purpose, we first established an assay system to evaluate the function of ventral iSPs. While the intact embryonic ureter at E12.5 grew and elongated in the organ culture setting, the manually excised ureteral epithelia without ureteral mesenchyme did not (Supplementary Fig. 4a). When such a naked epithelial tube was placed on the deposited sheet-like SPs (cell sheets) sorted from E12.5 Tbx18-GFP kidneys (see also Methods), only GFP+ ventral SPs, but not GFP− stromal cells, could reconstitute the ureteral structure after culture (Supplementary Fig. 4b–e). It is important to note that the reconstituted spheroids lost their structural integrity and gradually collapsed when placed directly on Transwell. Therefore, Matrigel was used to ensure that the shape of the reconstituted spheroids was maintained (Supplementary Fig. 4f). Differentiation of the ureteral epithelium into three layers was verified by the expressions of UPK1B, ΔNP63, and KRT5, which in combination mark superficial cells (UPK1B+/ΔNP63−/KRT5−), intermediate cells (UPK1B+/ΔNP63+/KRT5−), and basal cells (UPK1B−/ΔNP63+/KRT5+)17,24,33,34 (Supplementary Figs. 1a, 4g). The ureteral mesenchyme also differentiated into three layers, including lamina propria (ALDH1A2+, Tbx18-GFP+/ACTA2− adjacent to the ureteral epithelium)17,24,31,32,33, smooth muscle cells (SMCs, actin, alpha 2, smooth muscle, aorta: ACTA2+), and tunica adventitia (periostin: POSTN+)35 (Supplementary Figs. 1a, 4g). Therefore, ventral SPs in vivo can reconstitute the ureteral structure in vitro when combined with the embryonic ureteral epithelia. We then applied the same strategy to the mouse ESC-derived iSPs (Fig. 3a). When ESC-derived ventral iSPs were combined with ureteral epithelia, the ureter was reconstituted and elongated (Fig. 3b). The elongation ratio of the ventral iSPs was greater than that of the dorsal iSPs and comparable to that of the ventral SPs from embryonic kidneys (Fig. 3c). We also observed periodic contractions of the reconstituted ureter (Fig. 3d, Supplementary Movie 1), and the contraction intensity (analyzed by calculating the relative width of the cultured ureters during contraction and relaxation states, see also methods) of ventral iSPs was again greater than that of dorsal iSPs and comparable to their counterparts in vivo (Fig. 3e). Histochemical examination revealed that the three layers described above were formed in both epithelia and mesenchyme of the differentiating ureters, but these layers were not detected when dorsal iSPs were used (Fig. 3f, Supplementary Fig. 4h). These data indicate that ventral iSPs derived from mouse ESCs not only have the capacity to differentiate into multiple cell types of the ureteral mesenchyme but also induce differentiation of ureteral epithelia.
a Schematic diagram of a reconstitution method between in vivo ureteral epithelia and iSPs from mouse ESCs. E12.5 wild-type mouse embryonic ureteral epithelia and dorsal or ventral iSPs derived from G4-2 mouse ESC, which is ubiquitously expressed of GFP, are combined, and cultured at the air/liquid interface for 7 days. b Bright-field images of reconstituted ureters. The experiments were performed four times, with a minimum of three reconstituted spheroids per condition per experiment. 11 out of 13 samples showed the successful reconstitution under a stereo microscope. Scale bars: 100 μm. c Elongation ratio of reconstituted ureters using several types of SPs. Data are shown as mean ± SD (n = 5 biologically independent samples, respectively). The one-way ANOVA was performed. Tbx18-GFP⁻ vs Tbx18-GFP⁺; p < 0.0001, dorsal iSP vs Tbx18-GFP⁺; p < 0.0001. The source data are provided as a Source Data file. d Bright-field images of contraction and relaxation states in the reconstituted ureter using mouse ESC-derived ventral iSPs at day 8. All of the successfully reconstituted organoids from panel b showed contraction. The outlines of the ureter in relaxation and contraction states are shown by the dotted black and red lines, respectively. Scale bars: 100 μm. e Contraction intensity of reconstituted ureters using several types of SPs. Data are shown as mean ± SD (n = 5 biologically independent samples, respectively). The one-way ANOVA was performed. Tbx18-GFP⁻ vs Tbx18-GFP⁺; p < 0.0001, dorsal iSP vs Tbx18-GFP⁺; p < 0.0001. The source data are provided as a Source Data file. f Immunostaining of transverse sections of reconstituted ureters at day 8. Reconstituted ureters using mouse ESC-derived ventral iSPs show differentiation of the ureteral epithelium and mesenchyme. Scale bars: 100 μm. One or two samples from each condition in each experiment were subjected to histological analysis, and similar results were observed in all of the samples (n = 5). The representative images are shown (eventual success rate; 85%).
The reconstituted ureters mature to the postnatal stage
We performed scRNA-seq analysis of the reconstituted ureters using mouse ESC-derived ventral iSPs (day 8 after culture) and compared the data with those of in vivo kidneys connected to ureters at perinatal (E17.5 and newborn: P0) and postnatal (7 days after birth: P7) stages. The UMAP plots of in vivo samples showed the clusters for the urothelium and the ureteral mesenchyme in addition to those for nephrons, UBs, and intrarenal stroma (Fig. 4a). The reconstituted ureter also showed the presence of clusters for the urothelium (cluster 13, black arrows) and the ureteral mesenchyme (cluster 30, red arrows), both overlapping with those in vivo (Fig. 4a). We selected the representative (top 100) genes for on-target clusters and performed the unbiased hierarchical clustering analysis, which demonstrated that the expression profiles of the urothelium and the mesenchyme of the reconstituted ureters were more similar to the postnatal stage (P7) than to the perinatal stages (E17.5, P0) (Fig. 4b, c, Supplementary Fig. 5a, b, Supplementary Data 4, 5). Several known SMC-specific genes (Acta2, Actg2, Ckm, Cnn1, Myh11, Myocd, Pcp4l1, Tagln, Tnnt2)15,17,24,27,31,32,36 were identified in the ureteral mesenchyme (Fig. 4b, Supplementary Fig. 5a, c), while genes specific for the urothelium (Krt5, Krt14, Upk1b, Upk3a)14,17 were detected, in both the reconstituted and postnatal ureters (Fig. 4c, Supplementary Fig. 5b, c). Furthermore, novel maturation-related genes were detected in the urothelium (Cd200r4, Cyp4f15, Ly6k, Muc15, Tmem238l, Tmprss4) and the ureteral mesenchyme (Bank1, Casq1, Hmcn2), which were also expressed in the reconstituted ureter (Fig. 4b, c, Supplementary Fig. 5a, b, d, e, f, Supplementary Data 4, 5). Thus, our scRNA-seq analysis revealed that the reconstituted ureter using mouse ESC-derived ventral iSPs matured to the postnatal stage.
a UMAP plots of in vivo mouse kidneys with ureters (E17.5, P0, P7) and the reconstituted ureter combined mouse embryonic ureteral epithelia and mouse ESC-derived ventral iSPs. The clusters of the ureteral mesenchyme and the urothelium are indicated by red (cluster 30) and black (cluster 13) arrows, respectively. b Unbiased hierarchical clustering analysis of the ureteral mesenchyme (cluster 30 in Fig. 4a) of in vivo ureters (E17.5, P0, P7) and the reconstituted ureter (rUreter). c Unbiased hierarchical clustering analysis of the urothelium (cluster 13 in Fig. 4a) of in vivo ureters (E17.5, P0, P7) and the reconstituted ureter (rUreter). d Dot plots of genes related to mesenchymal-epithelial interactions during ureteral development in the ureteral mesenchyme (cluster 30 in Fig. 4a). e Dot plots of genes related to mesenchymal-epithelial interactions during ureteral development in the urothelium (cluster 13 in Fig. 4a). Pod podocytes, PT proximal tubule, LoH loop of Henle, DT distal tubule, UE ureteral epithelium, Ren renin cells, Mes mesangial cells, oMS outer medullary stroma, iMS inner medullary stroma, UM ureteral mesenchyme, CD collecting duct, NP nephron progenitor, SP stromal progenitor, UB ureteric bud, EC endothelial cell.
Importantly, we identified genes known to be involved in mesenchymal-epithelial interactions during ureteral development. FGF7 from the ureteral mesenchyme acts through FGFR2 to enhance SHH expression in the ureteral epithelium, leading to the SHH-FOXF1-BMP4 signaling axis in the ureteral mesenchyme, which stimulates proliferation and differentiation of both lineages24,31,32,37. In parallel, the WNT signaling from the ureteral epithelium radially patterns the adjacent mesenchyme and contributes to the formation of three-layered structures in the mesenchyme38,39 (Supplementary Fig. 5g). Genes related to these signaling pathways were detected in the ureteral mesenchyme (Fgf7, Foxf1, Hhip, Axin2) and the urothelium (Shh, Wnt7b) (Fig. 4b–e, Supplementary Fig. 5a, b, Supplementary Data 4, 5), highlighting that the molecular interactions between mesenchyme and epithelium manifest in the reconstituted ureter in a manner analogous to their in vivo behavior.
While reconstituted ureters contained some other clusters (clusters 34, 35), representative ventral SP markers (Tbx18, Foxf1, Col9a2, Syt1) were expressed throughout the mesenchyme of reconstituted ureters (Supplementary Fig. 5h). Splanchnic mesodermal markers (Gata4, Hand2, Tbx20) detected prior to reconstitution (Supplementary Figs. 2h, 3) remained in the mesenchyme (Supplementary Fig. 5i). These data suggest that not only the correctly induced ventral SPs but also the cells co-expressing other lineage markers were competent and may have contributed to ureteral mesenchymal formation during reconstitution.
Induction of ventral SPs from human iPSCs to model some pathological features caused by TBX18 deficiency
We applied our ventral SP induction protocol from mouse ESCs to human induced pluripotent stem cells (iPSCs). We have previously reported a protocol to induce NPs from human iPSCs via the posterior IM1,29. Therefore, we transferred the final step condition from the posterior IM in the mouse ESC protocol to the human protocol (Fig. 5a). The expression levels of representative ventral SP genes (TBX18, GATA2, and FOXF1) were higher in the presence of BMP4 than in the presence of LDN, while dorsal SP genes (FOXD1, NTN1) were higher in the presence of LDN (Fig. 5b), suggesting that the activation of BMP signaling shifts human iPSC-derived SPs to a ventral fate, as in mice. This trend was also observed when using a different human iPSC line, suggesting the robustness of our protocols (Supplementary Fig. 6a). Furthermore, FACS analysis showed that the ITGA8 expression was higher upon BMP4 treatment compared to LDN treatment, while both induction protocols resulted in the induction of over 90% of PDGFRA+ cells (Supplementary Fig. 6b). These data were consistent with the mouse ESC induction (see Fig. 2d) and suggest that the effect of BMP4 in SP induction is conserved in humans.
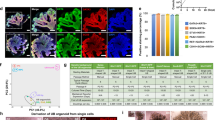
a Ventral SP induction method from human iPSCs. b Comparison of gene expression levels between LDN and BMP4 in the final step of the SP induction from human iPSCs. Data are shown as mean ± SD (n = 3 biologically independent experiments). Two-sided Student’s t-test was performed. The source data are provided as a Source Data file. c Contraction intensity of reconstituted ureters combined mouse embryonic ureteral epithelia with human iPSC-derived SPs induced by LDN or BMP4. Data are shown as mean ± SD (n = 3 biologically independent samples, respectively). Two-sided Student’s t-test was performed. The source data are provided as a Source Data file. d Immunostaining of transverse sections of reconstituted ureters on day 21. White arrowheads indicate the cells in which there are overlaps between ΔNP63 and UPK1B, which are identified as intermediate cells. Scale bars: 100 μm. Reconstitution experiments were performed four times, with a minimum of three reconstituted spheroids per condition per experiment. 9 out of 15 samples showed the successful outcome judged under a sterero microscope. One or two samples from each condition in each experiment were subjected to histological analysis. Similar results were observed in all of the samples (n = 5), and the representative images are shown (eventual success rate; 60%). e Mutation strategy of TBX18 in human iPSCs. The sequences of the resulting TBX18 mutant iPSCs are indicated on the bottom, showing 1 bp (T) and 2 bp (GT) insertion in each allele. f Comparison of the ventral SP and metanephric mesenchymal genes in ventral SPs induced by TBX18 mutant clones and a control clone, analyzed by qPCR (n = 3 biologically independent experiments). Relative mRNA expression levels normalized to β-actin gene expression are shown as mean ± SD. The one-way ANOVA was performed. The source data are provided as a Source Data file. g Contraction intensity of reconstituted ureters combined mouse embryonic ureteral epithelia with ventral SPs induced from TBX18 mutant human iPSC clones. Data are shown as mean ± SD (n = 3 biologically independent samples, respectively). The one-way ANOVA was performed. The source data are provided as a Source Data file. h Immunostaining of transverse sections of reconstituted ureters combined mouse embryonic ureteral epithelia with ventral iSPs induced by TBX18 mutant human iPSC lines on day 21. White arrowheads indicate the cells in which there are overlaps between ΔNP63 and UPK1B, which are identified as intermediate cells. Scale bars: 100 μm. Reconstitution experiments were performed four times, with a minimum of three reconstituted spheroids per condition per experiment. 8 out of 14 samples showed the successful outcome (contraction) judged under a sterero microscope. One or two samples from each condition in each experiment were subjected to histological analysis. Similar results were observed in all of the samples (n = 5), and the representative images are shown (eventual success rate; 57%).
We next performed scRNA-seq analysis of human iPSC-derived SPs induced by BMP4 and LDN. The resulting UMAP plots showed separate clusters of the stroma (PDGFRA+) and ECs (PECAM1+) (Supplementary Fig. 6c). Several clusters were identified within the stromal cluster, with cluster 0 being present only in the BMP4-induced SPs (Supplementary Fig. 6d). The exact similarity of these cells to their in vivo counterparts remained unclear, as the scRNA-seq data on developing human ureters are not available. While there is much room for improvement in our protocol, SPs induced by BMP4 exhibited robust expression of TBX18 throughout the SP, including cluster 0, and higher expression of other ventral markers (GATA2, FOXF1), while displaying weak expression of dorsal markers (FOXD1, NTN1) compared to SPs induced by LDN (Supplementary Fig. 6d, Supplementary Data 6). Several important markers of ureter development in mice (FGF7, GATA6, MYOCD)31,32,36 were expressed only in BMP4-induced SPs. These data suggest that our BMP4-based protocol can induce ventral SP-like cells from human iPSCs, as in the case of mouse ESCs.
We then investigated whether human iPSC-derived ventral iSPs could reconstitute the ureter when combined with the ureteral epithelia from mouse embryos (Supplementary Fig. 6e). Although human-mouse chimeric ureters did not form as smooth tubular structures as in the mouse alone, we were able to observe their contractions, and the contraction intensity using SPs induced by BMP4 was greater than that induced by LDN (Fig. 5c, Supplementary Fig. 6f, Supplementary Movie 2). Notably, the differentiation of ventral iSPs in humans took three weeks compared to one week in mice, likely reflecting the difference in the rate of differentiation between species. Furthermore, histochemical examination showed that SPs induced by BMP4, but not by LDN, could differentiate into multiple stromal layers, including SMCs (ACTA2+) and tunica adventitita (POSTN+), both of which were stained positively with the human-specific antibody (Numa) (Fig. 5d). The mouse embryo-derived ureteral epithelia also differentiated into three layers, suggesting that proper mesenchymal-epithelial interactions are present across species, at least partially. However, ALDH1A2 was only partially expressed in the innermost layer of the mesenchyme (Fig. 5d). These results may suggest a potential delay in the differentiation of ALDH1A2+ lamina propria cells in humans, similar to a report in mice40. This delay may be attributable to insufficient interactions across species or insufficient quality of human iSPs.
Next, we investigated whether ventral iSPs could be applied to disease models affecting the ureter. In humans, individuals with TBX18 mutations manifest a various degree of ureteropelvic junction obstruction and hydronephrosis, mainly caused by SMC impairment41. In Tbx18 knockout (KO) mice, the differentiation of the ventral SP into the ureteral mesenchyme, particularly SMCs, is impaired, leading to the development of short hydroureter and hydronephrosis19,20. In addition, ureteral epithelial differentiation is impaired20. Therefore, we generated TBX18 mutant human iPSC lines by CRISPR-Cas9 technology. We obtained mutant cell lines that harbored 1 bp (T) and 2 bp (GT) insertions in exon 5, which constitutes the majority of the T-box DNA-binding ___domain (Fig. 5e, Supplementary Fig. 6g, h). We applied our SP induction protocol to these mutant cell lines and found that the expressions of representative genes of ventral SPs (GATA2, FOXF1) were downregulated, whereas dorsal metanephric mesenchymal genes (FOXD1, WT1, WNT4) were upregulated compared to the control cell lines (Fig. 5f). These results indicate that TBX18 acts as a patterning factor that represses dorsal metanephric genes in both humans and mice30, thereby shifting the fate of SPs to a ventral side. When human ventral iSPs induced from TBX18 mutant iPSCs were combined with mouse embryonic ureteral epithelia, reconstituted ureters failed to form elongated tubular structures and contractions were significantly impaired (Fig. 5g, Supplementary Fig. 6i, j, Supplementary Movie 3). In addition, no differentiation into SMC-containing ureteral mesenchyme or multilayered epithelia was observed (Fig. 5h), highlighting the impairment of ureteral development caused by the TBX18 mutation. Thus, we have established the induction protocols for ventral SPs from human iPSCs and the authenticity of our methods was underscored by their potential applications in disease models affecting the ureter.
Generation of ureter-like organoids derived entirely from mouse and human PSCs
We next investigated whether ureteral organoids could be generated derived entirely from mouse and human PSCs. Instead of the in vivo ureteral epithelia, we induced UBs (iUBs) according to our previously established protocol6 and combined them with dorsal or ventral iSPs derived from mouse and human PSCs (Fig. 6a).
a Schematic diagram of reconstitution methods between iUBs and iSPs derived entirely from mouse ESCs or human iPSCs. Isolated UB tip and SPs derived from Hoxb7-GFP mouse ESC or human iPSCs are combined, and cultured at the air/liquid interface for 7 days (mouse) or 21 days (human). b Bright-field images of reconstituted organoids entirely derived from mouse ESCs. The experiments were performed three times, with a minimum of three reconstituted spheroids per condition per experiment. All of the 11 organoids showed spherical morphology. Scale bars: 100 μm. c Bright-field images of contraction and relaxation states in the reconstituted organoids using mouse ESC-derived iUBs and ventral iSPs at day 8. The outlines of the organoid in relaxation and contraction states are shown by the dotted black and red lines, respectively. 8 out of 11 spherical organoids showed contraction.d Contraction intensity of reconstituted organoids using several types of SPs from mouse embryos or mouse ESCs. Data are shown as mean ± SD (n = 3 biologically independent samples, respectively). The one-way ANOVA was performed. Tbx18-GFP⁻ vs Tbx18-GFP⁺; p < 0.0001, dorsal iSP vs Tbx18-GFP⁺; p < 0.0001. The source data are provided as a Source Data file. e Immunostaining of reconstituted organoids derived entirely from mouse ESCs at day 8. Scale bars: 100 μm. One or two samples from each condition in each experiment were subjected to histological analysis, and similar results were observed in all of the samples (n = 5). The representative images are shown (eventual success rate; 73%). f Bright-field images of reconstituted organoids entirely derived from human iPSCs. Scale bars: 100 μm. The experiments were performed three times, with a minimum of two reconstituted spheroids per condition per experiment. Out of 9 organoids, 5 showed spherical morphology, while the rest degenerated. g Immunostaining of reconstituted organoids entirely derived from human iPSCs at day 21. Scale bars: 100 μm. One or two samples from each condition in each experiment were subjected to histological analysis. Similar results were observed in all of the samples (n = 4), and the representative images are shown (eventual success rate; 56%).
We first investigated the generation of ureteral organoids derived entirely from mouse ESCs. Unlike the results obtained with in vivo ureteral epithelia, the reconstituted organoids did not elongate but formed a spherical morphology (Fig. 6b). Nevertheless, we could observe contractions of the reconstituted organoids, and the contraction intensity of the ventral iSPs was greater than that of the dorsal iSPs and comparable to their counterparts in vivo (Fig. 6c, d, Supplementary Movie 4). Histochemical examination showed that multiple layers of the epithelium and mesenchyme were formed, only when ventral iSPs were used (Fig. 6e). Therefore, we generated ureter-like organoids derived entirely from mouse ESCs. However, they were not perfect compared to the in vivo ureter. For example, although the clearly demarcated layer of lamina propria (ACTA2− cells underneath the urothelium) was observed, ALDH1A2, a marker of the lamina propria, was only partially expressed (Fig. 6e). This may indicate the delayed differentiation of ALDH1A2+ lamina propria cells40 in ureter-like organoids. The intermediate cells of the epithelia (UPK1B+/ΔNP63+/KRT5−, see also Supplementary Fig. 1a) were not identified (Fig. 6e). Since these phenomena such as the spherical morphology and partial differentiation of the ureter were also observed when embryonic ventral SPs were used (Supplementary Fig. 7a–d), they were more likely caused by the use of iUBs rather than iSPs. Although iUBs give rise to the collecting ducts within the kidney, their characteristics are different from those of the ureter that extends from the kidney16,42. To generate elongated and fully differentiated ureteral organoids from mouse ESCs, it would be necessary to establish an induction protocol for the ureteral epithelia from mouse ESCs and combine them with ventral iSPs.
We next examined ureteral organoids derived entirely from human iPSCs. Five out of nine reconstituted organoids exhibited a spherical morphology (Fig. 6f), while the rest degraded over time, possibly due to insufficient interaction between iUBs and iSPs. In addition, no contractions were detected even in organoids with spherical morphology. Nevertheless, histochemical examination of the spherical organoids revealed that only when the SP was induced by BMP4, ACTA2 and POSTN were uniformly expressed in the mesenchyme surrounding the epithelium, accompanied by UPK1B and ΔNP63 expression within the epithelial component (Fig. 6g), suggesting that differentiation of ventral iSPs into the ureteral mesenchyme and interactions with iUBs occurred, at least partially. However, ALDH1A2 expression was absent, and the expression of ACTA2 and POSTN was not segregated in the mesenchyme (Fig. 6g). In the epithelium, UPK1B and ΔNP63 were sporadically expressed, and KRT5 was absent (Fig. 6g). Therefore, distinct multilayered structures of the ureter were not formed, which may explain the lack of contraction in the organoids. Notably, these defects were not observed when human ventral iSPs were combined with mouse embryonic ureteral epithelia (Fig. 5d, h), suggesting that these defects may be primarily due to the use of human iUBs. Therefore, as described above, a new protocol for the induction of ureteral epithelium from human iPSCs would also be required.
Discussion
In this study, we established the induction protocols for ventral SPs from mouse and human PSCs by elucidating their origin and molecular features of the in vivo developmental process. When mouse ESC-derived ventral SPs were combined with mouse embryonic ureteral epithelia, contractile, elongated ureters with multilayered structures were reproduced. Additionally, the induction condition established for mouse ESCs can be applied to human iPSCs. Furthermore, the ureter-like spherical organoids derived entirely from mouse and human PSCs can also be generated by combining iUBs and ventral iSPs.
Lineage tracing analysis in mice revealed that the ventral SP originates from the Osr1+ posterior IM at E9.5. Although the localization of Osr1 and Tbx18 expression at E9.5 has been previously demonstrated20, we present here the first direct evidence that the Tbx18+ ventral SPs at E11.5 are derived from the Osr1+ posterior IM. Additionally, we have shown that BMP4 stimulation is effective for the induction of ventral SPs from the E9.5 posterior IM (this study), whereas BMP signaling inhibition results in the induction of dorsal SPs10. These findings are consistent with the in vivo studies showing that BMP4 is highly expressed in the stroma adjacent to the Wolffian duct in this stage43,44. This BMP4-dependent induction of ventral SPs applies not only to mouse PSCs but also to human PSCs, suggesting a conserved developmental mechanism across species.
Another feature of our study is the establishment of a reconstitution method for a ureter by combining the ventral iSP with the embryonic ureteral epithelia. This method serves as an assay system to evaluate the function of ventral iSPs. When mouse ESC-derived ventral iSPs were used, contractile elongated ureters with differentiated multilayered structures were reproduced, indicating that the ventral iSP has the function almost equivalent to that of the ventral SP in vivo, although some off-target cells are present. Furthermore, reconstituted ureters matured to the postnatal stage as revealed by scRNA-seq. Strikingly, peristaltic movement was observed and the degree of contraction of the reconstituted ureter using mouse iSPs is comparable to that using their embryonic counterparts, underscoring the functionality of the differentiated SMC layer derived from iSPs. This ureteral reconstitution is likely to be based on reciprocal interactions between the mesenchyme and epithelia mediated through the FGF7-SHH-FOXF1-BMP4 signaling and the WNT signaling. However, the reconstituted ureters lack unidirectional peristalsis, although they exhibit peristaltic contraction. Periodically coordinated unidirectional contractions of the ureter in vivo are controlled by two distinct urinary tract pacemaker cells (PMCs)45,46,47,48. PMCs at the pelvic-kidney junction are characterized by the expression of hyperpolarization-activated cyclic nucleotide-gated channel 3 (HCN3)46,47, whereas PMCs adjacent to the smooth muscle layer in the proximal ureter are termed interstitial cells of Cajal–like cells (ICC-LCs) which express cKIT45,48. Since these two types of PMCs have been shown to arise from neural crest cells49, it is unlikely that ventral iSP contain PMCs. Thus, it would be necessary to induce PMCs and incorporate them into the ureteral organoids.
Furthermore, when the induction protocol for ventral SPs in mouse ESCs was applied for human iPSCs and in reconstitution between human iSPs and mouse epithelia, mesenchymal-epithelial interactions occurred, at least in part. Human iSPs differentiated into a multilayered stroma, including SMCs in three weeks as opposed to one week for mouse iSPs, and the mouse epithelia also differentiated into multiple layers. These features were successfully applied to model some features of a ureteral disorder caused by TBX18 deficiency: TBX18 mutant PSCs failed to differentiate into SMCs and other stromal layers. In addition, we demonstrated that TBX18 acts as a patterning factor that represses dorsal metanephric genes and shift the fate of SPs to a ventral side in humans, as in mice30. The mouse epithelia wrapped by human iSPs also failed to differentiate in the absence of TBX18. These results underscore the authenticity of our induction and assembly protocols and open the door to the analysis of various human diseases affecting the ureter, such as renal coloboma syndrome (PAX2)50,51 and branchio-oto-renal syndrome (EYA1, SIX1)52,53.
Notably, we successfully generated ureteral organoids entirely from mouse ESCs and human iPSCs by combining iUBs and iSPs, which exhibited differentiation of the ureter. In mouse ESC-derived ureter-like organoids, multilayered mesenchyme and epithelia accompanied by contractions were observed. While the differentiation into ureteral epithelia has previously been demonstrated by placing mouse ESC-derived iUBs in the ventral stromal region of E11.5 mouse embryonic kidneys in culture54, this is the first report of the generation of the ureteral organoids derived entirely from mouse PSCs. We also generated ureter-like organoids entirely from human PSCs. However, the number of samples and the success rate are still low, and the quality is insufficient. The interaction between iUBs and iSPs was only partial, and no contraction was detected. Clearly, there is much room for improvement. Additionally, both mouse and human organoids do not elongate, resulting in spherical shapes. This may be due to the lack of a pre-formed elongated tubular structure and/or the use of UBs instead of the ureteral epithelium. For the former, a bioengineering approach may be required, such as the plating of epithelial cells in hydrogel within the inner lumen of the tunnel. For the latter, the behavior of UBs, which give rise to the collecting duct epithelia within the kidney, may be distinct from that of the ureteral epithelia that extend outward from the kidney16,42. It would be necessary to establish an induction protocol for the bona fide ureteral epithelia from PSCs and combine them with ventral iSPs.
To generate elongated, unidirectionally contractile ureteral organoids derived entirely from human PSCs, we need to find methods to induce ureteral epithelia instead of UBs, as well as PMCs, in humans, as discussed for the mouse organoids. In addition, the quality of iSPs and their progeny needs to be improved. Although scRNA-seq data for human embryonic kidneys are accumulating55,56, gene expression profiles for human embryonic ureters are still largely unknown. Thus, it is essential to obtain such data and to compare it with that of the iSPs.
For future transplant treatment, it is necessary to connect the ureter organoids with the kidney organoids. We have previously reported mouse ESC-derived organotypic mouse kidney organoids with multiply branched collecting ducts and numerous nephrons10. Generating such a structure in humans and connecting the collecting ducts and ureter by fusing the kidney and ureter organoids remains a challenge, but is worth pursuing.
In summary, we have revealed the developmental program for the ventral SPs and successfully induced them from PSCs in vitro. Reconstitution of ventral iSPs with mouse embryonic ureteral epithelia or iUBs recapitulated ureteral development. Furthermore, our ventral iSPs and reconstitution assay system enabled the modeling of pathological states affecting the ureter. Our study serves as a useful platform to elucidate the developmental and disease mechanisms of the ureter. In addition, the establishment of an induction protocol for ventral SPs will be an important step towards future functional kidney/ureter organoids with urine flow.
Methods
Mice
Tbx18-GFP mice were generated as described previously57. This mouse strain was maintained on a genetic background of C57BL/6. Osr1-CreER mice28 and ROSA26-CAG-tdTomato mice58 were purchased from Jackson Laboratory. These mouse strains were maintained on a mixed genetic background of C57BL/6 and ICR. The mice were housed in a specific pathogen-free animal facility. All animal experiments were performed in accordance with institutional ethical guidelines and approved by the licensing committee of Kumamoto University (approval numbers: A2021-008, A2023–009, and A2024-073).
Mouse ESC line maintenance
The Hoxb7-GFP mouse ESC line was previously described6,10 and maintained on mitotically inactivated murine embryonic fibroblasts. The G4-2 mouse ESC line with ubiquitous expression of GFP59 was kindly provided by Dr. Hitoshi Niwa (Kumamoto University) and cultured on gelatin-coated plates. The Hoxb7-GFP mouse ESC line was maintained in Glasgow’s minimal essential medium (Gibco #11710-35) supplemented with 14% knockout serum replacement (Gibco #10828-028), 1% fetal bovine serum (FBS) (Japan Bioserum #12303), nonessential amino acids (Gibco #11140-050), sodium pyruvate (Gibco #11360-070), GlutaMAX (Gibco #35050-061), penicillin/streptomycin (Gibco #15070-063), 0.1 mM 2-mercaptoethanol (Nakarai #21438-82), 1000 U/ml leukemia inhibitory factor (Millipore #ESG1106), 1.5 μM CHIR99021 (CHIR), and 0.5 μM PD0325901. The G4-2 mouse ESC was maintained in similar media except for the concentration of knockout serum replacement (10%), the source of leukemia inhibitory factor59, and the absence of CHIR and PD032590. The mouse ESC lines were cultured in a humidified atmosphere containing 5% CO2 at 37 °C. The Hoxb7-GFP mouse ESCs were passaged every other day, while the G4-2 mouse ESCs were passaged every 3 or 4 days.
SP induction from mouse ESCs
Differentiation of mouse ESCs was carried out in serum-free medium. ESCs were dissociated with Accutase (Millipore #SF006), aggregated, and induced until day 6.5 using the previously described NP induction protocol1,6. Initially, harvested cells were aggregated at 1000 cells/50 μl in 96-well U-bottom low cell-binding plates (Thermo Scientific #174925). After 48 h (at day 2), the spheres were dissociated with Accutase and reaggregated in differentiation medium with human Activin A (1 ng/ml). After 24 h (at day 3), the medium was switched to medium containing CHIR (10 μM). After 36 h (at day 4.5), the medium was replaced with medium containing Y27632 (10 μM) and CHIR (10 μM). After 24 h (at day 5.5), the medium was changed to medium containing ActivinA (10 ng/ml), BMP4 (3 ng/ml), CHIR (3 μM), RA (100 nM), and Y27632 (10 μM). At day 6.5, the spheres were dissociated with Accutase at 37 °C for 8 min and the ROBO2+/PDGFRA+ fraction was sorted. The sorted cells were resuspended in differentiation medium (20,000 cells/200 μl) containing several factors described below and seeded into 96-well low-cell-binding U-bottom plates, and cultured until day 9.510. The culture conditions from day 6.5 to day 9.5 for dorsal or ventral SPs were Y27632 (10 μM), RA (10 nM), FGF9 (10 ng/ml), SHH-N (1 μg/ml), and LDN193189 (25 nM) or BMP4 (10 ng/ml), respectively. The components of the mouse ESC differentiation medium were 75% Iscove’s modified Dulbecco’s medium (Gibco #21056-023) and 25% Ham’s F12 medium (Gibco #11765-054) supplemented with 1×B27 without RA (Gibco #12587-010), 0.5×N2 supplements (Gibco #17502-048), 0.05% BSA (Sigma #A4503), 0.5 mM ascorbic acid (Sigma #A4403), GlutaMAX (Gibco #35050-061), 0.5× penicillin/streptomycin (Gibco #15070-063), and 4.5×10−4M 1-thioglycerol (Sigma #M6145).
UB induction from mouse ESCs
The UB was induced from mouse ESCs based on our published protocol6. The serum-free medium described above was used as the differentiation medium. For the UB, 1000 cells/50 μl were aggregated in 96-well U-bottom low cell-binding plates. After 48 h (at day 2), the spheres were dissociated with Accutase and reaggregated in differentiation medium with human Activin A (10 ng/ml). After 24 h (at day 3), the medium was switched to medium containing human CHIR (10 μM). After 36 h (at day 4.5), the medium was changed to medium containing RA (100 nM), human FGF9 (200 ng/ml), and CHIR (2 μM). After 24 h (at day 5.5), the medium was changed to medium containing RA (100 nM), human FGF9 (100 ng/ml), and CHIR (3 μM). At day 6.25, spheres were dissociated by 0.25% trypsin/EDTA and sorted. A total of 3000 sorted CXCR4+/KIT+ cells were aggregated in V-bottom 96-well low cell-binding plates (Sumitomo Bakelite #MS-9096V), and cultured in the presence of Y27632 (10 μM), RA (100 nM), CHIR (1 μM), human FGF9 (5 ng/ml), and 10% Matrigel (growth factor reduced; Corning #356230). At day 7.25, the spheres were transferred to medium containing Y27632 (10 μM), RA (100 nM), CHIR (3 μM), human FGF9 (5 ng/ml), human GDNF (1 ng/ml), and 10% Matrigel. At day 8.25, the spheres were transferred to medium containing Y27632 (10 μM), RA (100 nM), CHIR (3 μM), human GDNF (2 ng/ml), and 10% Matrigel, and cultured for 24 h (day 9.25).
Ventral SP induction from the posterior IM in mouse embryos
We sorted the ROBO2+/PDGFRA+ fraction after removing the hindlimb buds including lateral plate mesoderm from E9.5 mouse embryos (caudal from the 26th somite; somite at the caudal end of the forelimb defined as somite 15), as previously described10. The sorted cells were resuspended in mouse ESC differentiation medium (10,000 cells/200 μl) described above containing Y27632 (10 μM), RA (10 nM), FGF9 (5 ng/ml), BMP4 (10 ng/ml) and seeded into 96-well low-cell-binding U-bottom plates, and cultured for 3 days.
Human iPSC line maintenance
The 201B7 and RN7 human iPSC line60,61 were maintained on iMatrix-511 (Nippi #892021) in StemFit AK02N medium (AJINOMOTO). The cells were cultured in a humidified atmosphere containing 5% CO2 at 37 °C and passaged every 5 days by treatment with Accutase. The cell culture medium was changed on days 2 and 4. All experiments using human iPSCs were performed in accordance with institutional guidelines and approved by the licensing committees at the Faculty of Life Science, Kumamoto University: Ethics Committee for Epidemiological and General Research (approval number: 1453) and Ethics Committee for Human Genome and Gene Analysis Research (approval number: 359).
Generation of TBX18 mutant human iPSCs
Two TBX18 mutant human iPSC lines were generated from the 201B7 line. A guide RNA (gRNA) was designed to target exon 5, (target sequence, 5’- CGAGTGCACGTCATCCGTAA-3’; PAM sequence, CGG). After preparing a single cell suspension of the 201B7 line, the gRNA and Cas9 protein (Integrated DNA Technologies, IDT) were electroporated into the dissociated cells, using 4D-Nucleofector (Lonza). The electroporated cells were subjected to single cell cloning by dilution. The target region of the genomic DNA was amplified by PCR, sequenced, and analyzed using the Synthego ICE Analysis Tool (v3) (https://ice.editco.bio/#/). The PCR fragments were also cloned into the pCR4-TOPO vector (Invitrogen), and the inserts of 13 individual plasmids per iPSC clone were sequenced. 1 bp (T) and 2 bp (GT) insertions were confirmed in both mutant iPSC clones, while wild-type sequences were detected only in the control clones. We further confirmed the same sequence mutations in PCR fragments amplified from cDNAs. Biologically independent induction/assembly experiments were performed four times (two mutant clones per experiment), and the results were consistent between the two mutant clones in all four experiments.
SP induction from human iPSCs
Differentiation of human iPSCs was carried out in serum-free medium consisting of DMEM/F12 (Gibco #11320-033) supplemented with Insulin-Transferrin-Selenium (ITS-G) (Gibco #41400-045), 2×B27 without RA (Gibco #12587-010), non-essential amino acids (Gibco #11140-050), sodium pyruvate (Gibco #11360-070), GlutaMAX (Gibco #35050-061), 0.5×penicillin/streptomycin (Gibco #15070-063), and 90 μM 2-mercaptoethanol (Nakarai #21438-82). iPSCs were induced to posterior IM until day 9 according to our published protocol1,6. At day 0, cells were aggregated in the medium containing human activin A (1 ng/ml), FGF2, and Y27632 in V-bottom 96-well low cell-binding plates (Sumitomo Bakelite #MS-9096V). At day 1, the spheres were stimulated with CHIR (10 μM) for 6 days, and then transferred at day 7 to the medium containing activin, BMP4, RA, CHIR, and Y27632. At day 9, the resulting posterior IM spheres were treated with Y27632 (10 μM), RA (1 μM), FGF9 (5 ng/ml), Purmorphamine (1 μM), and BMP4 (3 ng/ml) for 4 days to induce ventral SPs. In the comparative experiment of BMP signaling, the BMP inhibitor LDN193189 (50 nM) was used instead of BMP4.
UB induction from human iPSCs
UBs were induced from human iPSCs via anterior IM in the differentiation medium described above, based on our published protocol6. At day 0, iPSCs were aggregated in the medium containing human activin A (10 ng/ml), human BMP4, and Y27632 in V-bottom 96-well low cell-binding plates. At day 1, the spheres were stimulated with CHIR (10 μM) and human BMP4 for 36 h and then transferred at day 2.5 to the medium containing RA, human FGF9, LDN193189, and SB431542. At day 4.5, the resulting anterior IM spheres were treated with RA, CHIR, human FGF9, and LDN193189. At day 6.25, the spheres were dissociated with 0.25% trypsin/EDTA and sorted to isolate CXCR4+/KIT+ Wolffian duct progenitors. 5000 sorted cells per well were aggregated in 96-well low cell-binding plates, and cultured for 3 days to UBs in the three types of medium containing Y27632, RA, human FGF1, LDN193189, 10% Matrigel (growth factor reduced; Corning #356230), and varying concentrations of CHIR, FGF9 and GDNF.
Ureter reconstitution assay
SPs were isolated from E12.5 Tbx18-GFP+ mouse kidneys or induced from mouse ESCs or human iPSCs. SPs were sorted as the PDGFRA+ fraction and 30,000 SPs per well were seeded into 96-well low cell-binding U-bottom plates, followed by centrifugation (210 × g, 4 min). E12.5 wild-type ureteral epithelia, manually isolated by removing the ureteral mesenchyme with forceps, or single UBs, manually isolated with tungsten needles, were placed on the deposited sheet-like SPs (cell sheets). After culture at 37 °C overnight, the aggregated spheroids were transferred to Transwell inserts (Costar #3421) containing 100 μl of 10% Matrigel (growth factor reduced; Corning #356230) in culture medium described below, and cultured for 7 days (SPs in mice) or 21 days (SPs in humans) for in vitro analyses. The components of the culture medium were DMEM/F12 (Gibco #11320-033), 10% knockout serum replacement (Gibco #10828-028), nonessential amino acids (Gibco #11140-050), sodium pyruvate (Gibco #11360-070), GlutaMAX (Gibco #35050-061), penicillin/streptomycin (Gibco #15070-063). In the reconstitution assay between SPs sorted from mouse embryonic kidneys or iSPs from mouse ESCs and in vivo ureteral epithelia, BMP4 (10 ng/ml) was included for the first two days and the organoids were transferred into new Transwell inserts without Matrigel at day 4.
scRNA-seq analyses
scRNA-seq data of E15.5, E17.5, P0, and P7 kidneys were described previously62,63. scRNA-seq data of E11.5, E13.5, and dorsal iSPs from mouse ESCs were also described previously10. Ventral iSPs derived from mouse ESCs and the ureter reconstituted by assembling mouse ESC-derived ventral iSPs with mouse embryonic ureteral epithelia were newly analyzed in this study. Samples were digested in 0.25% trypsin/EDTA for 10 min. Trypsin was inactivated by the addition of DMEM (Sigma #D5796)/10% FBS containing 50 μg/ml DNase I (Worthington #LS002139), and the cells were washed with HEPES-buffered saline solution (HBSS) (Gibco #14185-052) containing 2% FBS, 50 μg/ml DNase I, 1 mM CaCl2 (Wako #031-00435), and 0.035% NaHCO3 (Wako # 191–01305). Cells were resuspended in 0.04% bovine serum albumin (BSA)/phosphate-buffered saline (PBS), filtered through a 40-μm-pore strainer (Falcon; Cat# 352340), and analyzed for their cell number and viability (>90%) using a Countess automated cell counter (Invitrogen # C10227).
Aliquots containing 5000 dissociated cells from each sample were applied to a Chromium Controller (10× Genomics). Chromium Single Cell 3′ Library & Gel Beads Kit v3 (10x Genomics) was used to generate cDNA libraries, which were then sequenced by an Illumina HiSeq X Ten (672,154,533 reads for E11.5; 311,733,555 reads for E13.5; 757,548,299 reads for E15.5; 259,180,950 reads for E17.5; 955,496,397 reads for P0; 370,196,520 reads for P7; 450,799,847 reads for dorsal iSPs from mouse ESCs; 344,602,471 reads for ventral iSPs from mouse ESCs; 327,869,474 reads for the ureter reconstituted by assembling mouse ESC-derived ventral iSPs with mouse embryonic ureteral epithelia; 563,112,735 reads for human iPSC-derived SPs induced by LDN; 361,480,084 reads for human iPSC-derived SPs induced by BMP4). The Q30 base RNA reads (Q-scores indicating sequencing quality) of the samples were 82.9% for E11.5, 88.5% for E13.5, 86.2% for E15.5, 63.8% for E17.5, 93.6% for P0, 91.9% for P7, 87.8% for dorsal iSPs from mouse ESCs, 89.1% for ventral iSPs from mouse ESCs, 95.4% for the reconstituted ureter, 96.4% for human iPSC-derived SPs induced by LDN, and 96.4% for human iPSC-derived SPs induced by BMP4.
The raw sequence data were processed using the cell ranger count command in Cell Ranger (version 7.0.0 for E11.5, E13.5, E15.5, dorsal iSPs from mouse ESCs, and ventral iSPs from mouse ESCs, 7.1.0 for E17.5, P0, P7, and the reconstituted ureter, 9.0.0 for human iPSC-derived SPs induced by LDN and BMP4 for integrated data; 10x Genomics). Subsequent analyses were performed using the R statistical programming language64 and the Seurat package (version 4.3.0 for E11.5, E13.5, E15.5, dorsal iSPs from mouse ESCs, and ventral iSPs from mouse ESCs integrated data, or version 5.0.2 for E17.5, P0, P7, reconstituted ureter, or version 5.2.1 for human iPSC-derived SPs induced by LDN and BMP4)65,66. A principal component analysis was applied for dimension reduction with dimension values of 50 for all integrated mouse data, determined by the JackStrawPlot function67. Uniform Manifold Approximation and Projection for Dimension Reduction (UMAP) plots were generated using the uwot package68.
Dot plot and unbiased hierarchal clustering analysis
We picked up representative genes in each cluster using the FindMarkers function in Seurat66 (Supplementary Data 1, 2, 4, 6). We compared each target cluster with other clusters and selected the genes with low P-values (P < 0.05). We then verified the expression of these genes in UMAP plots. Dot plot analysis was performed using the DotPlot function in Seurat. For unbiased hierarchal clustering analysis, we calculated the average expression of the representative genes using the AverageExpression function in Seurat66. Subsequently, specific genes in the ventral SP and the dorsal SP were selected from E13.5 mouse embryonic stroma (Fig. 2e: cluster 2; dorsal SPs, cluster 4; ventral SPs) (Supplementary Data 3). Then, the selected genes (dorsal SPs; 62, ventral SPs; 52, total; 114) were used for Fig. 2f and Supplementary Fig. 2g. Specific genes in the ureteral mesenchyme and the urothelium were selected from in vivo kidneys with ureters (E17.5, P0, P7) (Fig. 4a: cluster 30; the ureteral mesenchyme, cluster 13; the urothelium) (Supplementary Data 5). Subsequently, the selected genes (the ureteral mesenchyme; 100, the urothelium; 100, total; 200) were used for unbiased hierarchical clustering analysis for Fig. 4b, c, and Supplementary Fig. 5a, b. The pheatmap function69 and RNAseqChef70 were used to obtain heatmaps with hierarchy.
Flow cytometric analysis
Induced cell aggregates from embryonic tissues or mouse ESCs or human iPSCs were dissociated and blocked with normal mouse serum for 10 min on ice. Cell surface marker staining was carried out in HBSS containing 1% BSA and 0.035% NaHCO3 for 15 min on ice. Stained cells were analyzed using a FACS SORP Aria (BD Biosciences) or a FACS Aria III (BD Biosciences). The original FACS data sets were collected by using BD FACS Diva software (v8.0.1). Data analyses were performed with FlowJo software (ver 7.6.5, TreeStar). We performed three (Supplementary Fig. 6b) or four (Fig. 1g, Supplementary Fig. 1f, g) or six (Fig. 2d, Supplementary Fig. 4d) independent experiments for each condition. Representative images are shown for each condition. Quantification data were presented as mean ± SD. Detailed antibody information is provided in Supplementary Table 1.
Immunohistochemistry
For section immunostaining, paraffin sections were subjected to antigen retrieval in a citrate buffer, washed three times with PBS, and blocked by incubation with 1% BSA in PBS for 1 h at room temperature. The sections were incubated with primary antibodies at 4 °C overnight, followed by incubation with secondary antibodies conjugated with Alexa Fluor dyes at room temperature for 1 h. Fluorescence images were captured by confocal microscopy (TSC SP8; Leica). We performed at least three independent experiments for each condition, and collected, sectioned, and stained at least one sample per one condition. Representative images are shown for each condition. Detailed antibody information is provided in Supplementary Table 1.
Section in situ hybridization
RNAscope analysis of 10% formalin-fixed paraffin sections was performed using an RNAscope Multiplex Fluorescent Reagent Kit v2 (Advanced Cell Diagnostics; Cat# 323100). Signal amplification was performed with TSA plus fluorophores (Thermo Fisher Scientific). Details of the RNAscope probes are provided in Supplementary Table 1.
RNA extraction, reverse transcription, and quantitative RT-PCR
Harvested cells or spheroids were homogenized and total RNA was isolated using a RNeasy Plus Micro Kit (Qiagen #74034), and reverse-transcribed using Superscript VILO cDNA synthesis kit (Invitrogen #11754-250). Quantitative PCR was carried out using TB Green Fast qPCR Mix (Takara Bio #RR430A) and a Real-Time PCR System (Takara Bio). Relative mRNA expression levels were normalized to β-actin gene expression. We performed three independent experiments for each condition. Detailed primer information is provided in Supplementary Table 2.
Elongation ratio and contraction intensity
The length of ureteral epithelia before and after culture was measured using ImageJ software (1.54 g), and the ratio was calculated. To evaluate the contraction intensity of cultured ureters, 1 min videos of each ureter were taken using the Olympus IX73 microscope and analyzed by a frame rate of 14.7 per second in CellSens Standard software. In each 1 min movie, the width of cultured ureters was measured at three time points for contraction and relaxation states, respectively, using ImageJ software, and the average value was calculated. The contraction intensity was analyzed by calculating the relative width of the average value in cultured ureters during contraction and relaxation states. We performed three (Figs. 5c, g, 6d) or five (Fig. 3c, e) independent experiments for each condition.
Quantification and statistical analysis
Quantification data, including qRT-PCR and flow cytometric data, were presented as mean ± SD with plots. Bar charts with plots were generated by using Prism software (version 10). Two-sided Student’s t-test or the non-parametric Mann–Whitney U test (two-sided) was applied for statistical analysis of differences between two groups. For statistical analysis of more than two groups, one-way ANOVA was performed. If the ANOVA was significant, multiple comparisons between groups were performed by Dunnett’s multiple comparison test (two-sided). Differences with values of p < 0.05 were considered statistically significant. The p-values in the scRNA-seq data, calculated using the Seurat package, were two-sided, and adjustments were made for multiple comparisons (Supplementary Data 1–6).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data supporting the conclusions are present in the paper and the supplementary materials. A reporting summary for this article is available as a Supplementary Information file. Figures 1, 2, 4, Supplementary Figs. 1–6 have associated scRNA-seq raw data, which have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GSE282077 [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE282077]). The scRNA-seq data of E15.5, E17.5, P0 and P7 kidneys were described previously (GSE149134 [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE149134])62,63. The scRNA-seq data of E11.5, E13.5, dorsal iSPs from mouse ESCs were also described previously (GSE178263 [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE178263])10. The image data generated in this study have been deposited in the Figshare under the following link: https://doi.org/10.6084/m9.figshare.c.7823495. Source data are provided with this paper.
References
Taguchi, A. et al. Redefining the in vivo origin of metanephric nephron progenitors enables generation of complex kidney structures from pluripotent stem cells. Cell Stem Cell 14, 53–67 (2014).
Takasato, M. et al. Directing human embryonic stem cell differentiation towards a renal lineage generates a self-organizing kidney. Nat. Cell Biol. 16, 118–126 (2014).
Takasato, M. et al. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature 526, 564–568 (2015).
Morizane, R. et al. Nephron organoids derived from human pluripotent stem cells model kidney development and injury. Nat. Biotechnol. 33, 1193–1200 (2015).
Freedman, B. S. et al. Modelling kidney disease with CRISPR-mutant kidney organoids derived from human pluripotent epiblast spheroids. Nat. Commun. 6, 8715 (2015).
Taguchi, A. & Nishinakamura, R. Higher-order kidney organogenesis from pluripotent stem cells. Cell Stem Cell 21, 730–746.e6 (2017).
Mae, S. I. et al. Expansion of Human iPSC-derived ureteric bud organoids with repeated branching potential. Cell Rep. 32, 107963 (2020).
Uchimura, K. et al. Human pluripotent stem cell-derived kidney organoids with improved collecting duct maturation and injury modeling. Cell Rep. 33, 108514 (2020).
Zeng, Z. et al. Generation of patterned kidney organoids that recapitulate the adult kidney collecting duct system from expandable ureteric bud progenitors. Nat. Commun. 12, 3641 (2021).
Tanigawa, S. et al. Generation of the organotypic kidney structure by integrating pluripotent stem cell-derived renal stroma. Nat. Commun. 13, 611 (2022).
Shi, M. et al. Human ureteric bud organoids recapitulate branching morphogenesis and differentiate into functional collecting duct cell types. Nat. Biotechnol. 41, 252–261 (2023).
Howden, S. E. et al. Plasticity of distal nephron epithelia from human kidney organoids enables the induction of ureteric tip and stalk. Cell Stem Cell 28, 671–684.e6 (2021).
Wang, C. et al. Urothelial generation and regeneration in development, injury, and cancer. Dev. Dyn. 246, 336–343 (2017).
Dalghi, M. G. et al. The Urothelium: Life in a liquid environment. Physiol. Rev. 100, 1621–1705 (2020).
Gandhi, D. et al. Retinoid signaling in progenitors controls specification and regeneration of the urothelium. Dev. Cell 26, 469–482 (2013).
Bohnenpoll, T. & Kispert, A. Ureter growth and differentiation. Semi. Cell Dev. Biol. 36, 21–30 (2014).
Bohnenpoll, T. et al. Diversification of cell lineages in ureter development. J. Am. Soc. Nephrol. 28, 1792–1801 (2017).
Cunha, G. R. Epithelial-stromal interactions in development of the urogenital tract. Int. Rev. Cytol. 47, 137–194 (1976).
Airik, R. et al. Tbx18 regulates the development of the ureteral mesenchyme. J. Clin. Invest. 116, 663–674 (2006).
Bohnenpoll, T. et al. Tbx18 expression demarcates multipotent precursor populations in the developing urogenital system but is exclusively required within the ureteric mesenchymal lineage to suppress a renal stromal fate. Dev. Biol. 380, 25–36 (2013).
Combes, A. N. et al. Single cell analysis of the developing mouse kidney provides deeper insight into marker gene expression and ligand-receptor crosstalk. Development 146, dev178673 (2019).
England, A. R. et al. Identification and characterization of cellular heterogeneity within the developing renal interstitium. Development 147, dev190108 (2020).
Weiss, A. C. et al. Delayed onset of smooth muscle cell differentiation leads to hydroureter formation in mice with conditional loss of the zinc finger transcription factor gene Gata2 in the ureteric mesenchyme. J. Pathol. 248, 452–463 (2019).
Bohnenpoll, T. et al. A SHH-FOXF1-BMP4 signaling axis regulating growth and differentiation of epithelial and mesenchymal tissues in ureter development. PLoS Genet. 13, e1006951 (2017).
Itoh, F. et al. Promoting bone morphogenetic protein signaling through negative regulation of inhibitory Smads. EMBO J. 20, 4132–4142 (2001).
Mattei, M.-G. et al. Chromosomal assignment of retinoic acid receptor (RAR) genes in the human, mouse, and rat genomes. Genomics 10, 1061–1069 (1991).
Mamo, T. M. et al. BMP4 uses several different effector pathways to regulate proliferation and differentiation in the epithelial and mesenchymal tissue compartments of the developing mouse ureter. Hum. Mol. Genet. 26, 3553–3563 (2017).
Mugford, J. W. et al. Osr1 expression demarcates a multi-potent population of intermediate mesoderm that undergoes progressive restriction to an Osr1-dependent nephron progenitor compartment within the mammalian kidney. Dev. Biol. 324, 88–98 (2008).
Taguchi, A. & Nishinakamura, R. Nephron reconstitution from pluripotent stem cells. Kidney Int. 87, 894–900 (2015).
Weiss, A.-C. et al. Permissive ureter specification by TBX18-mediated repression of metanephric gene expression. Development 150, dev201048 (2023).
Meuser, M. et al. FGFR2 signaling enhances the SHH-BMP4 signaling axis in early ureter development. Development 149, dev200021 (2022).
Deuper, L. et al. Mesenchymal FGFR1 and FGFR2 control patterning of the ureteric mesenchyme by balancing SHH and BMP4 signaling. Development 149, dev200767 (2022).
Bohnenpoll, T. et al. Retinoic acid signaling maintains epithelial and mesenchymal progenitors in the developing mouse ureter. Sci. Rep. 7, 14803 (2017).
Yamany, T., Van Batavia, J. & Mendelsohn, C. Formation and regeneration of the urothelium. Curr. Opin. Organ Transpl. 19, 323–330 (2014).
Sorocos, K. et al. Expression patterns and roles of periostin during kidney and ureter development. J. Urol. 186, 1537–1544 (2011).
Kurz, J. et al. GATA6 is a crucial factor for Myocd expression in the visceral smooth muscle cell differentiation program of the murine ureter. Development 149, dev200522 (2022).
Yu, J. et al. Sonic hedgehog regulates proliferation and differentiation of mesenchymal cells in the mouse metanephric kidney. Development 129, 5301–5312 (2002).
Trowe, M. O. et al. Canonical wnt signaling regulates smooth muscle precursor development in the mouse ureter. Development 139, 3099–3108 (2012).
Aydoğdu, N. et al. TBX2 and TBX3 act downstream of canonical WNT signaling in patterning and differentiation of the mouse ureteric mesenchyme. Development 145, dev171827 (2018).
Straube, P. et al. Interplay of SHH, WNT and BMP4 signaling regulates the development of the lamina propria in the murine ureter. Development 152, dev204214. (2025).
Vivante, A. et al. Mutations in TBX18 cause dominant urinary tract malformations via transcriptional dysregulation of ureter development. Am. J. Hum. Genet. 97, 291–301 (2015).
Costantini, F. & Kopan, R. Patterning a complex organ: Branching morphogenesis and nephron segmentation in kidney development. Dev. Cell 18, 698–712 (2010).
Miyazaki, Y. et al. Bone morphogenetic protein 4 regulates the budding site and elongation of the mouse ureter. J. Clin. Invest. 105, 863–873 (2000).
Miyazaki, Y. et al. Evidence that bone morphogenetic protein 4 has multiple biological functions during kidney and urinary tract development. Kidney Int. 63, 835–844 (2003).
David, S. G. et al. C-Kit And Ureteral Peristalsis. J. Urol. 173, 292–295 (2005).
Hurtado, R. et al. The pelvis–kidney junction contains HCN3, a hyperpolarization-activated cation channel that triggers ureter peristalsis. Kidney Int. 77, 500–508 (2010).
Hurtado, R. et al. A molecular signature of tissues with pacemaker activity in the heart and upper urinary tract involves coexpressed hyperpolarization-activated cation and T-type Ca 2+ channels. FASEB J. 28, 730–739 (2014).
Pezzone, M. A. et al. Identification of c-kit-positive cells in the mouse ureter: the interstitial cells of Cajal of the urinary tract. Am. J. Physiol. Renal Physiol. 284, F925–F929 (2003).
Iskander, S. M. et al. Protein Kinase 2β is expressed in neural crest-derived urinary pacemaker cells and required for pyeloureteric contraction. J. Am. Soc. Nephrol. 29, 1198–1209 (2018).
Dureau, P. et al. Renal coloboma syndrome. Ophthalmology 108, 1912–1916 (2001).
Okumura, T. et al. Association of PAX2 and other gene mutations with the clinical manifestations of renal coloboma syndrome. PLoS One 10, e0142843 (2015).
Morisada, N. et al. Branchio-oto-renal syndrome: Comprehensive review based on nationwide surveillance in Japan. Pediatr. Int. 56, 309–314 (2014).
Unzaki, A. et al. Clinically diverse phenotypes and genotypes of patients with branchio-oto-renal syndrome. J. Hum. Genet. 63, 647–656 (2018).
Sallam, M. et al. Differentiation of a contractile, ureter-like tissue, from embryonic stem cell-derived ureteric bud and ex fetu mesenchyme. J. Am. Soc. Nephrol. 31, 2253–2262 (2020).
Lindström, N. O. et al. Conserved and divergent features of mesenchymal progenitor cell types within the cortical nephrogenic niche of the human and mouse kidney. J. Am. Soc. Nephrol. 29, 806–824 (2018).
Wang, P. et al. Dissecting the global dynamic molecular profiles of human fetal kidney development by single-cell RNA sequencing. Cell Rep. 24, 3554–3567.e3 (2018).
Cai, C.-L. et al. A myocardial lineage derives from Tbx18 epicardial cells. Nature 454, 104–108 (2008).
Madisen, L. et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140 (2010).
Niwa, H., Miyazaki, J. & Smith, A. G. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 24, 372–376 (2000).
Takahashi, K. et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 (2007).
Tanigawa, S. et al. Activin is superior to BMP7 for efficient maintenance of human IPSC-derived nephron progenitors. Stem Cell Reports 13, 322–337 (2019).
Naganuma, H. et al. Molecular detection of maturation stages in the developing kidney. Dev. Biol. 470, 62–73 (2021).
Ide, H. et al. Mouse embryonic kidney transplantation identifies maturation defects in the medulla. Sci. Rep. 14, 30293 (2024).
R Core Team R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/ (2024).
Butler, A. et al. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019).
Chung, N. C. & Storey, J. D. Statistical significance of variables driving systematic variation in high-dimensional data. Bioinformatics 31, 545–554 (2015).
Becht, E. et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 37, 38–44 (2019).
Kolde, R pheatmap: Pretty Heatmaps. R package version 1.0.13. https://github.com/raivokolde/pheatmap (2025).
Etoh, K. & Nakao, M. A web-based integrative transcriptome analysis, RNAseqChef, uncovers the cell/tissue type-dependent action of sulforaphane. J. Biol. Chem. 299, 104810 (2023).
Acknowledgements
We thank S. Fujimura and S. Usuki, members of the Liaison Laboratory Research Promotion Center, Institute of Molecular Embryology and Genetics, Kumamoto University, for experimental assistance. We also thank all members of the Nishinakamura laboratory for helpful discussions. This work was supported in part by JSPS KAKENHI Grant Number JP21H05050 (for R.N.), JSPS KAKENHI Grant Number JP24K23941 (International Leading Research: Creating a Kidney) (for R.N.), and the JST FOREST Program (Grant Number JPMJFR2266 for S.T.). This work was also supported by the MEXT-Supported Program for the Inter-University Research Network for High-Depth Omics, IMEG, Kumamoto University.
Author information
Authors and Affiliations
Contributions
Conceptualization, R.N.; methodology, Y.I.; investigation, Y.I., K.M., T.O., and S.T.; resources, R.N., and CL.C.; writing—original draft, Y.I., and R.N.; writing—review & editing, R.N.; funding acquisition, R.N.; project administration and supervision, R.N.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Kyle McCracken, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ibi, Y., Miike, K., Ohmori, T. et al. In vitro generation of a ureteral organoid from pluripotent stem cells. Nat Commun 16, 5309 (2025). https://doi.org/10.1038/s41467-025-60693-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-60693-6